Please be patient as the page loads
|
NPAS1_HUMAN
|
||||||
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NPAS1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
NPAS1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.994444 (rank : 2) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
NPAS3_MOUSE
|
||||||
| θ value | 4.03687e-117 (rank : 3) | NC score | 0.968659 (rank : 4) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_HUMAN
|
||||||
| θ value | 3.41741e-116 (rank : 4) | NC score | 0.970903 (rank : 3) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
SIM2_MOUSE
|
||||||
| θ value | 5.51964e-74 (rank : 5) | NC score | 0.951467 (rank : 6) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
SIM2_HUMAN
|
||||||
| θ value | 2.09745e-73 (rank : 6) | NC score | 0.953012 (rank : 5) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
SIM1_HUMAN
|
||||||
| θ value | 3.57772e-73 (rank : 7) | NC score | 0.945933 (rank : 7) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
SIM1_MOUSE
|
||||||
| θ value | 1.04096e-72 (rank : 8) | NC score | 0.945290 (rank : 8) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
HIF1A_HUMAN
|
||||||
| θ value | 6.56568e-51 (rank : 9) | NC score | 0.906437 (rank : 10) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
HIF1A_MOUSE
|
||||||
| θ value | 1.91031e-50 (rank : 10) | NC score | 0.908878 (rank : 9) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
EPAS1_HUMAN
|
||||||
| θ value | 2.3352e-48 (rank : 11) | NC score | 0.900224 (rank : 11) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
EPAS1_MOUSE
|
||||||
| θ value | 6.7944e-48 (rank : 12) | NC score | 0.899572 (rank : 12) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
BMAL1_HUMAN
|
||||||
| θ value | 3.17815e-21 (rank : 13) | NC score | 0.743139 (rank : 15) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| θ value | 7.0802e-21 (rank : 14) | NC score | 0.740541 (rank : 16) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
AHR_HUMAN
|
||||||
| θ value | 2.27762e-19 (rank : 15) | NC score | 0.805712 (rank : 13) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
AHR_MOUSE
|
||||||
| θ value | 2.7842e-17 (rank : 16) | NC score | 0.802056 (rank : 14) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
CLOCK_MOUSE
|
||||||
| θ value | 3.76295e-14 (rank : 17) | NC score | 0.701195 (rank : 18) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
NPAS4_HUMAN
|
||||||
| θ value | 6.41864e-14 (rank : 18) | NC score | 0.670872 (rank : 22) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
NPAS4_MOUSE
|
||||||
| θ value | 8.38298e-14 (rank : 19) | NC score | 0.657755 (rank : 26) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
CLOCK_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 20) | NC score | 0.702971 (rank : 17) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
ARNT2_HUMAN
|
||||||
| θ value | 2.69671e-12 (rank : 21) | NC score | 0.682830 (rank : 19) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 22) | NC score | 0.681275 (rank : 20) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
NCOA1_MOUSE
|
||||||
| θ value | 1.33837e-11 (rank : 23) | NC score | 0.603229 (rank : 27) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
NCOA1_HUMAN
|
||||||
| θ value | 1.74796e-11 (rank : 24) | NC score | 0.600679 (rank : 28) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
ARNT_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 25) | NC score | 0.669309 (rank : 23) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 1.9326e-10 (rank : 26) | NC score | 0.659219 (rank : 25) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
NPAS2_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 27) | NC score | 0.670979 (rank : 21) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
NCOA2_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 28) | NC score | 0.535100 (rank : 29) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA2_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 29) | NC score | 0.534650 (rank : 30) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NPAS2_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 30) | NC score | 0.661296 (rank : 24) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
NCOA3_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 31) | NC score | 0.527169 (rank : 31) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NCOA3_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 32) | NC score | 0.492031 (rank : 32) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
CTDP1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 33) | NC score | 0.036815 (rank : 55) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TSG2, Q7TSS7, Q9D4S8 | Gene names | Ctdp1, Fcp1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 0.163984 (rank : 34) | NC score | 0.003484 (rank : 95) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.022490 (rank : 62) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.021876 (rank : 63) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
CASC3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 37) | NC score | 0.026304 (rank : 58) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
BTBD7_MOUSE
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.018369 (rank : 68) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CFE5, Q80TC3, Q8BZY9 | Gene names | Btbd7, Kiaa1525 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 7. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.023031 (rank : 61) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.024196 (rank : 59) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.009962 (rank : 77) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
CGRE1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.027112 (rank : 57) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.020612 (rank : 65) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.033969 (rank : 56) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
BRD8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.021031 (rank : 64) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.017242 (rank : 69) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.009887 (rank : 78) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TCF8_MOUSE
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.002404 (rank : 96) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
CN032_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.023952 (rank : 60) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
DAXX_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.014604 (rank : 70) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35613, Q9QWT8, Q9QWV3 | Gene names | Daxx | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx). | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.019530 (rank : 67) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
CX039_MOUSE
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.014282 (rank : 72) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K2D0, Q3U3R2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein CXorf39 homolog. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.011885 (rank : 76) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MTSS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.020588 (rank : 66) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
NEST_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.003949 (rank : 93) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.014191 (rank : 73) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
BCOR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.006354 (rank : 86) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
PGCB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.004169 (rank : 91) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
FOXN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.004064 (rank : 92) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15353, O15352 | Gene names | FOXN1, WHN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.014520 (rank : 71) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
OGFR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.013943 (rank : 74) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.009207 (rank : 81) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
TFEB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.062053 (rank : 44) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.061330 (rank : 45) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
ZC11A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.009208 (rank : 80) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6NZF1, Q6NXW9, Q6PDR6, Q80TU7, Q8C5L5, Q99JN6 | Gene names | Zc3h11a, Kiaa0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
AKAP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.006961 (rank : 85) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.013261 (rank : 75) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
BCOR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.005304 (rank : 89) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
CLCB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.006104 (rank : 88) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09497, Q6FHW1 | Gene names | CLTB | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin light chain B (Lcb). | |||||
|
ECM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.008024 (rank : 83) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16610 | Gene names | ECM1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
HPS4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.007980 (rank : 84) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NQG7, Q5H8V6, Q96LX6, Q9BY93, Q9UH37, Q9UH38 | Gene names | HPS4, KIAA1667 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 4 protein (Light-ear protein homolog). | |||||
|
IL4RA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.008520 (rank : 82) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P16382, O54690, Q60583, Q8CBW5 | Gene names | Il4r, Il4ra | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (IL-4- binding protein) (IL4-BP)]. | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.004232 (rank : 90) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
SLIK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.000049 (rank : 97) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q810C1, Q9CXL0 | Gene names | Slitrk1, Sltk1 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT and NTRK-like protein 1 precursor. | |||||
|
SMAD4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.003852 (rank : 94) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13485 | Gene names | SMAD4, DPC4, MADH4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4) (hSMAD4). | |||||
|
TCF15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.009785 (rank : 79) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12870, Q9NQQ1 | Gene names | TCF15, BHLHEC2 | |||
|
Domain Architecture |
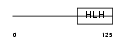
|
|||||
| Description | Transcription factor 15 (bHLH-EC2 protein) (Paraxis). | |||||
|
TULP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.006115 (rank : 87) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00294, O43536 | Gene names | TULP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
HES5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.052870 (rank : 51) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
HEY1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.074448 (rank : 42) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HEY1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.077762 (rank : 41) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.083841 (rank : 39) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.081527 (rank : 40) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
MITF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.061277 (rank : 46) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.062227 (rank : 43) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MLX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.052925 (rank : 50) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.053426 (rank : 49) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
PER1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.165747 (rank : 35) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PER1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.163256 (rank : 36) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
PER2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.166965 (rank : 34) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
PER2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.150724 (rank : 38) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
PER3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.160438 (rank : 37) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PER3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.180859 (rank : 33) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
TCFL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.060381 (rank : 47) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
TFE3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.059525 (rank : 48) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
USF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.052605 (rank : 52) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.052580 (rank : 53) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.051585 (rank : 54) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
NPAS1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
NPAS1_MOUSE
|
||||||
| NC score | 0.994444 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97459 | Gene names | Npas1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1). | |||||
|
NPAS3_HUMAN
|
||||||
| NC score | 0.970903 (rank : 3) | θ value | 3.41741e-116 (rank : 4) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IXF0, Q86US6, Q86US7, Q8IXF2, Q9BY81, Q9H323, Q9Y4L8 | Gene names | NPAS3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
NPAS3_MOUSE
|
||||||
| NC score | 0.968659 (rank : 4) | θ value | 4.03687e-117 (rank : 3) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QZQ0, Q9EQP4 | Gene names | Npas3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 3 (Neuronal PAS3) (Member of PAS protein 6) (MOP6). | |||||
|
SIM2_HUMAN
|
||||||
| NC score | 0.953012 (rank : 5) | θ value | 2.09745e-73 (rank : 6) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q14190, O60766, Q15470, Q15471, Q15472, Q15473, Q16532 | Gene names | SIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2. | |||||
|
SIM2_MOUSE
|
||||||
| NC score | 0.951467 (rank : 6) | θ value | 5.51964e-74 (rank : 5) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
SIM1_HUMAN
|
||||||
| NC score | 0.945933 (rank : 7) | θ value | 3.57772e-73 (rank : 7) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P81133, Q5TDP7 | Gene names | SIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1. | |||||
|
SIM1_MOUSE
|
||||||
| NC score | 0.945290 (rank : 8) | θ value | 1.04096e-72 (rank : 8) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61045, O70284, P70183 | Gene names | Sim1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 1 (mSIM1). | |||||
|
HIF1A_MOUSE
|
||||||
| NC score | 0.908878 (rank : 9) | θ value | 1.91031e-50 (rank : 10) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61221, O08741, O08993, Q61664, Q61665, Q8C681, Q8CC19, Q8CCB6, Q8R385, Q9CYA8 | Gene names | Hif1a | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein). | |||||
|
HIF1A_HUMAN
|
||||||
| NC score | 0.906437 (rank : 10) | θ value | 6.56568e-51 (rank : 9) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
EPAS1_HUMAN
|
||||||
| NC score | 0.900224 (rank : 11) | θ value | 2.3352e-48 (rank : 11) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99814, Q86VA2, Q99630 | Gene names | EPAS1, HIF2A | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Member of PAS protein 2) (MOP2) (Hypoxia-inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (HLF). | |||||
|
EPAS1_MOUSE
|
||||||
| NC score | 0.899572 (rank : 12) | θ value | 6.7944e-48 (rank : 12) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P97481, O08787, O55046 | Gene names | Epas1, Hif2a | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial PAS domain-containing protein 1 (EPAS-1) (Hypoxia- inducible factor 2 alpha) (HIF-2 alpha) (HIF2 alpha) (HIF-1 alpha-like factor) (MHLF) (HIF-related factor) (HRF). | |||||
|
AHR_HUMAN
|
||||||
| NC score | 0.805712 (rank : 13) | θ value | 2.27762e-19 (rank : 15) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P35869, Q13728, Q13803, Q13804 | Gene names | AHR | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
AHR_MOUSE
|
||||||
| NC score | 0.802056 (rank : 14) | θ value | 2.7842e-17 (rank : 16) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P30561, Q8QZX6, Q8R4S3, Q99P79, Q9QVY1 | Gene names | Ahr | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor precursor (Ah receptor) (AhR). | |||||
|
BMAL1_HUMAN
|
||||||
| NC score | 0.743139 (rank : 15) | θ value | 3.17815e-21 (rank : 13) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| NC score | 0.740541 (rank : 16) | θ value | 7.0802e-21 (rank : 14) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
CLOCK_HUMAN
|
||||||
| NC score | 0.702971 (rank : 17) | θ value | 1.86753e-13 (rank : 20) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
CLOCK_MOUSE
|
||||||
| NC score | 0.701195 (rank : 18) | θ value | 3.76295e-14 (rank : 17) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
ARNT2_HUMAN
|
||||||
| NC score | 0.682830 (rank : 19) | θ value | 2.69671e-12 (rank : 21) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| NC score | 0.681275 (rank : 20) | θ value | 4.59992e-12 (rank : 22) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
NPAS2_MOUSE
|
||||||
| NC score | 0.670979 (rank : 21) | θ value | 2.79066e-09 (rank : 27) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
NPAS4_HUMAN
|
||||||
| NC score | 0.670872 (rank : 22) | θ value | 6.41864e-14 (rank : 18) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.669309 (rank : 23) | θ value | 1.9326e-10 (rank : 25) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
NPAS2_HUMAN
|
||||||
| NC score | 0.661296 (rank : 24) | θ value | 2.36244e-08 (rank : 30) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.659219 (rank : 25) | θ value | 1.9326e-10 (rank : 26) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
NPAS4_MOUSE
|
||||||
| NC score | 0.657755 (rank : 26) | θ value | 8.38298e-14 (rank : 19) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
NCOA1_MOUSE
|
||||||
| NC score | 0.603229 (rank : 27) | θ value | 1.33837e-11 (rank : 23) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
NCOA1_HUMAN
|
||||||
| NC score | 0.600679 (rank : 28) | θ value | 1.74796e-11 (rank : 24) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
NCOA2_HUMAN
|
||||||
| NC score | 0.535100 (rank : 29) | θ value | 1.38499e-08 (rank : 28) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15596 | Gene names | NCOA2, TIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2). | |||||
|
NCOA2_MOUSE
|
||||||
| NC score | 0.534650 (rank : 30) | θ value | 1.38499e-08 (rank : 29) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61026, O09001, P97759 | Gene names | Ncoa2, Grip1, Tif2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 2 (NCoA-2) (Transcriptional intermediary factor 2) (Glucocorticoid receptor-interacting protein 1) (GRIP-1). | |||||
|
NCOA3_HUMAN
|
||||||
| NC score | 0.527169 (rank : 31) | θ value | 2.61198e-07 (rank : 31) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y6Q9, Q5JYD9, Q5JYE0, Q9BR49, Q9UPC9, Q9UPG4, Q9UPG7 | Gene names | NCOA3, AIB1, RAC3, TRAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP-interacting protein) (pCIP). | |||||
|
NCOA3_MOUSE
|
||||||
| NC score | 0.492031 (rank : 32) | θ value | 1.09739e-05 (rank : 32) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
PER3_MOUSE
|
||||||
| NC score | 0.180859 (rank : 33) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
PER2_HUMAN
|
||||||
| NC score | 0.166965 (rank : 34) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15055 | Gene names | PER2, KIAA0347 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2. | |||||
|
PER1_HUMAN
|
||||||
| NC score | 0.165747 (rank : 35) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PER1_MOUSE
|
||||||
| NC score | 0.163256 (rank : 36) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O35973 | Gene names | Per1, Per, Rigui | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (mPER) (M-Rigui). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.160438 (rank : 37) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PER2_MOUSE
|
||||||
| NC score | 0.150724 (rank : 38) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O54943, O54954 | Gene names | Per2 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 2 (mPER2). | |||||
|
HEY2_HUMAN
|
||||||
| NC score | 0.083841 (rank : 39) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| NC score | 0.081527 (rank : 40) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
HEY1_MOUSE
|
||||||
| NC score | 0.077762 (rank : 41) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY1_HUMAN
|
||||||
| NC score | 0.074448 (rank : 42) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.062227 (rank : 43) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.062053 (rank : 44) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.061330 (rank : 45) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.061277 (rank : 46) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
TCFL5_HUMAN
|
||||||
| NC score | 0.060381 (rank : 47) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.059525 (rank : 48) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
MLX_MOUSE
|
||||||
| NC score | 0.053426 (rank : 49) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_HUMAN
|
||||||
| NC score | 0.052925 (rank : 50) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
HES5_MOUSE
|
||||||
| NC score | 0.052870 (rank : 51) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
USF1_HUMAN
|
||||||
| NC score | 0.052605 (rank : 52) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| NC score | 0.052580 (rank : 53) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_HUMAN
|
||||||
| NC score | 0.051585 (rank : 54) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
CTDP1_MOUSE
|
||||||
| NC score | 0.036815 (rank : 55) | θ value | 0.0736092 (rank : 33) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7TSG2, Q7TSS7, Q9D4S8 | Gene names | Ctdp1, Fcp1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.033969 (rank : 56) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
CGRE1_HUMAN
|
||||||
| NC score | 0.027112 (rank : 57) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
CASC3_MOUSE
|
||||||
| NC score | 0.026304 (rank : 58) | θ value | 0.62314 (rank : 37) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.024196 (rank : 59) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.023952 (rank : 60) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.023031 (rank : 61) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.022490 (rank : 62) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.021876 (rank : 63) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
BRD8_HUMAN
|
||||||
| NC score | 0.021031 (rank : 64) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.020612 (rank : 65) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
MTSS1_HUMAN
|
||||||
| NC score | 0.020588 (rank : 66) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43312, Q8TCA2, Q96RX2 | Gene names | MTSS1, KIAA0429, MIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein) (Metastasis suppressor YGL-1). | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.019530 (rank : 67) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
BTBD7_MOUSE
|
||||||
| NC score | 0.018369 (rank : 68) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CFE5, Q80TC3, Q8BZY9 | Gene names | Btbd7, Kiaa1525 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 7. | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.017242 (rank : 69) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
DAXX_MOUSE
|
||||||
| NC score | 0.014604 (rank : 70) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35613, Q9QWT8, Q9QWV3 | Gene names | Daxx | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.014520 (rank : 71) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
CX039_MOUSE
|
||||||
| NC score | 0.014282 (rank : 72) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K2D0, Q3U3R2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein CXorf39 homolog. | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.014191 (rank : 73) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.013943 (rank : 74) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.013261 (rank : 75) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.011885 (rank : 76) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.009962 (rank : 77) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.009887 (rank : 78) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TCF15_HUMAN
|
||||||
| NC score | 0.009785 (rank : 79) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12870, Q9NQQ1 | Gene names | TCF15, BHLHEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 15 (bHLH-EC2 protein) (Paraxis). | |||||
|
ZC11A_MOUSE
|
||||||
| NC score | 0.009208 (rank : 80) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6NZF1, Q6NXW9, Q6PDR6, Q80TU7, Q8C5L5, Q99JN6 | Gene names | Zc3h11a, Kiaa0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.009207 (rank : 81) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
IL4RA_MOUSE
|
||||||
| NC score | 0.008520 (rank : 82) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P16382, O54690, Q60583, Q8CBW5 | Gene names | Il4r, Il4ra | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (IL-4- binding protein) (IL4-BP)]. | |||||
|
ECM1_HUMAN
|
||||||
| NC score | 0.008024 (rank : 83) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16610 | Gene names | ECM1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular matrix protein 1 precursor (Secretory component p85). | |||||
|
HPS4_HUMAN
|
||||||
| NC score | 0.007980 (rank : 84) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NQG7, Q5H8V6, Q96LX6, Q9BY93, Q9UH37, Q9UH38 | Gene names | HPS4, KIAA1667 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 4 protein (Light-ear protein homolog). | |||||
|
AKAP1_MOUSE
|
||||||
| NC score | 0.006961 (rank : 85) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08715, O08714, P97488 | Gene names | Akap1, Akap | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 1, mitochondrial precursor (Protein kinase A- anchoring protein 1) (PRKA1) (Dual specificity A-kinase-anchoring protein 1) (D-AKAP-1) (Spermatid A-kinase anchor protein) (S-AKAP). | |||||
|
BCOR_HUMAN
|
||||||
| NC score | 0.006354 (rank : 86) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
TULP1_HUMAN
|
||||||
| NC score | 0.006115 (rank : 87) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00294, O43536 | Gene names | TULP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-related protein 1 (Tubby-like protein 1). | |||||
|
CLCB_HUMAN
|
||||||
| NC score | 0.006104 (rank : 88) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09497, Q6FHW1 | Gene names | CLTB | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin light chain B (Lcb). | |||||
|
BCOR_MOUSE
|
||||||
| NC score | 0.005304 (rank : 89) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.004232 (rank : 90) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
PGCB_HUMAN
|
||||||
| NC score | 0.004169 (rank : 91) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
FOXN1_HUMAN
|
||||||
| NC score | 0.004064 (rank : 92) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15353, O15352 | Gene names | FOXN1, WHN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude). | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.003949 (rank : 93) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
SMAD4_HUMAN
|
||||||
| NC score | 0.003852 (rank : 94) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13485 | Gene names | SMAD4, DPC4, MADH4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4) (hSMAD4). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.003484 (rank : 95) | θ value | 0.163984 (rank : 34) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TCF8_MOUSE
|
||||||
| NC score | 0.002404 (rank : 96) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q64318, Q62519 | Gene names | Tcf8, Zfhx1a, Zfx1a, Zfx1ha | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (Zinc finger homeobox protein 1a) (MEB1) (Delta EF1). | |||||
|
SLIK1_MOUSE
|
||||||
| NC score | 0.000049 (rank : 97) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q810C1, Q9CXL0 | Gene names | Slitrk1, Sltk1 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT and NTRK-like protein 1 precursor. | |||||