Please be patient as the page loads
|
MITF_HUMAN
|
||||||
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MITF_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.996538 (rank : 2) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
TFE3_HUMAN
|
||||||
| θ value | 4.19693e-98 (rank : 3) | NC score | 0.941676 (rank : 3) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
TFEB_HUMAN
|
||||||
| θ value | 4.66183e-81 (rank : 4) | NC score | 0.939925 (rank : 5) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| θ value | 6.08852e-81 (rank : 5) | NC score | 0.940577 (rank : 4) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
USF2_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 6) | NC score | 0.517998 (rank : 6) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 7) | NC score | 0.512534 (rank : 7) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
USF1_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 8) | NC score | 0.500346 (rank : 9) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| θ value | 8.97725e-08 (rank : 9) | NC score | 0.501193 (rank : 8) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 10) | NC score | 0.380670 (rank : 12) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
SRBP1_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 11) | NC score | 0.395065 (rank : 11) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
SRBP2_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 12) | NC score | 0.409881 (rank : 10) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
CLOCK_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 13) | NC score | 0.226680 (rank : 40) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
MYCN_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 14) | NC score | 0.288869 (rank : 16) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
CLOCK_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 15) | NC score | 0.223297 (rank : 42) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
ARNT_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 16) | NC score | 0.255650 (rank : 26) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 17) | NC score | 0.251485 (rank : 28) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
RNC_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 18) | NC score | 0.133034 (rank : 58) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NRR4, Q7Z5V2, Q86YH0, Q9NW73, Q9Y2V9, Q9Y4Y0 | Gene names | RNASEN, RN3, RNASE3L | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease III (EC 3.1.26.3) (RNase III) (Drosha) (p241). | |||||
|
BMAL1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 19) | NC score | 0.235612 (rank : 33) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 20) | NC score | 0.235139 (rank : 34) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
MAD4_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 21) | NC score | 0.303060 (rank : 15) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MYCN_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 22) | NC score | 0.276061 (rank : 18) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
TFAP4_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 23) | NC score | 0.257051 (rank : 24) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
HES1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 24) | NC score | 0.232891 (rank : 37) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
HES1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 25) | NC score | 0.233812 (rank : 36) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
MYCS_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 26) | NC score | 0.263777 (rank : 22) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
MNT_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 27) | NC score | 0.218628 (rank : 44) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MAD3_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 28) | NC score | 0.255856 (rank : 25) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
MYC_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 29) | NC score | 0.261094 (rank : 23) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
ARNT2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 30) | NC score | 0.226931 (rank : 39) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 31) | NC score | 0.227230 (rank : 38) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
HEY1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 32) | NC score | 0.264179 (rank : 21) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
MNT_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 33) | NC score | 0.224470 (rank : 41) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
WBS14_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 34) | NC score | 0.170512 (rank : 48) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
HEY2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 35) | NC score | 0.271921 (rank : 19) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 36) | NC score | 0.269763 (rank : 20) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
MXI1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 37) | NC score | 0.243252 (rank : 32) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MXI1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 38) | NC score | 0.243347 (rank : 31) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
WBS14_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 39) | NC score | 0.155183 (rank : 55) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
HES4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 40) | NC score | 0.213779 (rank : 46) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HCC6 | Gene names | HES4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-4 (Hairy and enhancer of split 4) (bHLH factor Hes4). | |||||
|
MYCL2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 41) | NC score | 0.195983 (rank : 47) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 42) | NC score | 0.031885 (rank : 108) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
HEY1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 43) | NC score | 0.252974 (rank : 27) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
MAD3_MOUSE
|
||||||
| θ value | 0.125558 (rank : 44) | NC score | 0.234482 (rank : 35) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
MAX_MOUSE
|
||||||
| θ value | 0.163984 (rank : 45) | NC score | 0.345385 (rank : 14) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
MLX_MOUSE
|
||||||
| θ value | 0.163984 (rank : 46) | NC score | 0.280055 (rank : 17) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MYC_MOUSE
|
||||||
| θ value | 0.163984 (rank : 47) | NC score | 0.250261 (rank : 29) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 48) | NC score | 0.032641 (rank : 107) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
MAX_HUMAN
|
||||||
| θ value | 0.21417 (rank : 49) | NC score | 0.347666 (rank : 13) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MLX_HUMAN
|
||||||
| θ value | 0.279714 (rank : 50) | NC score | 0.248498 (rank : 30) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 0.279714 (rank : 51) | NC score | 0.023156 (rank : 112) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
NPAS2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 52) | NC score | 0.170197 (rank : 50) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
NPAS2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 53) | NC score | 0.170312 (rank : 49) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
NCOA1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 54) | NC score | 0.099486 (rank : 66) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 55) | NC score | 0.099038 (rank : 67) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
HES6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.122602 (rank : 60) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JHE6, Q3U2D7 | Gene names | Hes6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6). | |||||
|
MAD1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.157124 (rank : 54) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q05195 | Gene names | MXD1, MAD | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
MYCL1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.220245 (rank : 43) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 0.813845 (rank : 59) | NC score | 0.019500 (rank : 119) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
TAL1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 60) | NC score | 0.081270 (rank : 71) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P17542 | Gene names | TAL1, SCL, TCL5 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein (TAL-1 protein) (Stem cell protein) (T-cell leukemia/lymphoma-5 protein). | |||||
|
TAL1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 61) | NC score | 0.079633 (rank : 73) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P22091 | Gene names | Tal1, Scl, Tal-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein homolog (TAL-1 protein) (Stem cell protein). | |||||
|
TSK_HUMAN
|
||||||
| θ value | 0.813845 (rank : 62) | NC score | 0.020957 (rank : 115) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WUA8, Q6UXK1, Q9UG10, Q9UJX9 | Gene names | LRRC54, E2IG4, TSK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tsukushi precursor (Leucine-rich repeat-containing protein 54) (E2- induced gene 4 protein). | |||||
|
PRPF3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 63) | NC score | 0.053505 (rank : 94) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q922U1, Q3TJH4, Q9D6C6 | Gene names | Prpf3 | |||
|
Domain Architecture |

|
|||||
| Description | U4/U6 small nuclear ribonucleoprotein Prp3 (Pre-mRNA-splicing factor 3). | |||||
|
XE7_HUMAN
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.060523 (rank : 88) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.035935 (rank : 105) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.029707 (rank : 109) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
HES6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.119460 (rank : 61) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96HZ4, Q8N2J2, Q96T93, Q9P2S3 | Gene names | HES6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6) (C- HAIRY1). | |||||
|
MAD1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.161499 (rank : 51) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
MYCL1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.217058 (rank : 45) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
PRPF3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.049396 (rank : 98) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43395, O43446 | Gene names | PRPF3, HPRP3, PRP3 | |||
|
Domain Architecture |
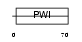
|
|||||
| Description | U4/U6 small nuclear ribonucleoprotein Prp3 (Pre-mRNA-splicing factor 3) (U4/U6 snRNP 90 kDa protein) (hPrp3). | |||||
|
BHLH3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.159342 (rank : 52) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
CEP63_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.020491 (rank : 117) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
DLG7_MOUSE
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.037411 (rank : 103) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K4R9, Q6ZQL3, Q80VS7, Q8BUC8, Q8BZ79 | Gene names | Dlg7, Kiaa0008 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discs large homolog 7 (Hepatoma up-regulated protein homolog) (HURP). | |||||
|
LYL1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.047623 (rank : 100) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P12980, O76102 | Gene names | LYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
BHLH3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.154491 (rank : 56) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
SON_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.026785 (rank : 110) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
ASCL2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.051917 (rank : 95) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35885, Q9WUJ7 | Gene names | Ascl2, Mash2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash-2). | |||||
|
CCD39_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.024549 (rank : 111) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
HES7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.110038 (rank : 63) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BYE0 | Gene names | HES7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
HES7_MOUSE
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.106530 (rank : 64) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BKT2, Q99JA6 | Gene names | Hes7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
IF3A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.023046 (rank : 113) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
INVO_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.020672 (rank : 116) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
MAML3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.018983 (rank : 120) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
AHR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.080537 (rank : 72) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
DDX20_MOUSE
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.004207 (rank : 139) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
DESP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.018074 (rank : 124) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
HDAC5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.009269 (rank : 137) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UQL6, O60340, O60528, Q96DY4 | Gene names | HDAC5, KIAA0600 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Antigen NY-CO-9). | |||||
|
ITF2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.040632 (rank : 102) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P15884, Q15439, Q15440 | Gene names | TCF4, ITF2, SEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (SL3-3 enhancer factor 2) (SEF-2). | |||||
|
ITF2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.035257 (rank : 106) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60722, Q60721, Q62211, Q64072, Q80UE8 | Gene names | Tcf4, Itf2, Sef2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (MITF-2) (SL3-3 enhancer factor 2) (SEF-2) (Class A helix-loop-helix transcription factor ME2). | |||||
|
MYF6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.048296 (rank : 99) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P23409, Q53X80, Q6FHI9 | Gene names | MYF6 | |||
|
Domain Architecture |
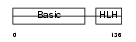
|
|||||
| Description | Myogenic factor 6 (Myf-6). | |||||
|
MYF6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.045499 (rank : 101) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15375 | Gene names | Myf6, Myf-6 | |||
|
Domain Architecture |
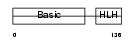
|
|||||
| Description | Myogenic factor 6 (Myf-6) (Herculin). | |||||
|
SRBS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.010557 (rank : 134) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62417, Q80TF8, Q8BZI3, Q8K3Y2, Q921F8, Q9Z0Z8, Q9Z0Z9 | Gene names | Sorbs1, Kiaa1296, Sh3d5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
TFE2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.036205 (rank : 104) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P15923, P15883, Q14208, Q14635, Q14636, Q9UPI9 | Gene names | TCF3, E2A, ITF1 | |||
|
Domain Architecture |
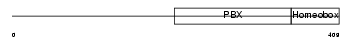
|
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Immunoglobulin transcription factor 1) (Transcription factor ITF-1) (Kappa-E2-binding factor). | |||||
|
TLE1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.009984 (rank : 135) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q04724, Q5T3G4, Q969V9 | Gene names | TLE1 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 1 (ESG1) (E(Sp1) homolog). | |||||
|
TLE1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.009795 (rank : 136) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62440 | Gene names | Tle1, Grg1 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 1 (Groucho-related protein 1) (Grg- 1). | |||||
|
CNOT3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.013121 (rank : 131) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CNTRB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.020059 (rank : 118) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N137, Q331K3, Q69YV7, Q8NCB8, Q8WXV3, Q96CQ7, Q9C060 | Gene names | CNTROB, LIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
ENAH_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.018125 (rank : 123) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
ITSN2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.007996 (rank : 138) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.022528 (rank : 114) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SVIL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.014073 (rank : 129) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.018335 (rank : 122) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TNIK_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.004105 (rank : 140) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
BHLH2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.130502 (rank : 59) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.013489 (rank : 130) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CCD1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.011222 (rank : 133) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P1N0, Q7Z435, Q86XV0, Q8NF89, Q9H603, Q9NXI1 | Gene names | CC2D1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil and C2 domain-containing protein 1A (Five repressor element under dual repression-binding protein 1) (FRE under dual repression-binding protein 1) (Freud-1) (Putative NF-kappa-B- activating protein 023N). | |||||
|
CRTC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.018652 (rank : 121) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
MKL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.014356 (rank : 128) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59759 | Gene names | Mkl2, Mrtfb | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B). | |||||
|
PESC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.016054 (rank : 127) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EQ61, Q542F0 | Gene names | Pes1 | |||
|
Domain Architecture |

|
|||||
| Description | Pescadillo homolog 1. | |||||
|
RASF7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.012391 (rank : 132) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02833 | Gene names | RASSF7, C11orf13, HRC1 | |||
|
Domain Architecture |
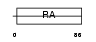
|
|||||
| Description | Ras association domain-containing protein 7 (HRAS1-related cluster protein 1). | |||||
|
TACC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.016533 (rank : 126) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6Y685, Q6Y686 | Gene names | Tacc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming acidic coiled-coil-containing protein 1. | |||||
|
TACC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.016609 (rank : 125) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
TCFL5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.158218 (rank : 53) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
AHR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.069238 (rank : 79) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
BHLH2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.117714 (rank : 62) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
BHLH8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.050542 (rank : 97) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7RTS1 | Gene names | BHLHB8, MIST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
BHLH8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.050954 (rank : 96) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QYC3, Q9QYE4 | Gene names | Bhlhb8, Mist1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
EPAS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.075181 (rank : 75) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
EPAS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.074516 (rank : 76) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
HES2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.083810 (rank : 69) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y543, Q96EN4, Q9Y542 | Gene names | HES2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
HES2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.087915 (rank : 68) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O54792 | Gene names | Hes2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
HES3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.076704 (rank : 74) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61657, O09083, O09150 | Gene names | Hes3, Hes-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-3 (Hairy and enhancer of split 3). | |||||
|
HES5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.103555 (rank : 65) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
HIF1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.073595 (rank : 77) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
HIF1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.072666 (rank : 78) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
MAD4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.144163 (rank : 57) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MYCB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.083275 (rank : 70) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P8Z1, Q9D6E3 | Gene names | Mycb, Bmyc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein B-Myc. | |||||
|
NCOA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.058597 (rank : 91) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.058923 (rank : 90) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NCOA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.066361 (rank : 84) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NCOA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.060252 (rank : 89) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
NPAS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.061277 (rank : 87) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
NPAS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.058166 (rank : 92) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
NPAS3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.062481 (rank : 86) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.063733 (rank : 85) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
PER2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.055490 (rank : 93) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
SIM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.068293 (rank : 83) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
SIM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.068444 (rank : 82) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
SIM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.069012 (rank : 80) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
SIM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.068644 (rank : 81) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.996538 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.941676 (rank : 3) | θ value | 4.19693e-98 (rank : 3) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.940577 (rank : 4) | θ value | 6.08852e-81 (rank : 5) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.939925 (rank : 5) | θ value | 4.66183e-81 (rank : 4) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
USF2_HUMAN
|
||||||
| NC score | 0.517998 (rank : 6) | θ value | 1.38499e-08 (rank : 6) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| NC score | 0.512534 (rank : 7) | θ value | 5.26297e-08 (rank : 7) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
USF1_MOUSE
|
||||||
| NC score | 0.501193 (rank : 8) | θ value | 8.97725e-08 (rank : 9) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_HUMAN
|
||||||
| NC score | 0.500346 (rank : 9) | θ value | 8.97725e-08 (rank : 8) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
SRBP2_HUMAN
|
||||||
| NC score | 0.409881 (rank : 10) | θ value | 1.69304e-06 (rank : 12) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
SRBP1_HUMAN
|
||||||
| NC score | 0.395065 (rank : 11) | θ value | 7.59969e-07 (rank : 11) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.380670 (rank : 12) | θ value | 5.81887e-07 (rank : 10) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
MAX_HUMAN
|
||||||
| NC score | 0.347666 (rank : 13) | θ value | 0.21417 (rank : 49) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MAX_MOUSE
|
||||||
| NC score | 0.345385 (rank : 14) | θ value | 0.163984 (rank : 45) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
MAD4_HUMAN
|
||||||
| NC score | 0.303060 (rank : 15) | θ value | 0.00228821 (rank : 21) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MYCN_HUMAN
|
||||||
| NC score | 0.288869 (rank : 16) | θ value | 0.000602161 (rank : 14) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MLX_MOUSE
|
||||||
| NC score | 0.280055 (rank : 17) | θ value | 0.163984 (rank : 46) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MYCN_MOUSE
|
||||||
| NC score | 0.276061 (rank : 18) | θ value | 0.00228821 (rank : 22) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
HEY2_HUMAN
|
||||||
| NC score | 0.271921 (rank : 19) | θ value | 0.0563607 (rank : 35) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| NC score | 0.269763 (rank : 20) | θ value | 0.0563607 (rank : 36) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
HEY1_MOUSE
|
||||||
| NC score | 0.264179 (rank : 21) | θ value | 0.0431538 (rank : 32) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
MYCS_MOUSE
|
||||||
| NC score | 0.263777 (rank : 22) | θ value | 0.0113563 (rank : 26) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
MYC_HUMAN
|
||||||
| NC score | 0.261094 (rank : 23) | θ value | 0.0252991 (rank : 29) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
TFAP4_HUMAN
|
||||||
| NC score | 0.257051 (rank : 24) | θ value | 0.00665767 (rank : 23) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
MAD3_HUMAN
|
||||||
| NC score | 0.255856 (rank : 25) | θ value | 0.0252991 (rank : 28) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.255650 (rank : 26) | θ value | 0.00134147 (rank : 16) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
HEY1_HUMAN
|
||||||
| NC score | 0.252974 (rank : 27) | θ value | 0.0961366 (rank : 43) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.251485 (rank : 28) | θ value | 0.00134147 (rank : 17) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
MYC_MOUSE
|
||||||
| NC score | 0.250261 (rank : 29) | θ value | 0.163984 (rank : 47) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MLX_HUMAN
|
||||||
| NC score | 0.248498 (rank : 30) | θ value | 0.279714 (rank : 50) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MXI1_MOUSE
|
||||||
| NC score | 0.243347 (rank : 31) | θ value | 0.0563607 (rank : 38) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MXI1_HUMAN
|
||||||
| NC score | 0.243252 (rank : 32) | θ value | 0.0563607 (rank : 37) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
BMAL1_HUMAN
|
||||||
| NC score | 0.235612 (rank : 33) | θ value | 0.00228821 (rank : 19) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| NC score | 0.235139 (rank : 34) | θ value | 0.00228821 (rank : 20) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
MAD3_MOUSE
|
||||||
| NC score | 0.234482 (rank : 35) | θ value | 0.125558 (rank : 44) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
HES1_MOUSE
|
||||||
| NC score | 0.233812 (rank : 36) | θ value | 0.0113563 (rank : 25) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
HES1_HUMAN
|
||||||
| NC score | 0.232891 (rank : 37) | θ value | 0.0113563 (rank : 24) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
ARNT2_MOUSE
|
||||||
| NC score | 0.227230 (rank : 38) | θ value | 0.0431538 (rank : 31) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_HUMAN
|
||||||
| NC score | 0.226931 (rank : 39) | θ value | 0.0431538 (rank : 30) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
CLOCK_MOUSE
|
||||||
| NC score | 0.226680 (rank : 40) | θ value | 0.000461057 (rank : 13) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.224470 (rank : 41) | θ value | 0.0431538 (rank : 33) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
CLOCK_HUMAN
|
||||||
| NC score | 0.223297 (rank : 42) | θ value | 0.000786445 (rank : 15) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
MYCL1_HUMAN
|
||||||
| NC score | 0.220245 (rank : 43) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.218628 (rank : 44) | θ value | 0.0193708 (rank : 27) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MYCL1_MOUSE
|
||||||
| NC score | 0.217058 (rank : 45) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
HES4_HUMAN
|
||||||
| NC score | 0.213779 (rank : 46) | θ value | 0.0736092 (rank : 40) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HCC6 | Gene names | HES4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-4 (Hairy and enhancer of split 4) (bHLH factor Hes4). | |||||
|
MYCL2_HUMAN
|
||||||
| NC score | 0.195983 (rank : 47) | θ value | 0.0736092 (rank : 41) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
WBS14_HUMAN
|
||||||
| NC score | 0.170512 (rank : 48) | θ value | 0.0431538 (rank : 34) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
NPAS2_MOUSE
|
||||||
| NC score | 0.170312 (rank : 49) | θ value | 0.279714 (rank : 53) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
NPAS2_HUMAN
|
||||||
| NC score | 0.170197 (rank : 50) | θ value | 0.279714 (rank : 52) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
MAD1_MOUSE
|
||||||
| NC score | 0.161499 (rank : 51) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
BHLH3_HUMAN
|
||||||
| NC score | 0.159342 (rank : 52) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
TCFL5_HUMAN
|
||||||
| NC score | 0.158218 (rank : 53) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
MAD1_HUMAN
|
||||||
| NC score | 0.157124 (rank : 54) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q05195 | Gene names | MXD1, MAD | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
WBS14_MOUSE
|
||||||
| NC score | 0.155183 (rank : 55) | θ value | 0.0563607 (rank : 39) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
BHLH3_MOUSE
|
||||||
| NC score | 0.154491 (rank : 56) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
MAD4_MOUSE
|
||||||
| NC score | 0.144163 (rank : 57) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
RNC_HUMAN
|
||||||
| NC score | 0.133034 (rank : 58) | θ value | 0.00134147 (rank : 18) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NRR4, Q7Z5V2, Q86YH0, Q9NW73, Q9Y2V9, Q9Y4Y0 | Gene names | RNASEN, RN3, RNASE3L | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease III (EC 3.1.26.3) (RNase III) (Drosha) (p241). | |||||
|
BHLH2_HUMAN
|
||||||
| NC score | 0.130502 (rank : 59) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
HES6_MOUSE
|
||||||
| NC score | 0.122602 (rank : 60) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JHE6, Q3U2D7 | Gene names | Hes6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6). | |||||
|
HES6_HUMAN
|
||||||
| NC score | 0.119460 (rank : 61) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96HZ4, Q8N2J2, Q96T93, Q9P2S3 | Gene names | HES6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6) (C- HAIRY1). | |||||
|
BHLH2_MOUSE
|
||||||
| NC score | 0.117714 (rank : 62) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
HES7_HUMAN
|
||||||
| NC score | 0.110038 (rank : 63) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BYE0 | Gene names | HES7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
HES7_MOUSE
|
||||||
| NC score | 0.106530 (rank : 64) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BKT2, Q99JA6 | Gene names | Hes7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
HES5_MOUSE
|
||||||
| NC score | 0.103555 (rank : 65) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
NCOA1_HUMAN
|
||||||
| NC score | 0.099486 (rank : 66) | θ value | 0.62314 (rank : 54) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA1_MOUSE
|
||||||
| NC score | 0.099038 (rank : 67) | θ value | 0.62314 (rank : 55) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
HES2_MOUSE
|
||||||
| NC score | 0.087915 (rank : 68) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O54792 | Gene names | Hes2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
HES2_HUMAN
|
||||||
| NC score | 0.083810 (rank : 69) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y543, Q96EN4, Q9Y542 | Gene names | HES2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
MYCB_MOUSE
|
||||||
| NC score | 0.083275 (rank : 70) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P8Z1, Q9D6E3 | Gene names | Mycb, Bmyc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein B-Myc. | |||||
|
TAL1_HUMAN
|
||||||
| NC score | 0.081270 (rank : 71) | θ value | 0.813845 (rank : 60) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P17542 | Gene names | TAL1, SCL, TCL5 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein (TAL-1 protein) (Stem cell protein) (T-cell leukemia/lymphoma-5 protein). | |||||
|
AHR_HUMAN
|
||||||
| NC score | 0.080537 (rank : 72) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
TAL1_MOUSE
|
||||||
| NC score | 0.079633 (rank : 73) | θ value | 0.813845 (rank : 61) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P22091 | Gene names | Tal1, Scl, Tal-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein homolog (TAL-1 protein) (Stem cell protein). | |||||
|
HES3_MOUSE
|
||||||
| NC score | 0.076704 (rank : 74) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61657, O09083, O09150 | Gene names | Hes3, Hes-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-3 (Hairy and enhancer of split 3). | |||||
|
EPAS1_HUMAN
|
||||||
| NC score | 0.075181 (rank : 75) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
EPAS1_MOUSE
|
||||||
| NC score | 0.074516 (rank : 76) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
HIF1A_HUMAN
|
||||||
| NC score | 0.073595 (rank : 77) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
HIF1A_MOUSE
|
||||||
| NC score | 0.072666 (rank : 78) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
AHR_MOUSE
|
||||||
| NC score | 0.069238 (rank : 79) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
SIM2_HUMAN
|
||||||
| NC score | 0.069012 (rank : 80) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
SIM2_MOUSE
|
||||||
| NC score | 0.068644 (rank : 81) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
SIM1_MOUSE
|
||||||
| NC score | 0.068444 (rank : 82) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
SIM1_HUMAN
|
||||||
| NC score | 0.068293 (rank : 83) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
NCOA3_HUMAN
|
||||||
| NC score | 0.066361 (rank : 84) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NPAS3_MOUSE
|
||||||
| NC score | 0.063733 (rank : 85) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_HUMAN
|
||||||
| NC score | 0.062481 (rank : 86) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS1_HUMAN
|
||||||
| NC score | 0.061277 (rank : 87) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
XE7_HUMAN
|
||||||
| NC score | 0.060523 (rank : 88) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
NCOA3_MOUSE
|
||||||
| NC score | 0.060252 (rank : 89) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
NCOA2_MOUSE
|
||||||
| NC score | 0.058923 (rank : 90) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NCOA2_HUMAN
|
||||||
| NC score | 0.058597 (rank : 91) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NPAS1_MOUSE
|
||||||
| NC score | 0.058166 (rank : 92) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.055490 (rank : 93) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
PRPF3_MOUSE
|
||||||
| NC score | 0.053505 (rank : 94) | θ value | 1.06291 (rank : 63) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q922U1, Q3TJH4, Q9D6C6 | Gene names | Prpf3 | |||
|
Domain Architecture |

|
|||||
| Description | U4/U6 small nuclear ribonucleoprotein Prp3 (Pre-mRNA-splicing factor 3). | |||||
|
ASCL2_MOUSE
|
||||||
| NC score | 0.051917 (rank : 95) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35885, Q9WUJ7 | Gene names | Ascl2, Mash2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash-2). | |||||
|
BHLH8_MOUSE
|
||||||
| NC score | 0.050954 (rank : 96) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QYC3, Q9QYE4 | Gene names | Bhlhb8, Mist1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
BHLH8_HUMAN
|
||||||
| NC score | 0.050542 (rank : 97) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7RTS1 | Gene names | BHLHB8, MIST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
PRPF3_HUMAN
|
||||||
| NC score | 0.049396 (rank : 98) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43395, O43446 | Gene names | PRPF3, HPRP3, PRP3 | |||
|
Domain Architecture |

|
|||||
| Description | U4/U6 small nuclear ribonucleoprotein Prp3 (Pre-mRNA-splicing factor 3) (U4/U6 snRNP 90 kDa protein) (hPrp3). | |||||
|
MYF6_HUMAN
|
||||||
| NC score | 0.048296 (rank : 99) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P23409, Q53X80, Q6FHI9 | Gene names | MYF6 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 6 (Myf-6). | |||||
|
LYL1_HUMAN
|
||||||
| NC score | 0.047623 (rank : 100) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P12980, O76102 | Gene names | LYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
MYF6_MOUSE
|
||||||
| NC score | 0.045499 (rank : 101) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15375 | Gene names | Myf6, Myf-6 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 6 (Myf-6) (Herculin). | |||||
|
ITF2_HUMAN
|
||||||
| NC score | 0.040632 (rank : 102) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P15884, Q15439, Q15440 | Gene names | TCF4, ITF2, SEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (SL3-3 enhancer factor 2) (SEF-2). | |||||
|
DLG7_MOUSE
|
||||||
| NC score | 0.037411 (rank : 103) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K4R9, Q6ZQL3, Q80VS7, Q8BUC8, Q8BZ79 | Gene names | Dlg7, Kiaa0008 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discs large homolog 7 (Hepatoma up-regulated protein homolog) (HURP). | |||||
|
TFE2_HUMAN
|
||||||
| NC score | 0.036205 (rank : 104) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P15923, P15883, Q14208, Q14635, Q14636, Q9UPI9 | Gene names | TCF3, E2A, ITF1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Immunoglobulin transcription factor 1) (Transcription factor ITF-1) (Kappa-E2-binding factor). | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.035935 (rank : 105) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
ITF2_MOUSE
|
||||||
| NC score | 0.035257 (rank : 106) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60722, Q60721, Q62211, Q64072, Q80UE8 | Gene names | Tcf4, Itf2, Sef2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 4 (Immunoglobulin transcription factor 2) (ITF-2) (MITF-2) (SL3-3 enhancer factor 2) (SEF-2) (Class A helix-loop-helix transcription factor ME2). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.032641 (rank : 107) | θ value | 0.21417 (rank : 48) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.031885 (rank : 108) | θ value | 0.0736092 (rank : 42) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.029707 (rank : 109) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.026785 (rank : 110) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CCD39_HUMAN
|
||||||
| NC score | 0.024549 (rank : 111) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.023156 (rank : 112) | θ value | 0.279714 (rank : 51) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
IF3A_HUMAN
|
||||||
| NC score | 0.023046 (rank : 113) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.022528 (rank : 114) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
TSK_HUMAN
|
||||||
| NC score | 0.020957 (rank : 115) | θ value | 0.813845 (rank : 62) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WUA8, Q6UXK1, Q9UG10, Q9UJX9 | Gene names | LRRC54, E2IG4, TSK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tsukushi precursor (Leucine-rich repeat-containing protein 54) (E2- induced gene 4 protein). | |||||
|
INVO_HUMAN
|
||||||
| NC score | 0.020672 (rank : 116) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
CEP63_HUMAN
|
||||||
| NC score | 0.020491 (rank : 117) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
CNTRB_HUMAN
|
||||||
| NC score | 0.020059 (rank : 118) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8N137, Q331K3, Q69YV7, Q8NCB8, Q8WXV3, Q96CQ7, Q9C060 | Gene names | CNTROB, LIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.019500 (rank : 119) | θ value | 0.813845 (rank : 59) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MAML3_HUMAN
|
||||||
| NC score | 0.018983 (rank : 120) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
CRTC1_MOUSE
|
||||||
| NC score | 0.018652 (rank : 121) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.018335 (rank : 122) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
ENAH_HUMAN
|
||||||
| NC score | 0.018125 (rank : 123) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.018074 (rank : 124) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
TACC2_HUMAN
|
||||||
| NC score | 0.016609 (rank : 125) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
TACC1_MOUSE
|
||||||
| NC score | 0.016533 (rank : 126) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6Y685, Q6Y686 | Gene names | Tacc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transforming acidic coiled-coil-containing protein 1. | |||||
|
PESC_MOUSE
|
||||||
| NC score | 0.016054 (rank : 127) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EQ61, Q542F0 | Gene names | Pes1 | |||
|
Domain Architecture |

|
|||||
| Description | Pescadillo homolog 1. | |||||
|
MKL2_MOUSE
|
||||||
| NC score | 0.014356 (rank : 128) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59759 | Gene names | Mkl2, Mrtfb | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 2 (Myocardin-related transcription factor B) (MRTF-B). | |||||
|
SVIL_MOUSE
|
||||||
| NC score | 0.014073 (rank : 129) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.013489 (rank : 130) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CNOT3_MOUSE
|
||||||
| NC score | 0.013121 (rank : 131) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
RASF7_HUMAN
|
||||||
| NC score | 0.012391 (rank : 132) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02833 | Gene names | RASSF7, C11orf13, HRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras association domain-containing protein 7 (HRAS1-related cluster protein 1). | |||||
|
CCD1A_HUMAN
|
||||||
| NC score | 0.011222 (rank : 133) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P1N0, Q7Z435, Q86XV0, Q8NF89, Q9H603, Q9NXI1 | Gene names | CC2D1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil and C2 domain-containing protein 1A (Five repressor element under dual repression-binding protein 1) (FRE under dual repression-binding protein 1) (Freud-1) (Putative NF-kappa-B- activating protein 023N). | |||||
|
SRBS1_MOUSE
|
||||||
| NC score | 0.010557 (rank : 134) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62417, Q80TF8, Q8BZI3, Q8K3Y2, Q921F8, Q9Z0Z8, Q9Z0Z9 | Gene names | Sorbs1, Kiaa1296, Sh3d5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
TLE1_HUMAN
|
||||||
| NC score | 0.009984 (rank : 135) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q04724, Q5T3G4, Q969V9 | Gene names | TLE1 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 1 (ESG1) (E(Sp1) homolog). | |||||
|
TLE1_MOUSE
|
||||||
| NC score | 0.009795 (rank : 136) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62440 | Gene names | Tle1, Grg1 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 1 (Groucho-related protein 1) (Grg- 1). | |||||
|
HDAC5_HUMAN
|
||||||
| NC score | 0.009269 (rank : 137) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UQL6, O60340, O60528, Q96DY4 | Gene names | HDAC5, KIAA0600 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Antigen NY-CO-9). | |||||
|
ITSN2_MOUSE
|
||||||
| NC score | 0.007996 (rank : 138) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
DDX20_MOUSE
|
||||||
| NC score | 0.004207 (rank : 139) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJY4, Q9JIK4 | Gene names | Ddx20, Dp103, Gemin3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3) (Regulator of steroidogenic factor 1) (ROSF-1). | |||||
|
TNIK_HUMAN
|
||||||
| NC score | 0.004105 (rank : 140) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||