Please be patient as the page loads
|
BHLH3_HUMAN
|
||||||
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
BHLH3_HUMAN
|
||||||
| θ value | 1.29261e-131 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
BHLH3_MOUSE
|
||||||
| θ value | 1.34075e-120 (rank : 2) | NC score | 0.981441 (rank : 2) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
BHLH2_MOUSE
|
||||||
| θ value | 1.61343e-57 (rank : 3) | NC score | 0.926259 (rank : 3) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
BHLH2_HUMAN
|
||||||
| θ value | 9.78833e-55 (rank : 4) | NC score | 0.917906 (rank : 4) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
HES1_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 5) | NC score | 0.588307 (rank : 6) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
HES1_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 6) | NC score | 0.585492 (rank : 7) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
HES4_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 7) | NC score | 0.609596 (rank : 5) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9HCC6 | Gene names | HES4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-4 (Hairy and enhancer of split 4) (bHLH factor Hes4). | |||||
|
HEY2_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 8) | NC score | 0.558761 (rank : 9) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
HEY2_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 9) | NC score | 0.554782 (rank : 10) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY1_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 10) | NC score | 0.549050 (rank : 11) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY1_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 11) | NC score | 0.541326 (rank : 12) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HES5_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 12) | NC score | 0.575593 (rank : 8) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
HES2_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 13) | NC score | 0.504382 (rank : 13) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O54792 | Gene names | Hes2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
HES2_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 14) | NC score | 0.501199 (rank : 14) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y543, Q96EN4, Q9Y542 | Gene names | HES2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
HES6_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 15) | NC score | 0.450532 (rank : 16) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96HZ4, Q8N2J2, Q96T93, Q9P2S3 | Gene names | HES6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6) (C- HAIRY1). | |||||
|
HES3_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 16) | NC score | 0.475450 (rank : 15) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61657, O09083, O09150 | Gene names | Hes3, Hes-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-3 (Hairy and enhancer of split 3). | |||||
|
HES6_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 17) | NC score | 0.428965 (rank : 17) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JHE6, Q3U2D7 | Gene names | Hes6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6). | |||||
|
BMAL1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 18) | NC score | 0.182104 (rank : 35) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 19) | NC score | 0.183231 (rank : 34) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
MXI1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 20) | NC score | 0.193074 (rank : 29) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MXI1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 21) | NC score | 0.194422 (rank : 28) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MAD1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 22) | NC score | 0.202430 (rank : 24) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
EGR4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 23) | NC score | 0.009550 (rank : 111) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
SOX10_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 24) | NC score | 0.042656 (rank : 68) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q04888, O08518, O09141, O54856, P70416 | Gene names | Sox10, Sox-10, Sox21 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10 (SOX-21) (Transcription factor SOX-M). | |||||
|
MAD1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 25) | NC score | 0.192116 (rank : 30) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q05195 | Gene names | MXD1, MAD | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
SOX10_HUMAN
|
||||||
| θ value | 0.125558 (rank : 26) | NC score | 0.041569 (rank : 69) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P56693 | Gene names | SOX10 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10. | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 27) | NC score | 0.191085 (rank : 32) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
USF1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 28) | NC score | 0.232951 (rank : 21) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 29) | NC score | 0.233014 (rank : 20) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
HES7_MOUSE
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.386499 (rank : 18) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BKT2, Q99JA6 | Gene names | Hes7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
SRBP1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 31) | NC score | 0.197899 (rank : 27) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
HES7_HUMAN
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.372051 (rank : 19) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BYE0 | Gene names | HES7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
MLX_HUMAN
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.203244 (rank : 23) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| θ value | 0.279714 (rank : 34) | NC score | 0.205543 (rank : 22) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.134477 (rank : 42) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
CLOCK_HUMAN
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.120811 (rank : 47) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
CO8A1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 37) | NC score | 0.028493 (rank : 81) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q00780, Q9D2V4 | Gene names | Col8a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VIII) chain precursor. | |||||
|
MYCL1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 38) | NC score | 0.111033 (rank : 51) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCL1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 39) | NC score | 0.115026 (rank : 49) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.043068 (rank : 67) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
TFE3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.186419 (rank : 33) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
USF2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.201897 (rank : 25) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
FOXD4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.012073 (rank : 110) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60688, Q61573 | Gene names | Foxd4, Fkh2, Fkhl9, Freac5 | |||
|
Domain Architecture |
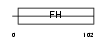
|
|||||
| Description | Forkhead box protein D4 (Forkhead-related protein FKHL9) (Forkhead- related transcription factor 5) (FREAC-5) (Transcription factor FKH- 2). | |||||
|
FUBP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.039770 (rank : 71) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96AE4, Q12828 | Gene names | FUBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP) (DNA helicase V) (HDH V). | |||||
|
FUBP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | 0.039881 (rank : 70) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91WJ8, Q8C0Y8 | Gene names | Fubp1, D3Ertd330e | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.032891 (rank : 80) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
SRBP2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.191465 (rank : 31) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
USF2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.197948 (rank : 26) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
ABRA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.034528 (rank : 76) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N0Z2 | Gene names | ABRA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding Rho-activating protein (Striated muscle activator of Rho-dependent signaling) (STARS). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.039455 (rank : 72) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CLOCK_MOUSE
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.117387 (rank : 48) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.023115 (rank : 95) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
MAD3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.151486 (rank : 39) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.046697 (rank : 66) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
CJ047_HUMAN
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.038084 (rank : 73) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86WR7, Q5W0J9, Q5W0K0, Q5W0K1, Q5W0K2, Q6PJC8, Q8N317 | Gene names | C10orf47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47. | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.034850 (rank : 75) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
FBSH_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.033766 (rank : 78) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HAH7, Q96CI9, Q9H9X4 | Gene names | FBS1, FBS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable fibrosin-1 long transcript protein. | |||||
|
NPAS2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.110389 (rank : 53) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
NPAS2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.110704 (rank : 52) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
ARNT2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.126356 (rank : 46) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.130895 (rank : 43) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
GASP2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.016680 (rank : 104) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
MITF_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.159342 (rank : 37) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.006463 (rank : 114) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
PRP2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.037758 (rank : 74) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
ARNT_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.129974 (rank : 44) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.025255 (rank : 86) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CNOT3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.023193 (rank : 93) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
FOSL2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.018591 (rank : 102) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P47930 | Gene names | Fosl2, Fra-2, Fra2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
FYB_MOUSE
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.023135 (rank : 94) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35601, Q9Z2H3 | Gene names | Fyb | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.033969 (rank : 77) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MITF_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.161648 (rank : 36) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.024461 (rank : 88) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
MYC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.092008 (rank : 56) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.032983 (rank : 79) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.026807 (rank : 83) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
RFIP3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.013682 (rank : 106) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
TFEB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.144679 (rank : 41) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.144989 (rank : 40) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
CO5A1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.021063 (rank : 100) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P20908, Q15094, Q5SUX4 | Gene names | COL5A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
CO5A1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.021391 (rank : 99) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O88207 | Gene names | Col5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
CO8A1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.025505 (rank : 85) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P27658, Q96D07 | Gene names | COL8A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VIII) chain precursor (Endothelial collagen). | |||||
|
COAA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.024600 (rank : 87) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q03692 | Gene names | COL10A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(X) chain precursor. | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.023401 (rank : 92) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MYBPH_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.008212 (rank : 112) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70402 | Gene names | Mybph | |||
|
Domain Architecture |
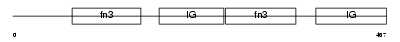
|
|||||
| Description | Myosin-binding protein H (MyBP-H) (H-protein). | |||||
|
SF3B4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.015354 (rank : 105) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
SUHW3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.002962 (rank : 116) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8ND82, Q9NXR3 | Gene names | SUHW3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 3. | |||||
|
TAF6L_MOUSE
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.023529 (rank : 91) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R2K4, Q8C5T2 | Gene names | Taf6l, Paf65a | |||
|
Domain Architecture |
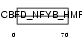
|
|||||
| Description | TAF6-like RNA polymerase II p300/CBP-associated factor-associated factor 65 kDa subunit 6L (PCAF-associated factor 65 alpha) (PAF65- alpha). | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.022393 (rank : 96) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.022010 (rank : 97) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
H6ST3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.013039 (rank : 108) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QYK4 | Gene names | Hs6st3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3) (mHS6ST- 3). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.012981 (rank : 109) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.023945 (rank : 90) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
SYT3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.004815 (rank : 115) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQG1, Q8N5Z1, Q8N640 | Gene names | SYT3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
TMPS9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.002127 (rank : 118) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z410, Q6ZND6, Q7Z411 | Gene names | TMPRSS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 9 (EC 3.4.21.-) (Polyserase-1) (Polyserase-I) (Polyserine protease 1) [Contains: Serase-1; Serase-2; Serase-3]. | |||||
|
CBS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.021491 (rank : 98) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35520, Q99425, Q9BWC5 | Gene names | CBS | |||
|
Domain Architecture |
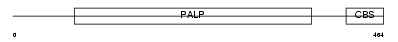
|
|||||
| Description | Cystathionine beta-synthase (EC 4.2.1.22) (Serine sulfhydrase) (Beta- thionase). | |||||
|
MAD3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.128418 (rank : 45) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
MAD4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.156492 (rank : 38) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.013667 (rank : 107) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.018791 (rank : 101) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.018557 (rank : 103) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.025547 (rank : 84) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.002578 (rank : 117) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
PCGF4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.007488 (rank : 113) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35226, Q16030, Q96F37 | Gene names | PCGF4, BMI1, RNF51 | |||
|
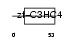
Domain Architecture |
|
|||||
| Description | Polycomb group RING finger protein 4 (Polycomb complex protein BMI-1) (RING finger protein 51). | |||||
|
PRPC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.027983 (rank : 82) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
TMAP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.024001 (rank : 89) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
MAD4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.113384 (rank : 50) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MAX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.092165 (rank : 55) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MAX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.092692 (rank : 54) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
MNT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.058483 (rank : 60) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MNT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.060191 (rank : 59) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MYCL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.053577 (rank : 62) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
MYCN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.051119 (rank : 63) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.060665 (rank : 58) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.050597 (rank : 64) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
MYC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.063686 (rank : 57) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
NCOA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.050289 (rank : 65) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
TCFL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.056871 (rank : 61) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
BHLH3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.29261e-131 (rank : 1) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
BHLH3_MOUSE
|
||||||
| NC score | 0.981441 (rank : 2) | θ value | 1.34075e-120 (rank : 2) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
BHLH2_MOUSE
|
||||||
| NC score | 0.926259 (rank : 3) | θ value | 1.61343e-57 (rank : 3) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
BHLH2_HUMAN
|
||||||
| NC score | 0.917906 (rank : 4) | θ value | 9.78833e-55 (rank : 4) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
HES4_HUMAN
|
||||||
| NC score | 0.609596 (rank : 5) | θ value | 8.97725e-08 (rank : 7) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9HCC6 | Gene names | HES4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-4 (Hairy and enhancer of split 4) (bHLH factor Hes4). | |||||
|
HES1_HUMAN
|
||||||
| NC score | 0.588307 (rank : 6) | θ value | 4.76016e-09 (rank : 5) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
HES1_MOUSE
|
||||||
| NC score | 0.585492 (rank : 7) | θ value | 4.76016e-09 (rank : 6) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
HES5_MOUSE
|
||||||
| NC score | 0.575593 (rank : 8) | θ value | 8.40245e-06 (rank : 12) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
HEY2_MOUSE
|
||||||
| NC score | 0.558761 (rank : 9) | θ value | 3.41135e-07 (rank : 8) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
HEY2_HUMAN
|
||||||
| NC score | 0.554782 (rank : 10) | θ value | 5.81887e-07 (rank : 9) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY1_MOUSE
|
||||||
| NC score | 0.549050 (rank : 11) | θ value | 9.92553e-07 (rank : 10) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY1_HUMAN
|
||||||
| NC score | 0.541326 (rank : 12) | θ value | 1.69304e-06 (rank : 11) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HES2_MOUSE
|
||||||
| NC score | 0.504382 (rank : 13) | θ value | 0.00228821 (rank : 13) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O54792 | Gene names | Hes2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
HES2_HUMAN
|
||||||
| NC score | 0.501199 (rank : 14) | θ value | 0.00390308 (rank : 14) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y543, Q96EN4, Q9Y542 | Gene names | HES2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
HES3_MOUSE
|
||||||
| NC score | 0.475450 (rank : 15) | θ value | 0.00509761 (rank : 16) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61657, O09083, O09150 | Gene names | Hes3, Hes-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-3 (Hairy and enhancer of split 3). | |||||
|
HES6_HUMAN
|
||||||
| NC score | 0.450532 (rank : 16) | θ value | 0.00390308 (rank : 15) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96HZ4, Q8N2J2, Q96T93, Q9P2S3 | Gene names | HES6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6) (C- HAIRY1). | |||||
|
HES6_MOUSE
|
||||||
| NC score | 0.428965 (rank : 17) | θ value | 0.0193708 (rank : 17) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JHE6, Q3U2D7 | Gene names | Hes6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6). | |||||
|
HES7_MOUSE
|
||||||
| NC score | 0.386499 (rank : 18) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BKT2, Q99JA6 | Gene names | Hes7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
HES7_HUMAN
|
||||||
| NC score | 0.372051 (rank : 19) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BYE0 | Gene names | HES7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
USF1_MOUSE
|
||||||
| NC score | 0.233014 (rank : 20) | θ value | 0.125558 (rank : 29) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_HUMAN
|
||||||
| NC score | 0.232951 (rank : 21) | θ value | 0.125558 (rank : 28) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
MLX_MOUSE
|
||||||
| NC score | 0.205543 (rank : 22) | θ value | 0.279714 (rank : 34) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_HUMAN
|
||||||
| NC score | 0.203244 (rank : 23) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MAD1_MOUSE
|
||||||
| NC score | 0.202430 (rank : 24) | θ value | 0.0431538 (rank : 22) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
USF2_MOUSE
|
||||||
| NC score | 0.201897 (rank : 25) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
USF2_HUMAN
|
||||||
| NC score | 0.197948 (rank : 26) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
SRBP1_HUMAN
|
||||||
| NC score | 0.197899 (rank : 27) | θ value | 0.21417 (rank : 31) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
MXI1_HUMAN
|
||||||
| NC score | 0.194422 (rank : 28) | θ value | 0.0330416 (rank : 21) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MXI1_MOUSE
|
||||||
| NC score | 0.193074 (rank : 29) | θ value | 0.0252991 (rank : 20) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MAD1_HUMAN
|
||||||
| NC score | 0.192116 (rank : 30) | θ value | 0.125558 (rank : 25) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q05195 | Gene names | MXD1, MAD | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
SRBP2_HUMAN
|
||||||
| NC score | 0.191465 (rank : 31) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.191085 (rank : 32) | θ value | 0.125558 (rank : 27) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.186419 (rank : 33) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
BMAL1_MOUSE
|
||||||
| NC score | 0.183231 (rank : 34) | θ value | 0.0252991 (rank : 19) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
BMAL1_HUMAN
|
||||||
| NC score | 0.182104 (rank : 35) | θ value | 0.0252991 (rank : 18) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.161648 (rank : 36) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.159342 (rank : 37) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MAD4_HUMAN
|
||||||
| NC score | 0.156492 (rank : 38) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MAD3_HUMAN
|
||||||
| NC score | 0.151486 (rank : 39) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.144989 (rank : 40) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.144679 (rank : 41) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.134477 (rank : 42) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT2_MOUSE
|
||||||
| NC score | 0.130895 (rank : 43) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.129974 (rank : 44) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
MAD3_MOUSE
|
||||||
| NC score | 0.128418 (rank : 45) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
ARNT2_HUMAN
|
||||||
| NC score | 0.126356 (rank : 46) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
CLOCK_HUMAN
|
||||||
| NC score | 0.120811 (rank : 47) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
CLOCK_MOUSE
|
||||||
| NC score | 0.117387 (rank : 48) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
MYCL1_MOUSE
|
||||||
| NC score | 0.115026 (rank : 49) | θ value | 0.62314 (rank : 39) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MAD4_MOUSE
|
||||||
| NC score | 0.113384 (rank : 50) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MYCL1_HUMAN
|
||||||
| NC score | 0.111033 (rank : 51) | θ value | 0.62314 (rank : 38) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
NPAS2_MOUSE
|
||||||
| NC score | 0.110704 (rank : 52) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
NPAS2_HUMAN
|
||||||
| NC score | 0.110389 (rank : 53) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
MAX_MOUSE
|
||||||
| NC score | 0.092692 (rank : 54) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
MAX_HUMAN
|
||||||
| NC score | 0.092165 (rank : 55) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MYC_HUMAN
|
||||||
| NC score | 0.092008 (rank : 56) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYC_MOUSE
|
||||||
| NC score | 0.063686 (rank : 57) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYCN_MOUSE
|
||||||
| NC score | 0.060665 (rank : 58) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.060191 (rank : 59) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.058483 (rank : 60) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
TCFL5_HUMAN
|
||||||
| NC score | 0.056871 (rank : 61) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
MYCL2_HUMAN
|
||||||
| NC score | 0.053577 (rank : 62) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
MYCN_HUMAN
|
||||||
| NC score | 0.051119 (rank : 63) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCS_MOUSE
|
||||||
| NC score | 0.050597 (rank : 64) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
NCOA1_HUMAN
|
||||||
| NC score | 0.050289 (rank : 65) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.046697 (rank : 66) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.043068 (rank : 67) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
SOX10_MOUSE
|
||||||
| NC score | 0.042656 (rank : 68) | θ value | 0.0736092 (rank : 24) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q04888, O08518, O09141, O54856, P70416 | Gene names | Sox10, Sox-10, Sox21 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10 (SOX-21) (Transcription factor SOX-M). | |||||
|
SOX10_HUMAN
|
||||||
| NC score | 0.041569 (rank : 69) | θ value | 0.125558 (rank : 26) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P56693 | Gene names | SOX10 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-10. | |||||
|
FUBP1_MOUSE
|
||||||
| NC score | 0.039881 (rank : 70) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91WJ8, Q8C0Y8 | Gene names | Fubp1, D3Ertd330e | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP). | |||||
|
FUBP1_HUMAN
|
||||||
| NC score | 0.039770 (rank : 71) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96AE4, Q12828 | Gene names | FUBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 1 (FUSE-binding protein 1) (FBP) (DNA helicase V) (HDH V). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.039455 (rank : 72) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CJ047_HUMAN
|
||||||
| NC score | 0.038084 (rank : 73) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86WR7, Q5W0J9, Q5W0K0, Q5W0K1, Q5W0K2, Q6PJC8, Q8N317 | Gene names | C10orf47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47. | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.037758 (rank : 74) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.034850 (rank : 75) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
ABRA_HUMAN
|
||||||
| NC score | 0.034528 (rank : 76) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N0Z2 | Gene names | ABRA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding Rho-activating protein (Striated muscle activator of Rho-dependent signaling) (STARS). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.033969 (rank : 77) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
FBSH_HUMAN
|
||||||
| NC score | 0.033766 (rank : 78) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HAH7, Q96CI9, Q9H9X4 | Gene names | FBS1, FBS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable fibrosin-1 long transcript protein. | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.032983 (rank : 79) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.032891 (rank : 80) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
CO8A1_MOUSE
|
||||||
| NC score | 0.028493 (rank : 81) | θ value | 0.62314 (rank : 37) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q00780, Q9D2V4 | Gene names | Col8a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VIII) chain precursor. | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.027983 (rank : 82) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.026807 (rank : 83) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.025547 (rank : 84) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CO8A1_HUMAN
|
||||||
| NC score | 0.025505 (rank : 85) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P27658, Q96D07 | Gene names | COL8A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VIII) chain precursor (Endothelial collagen). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.025255 (rank : 86) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
COAA1_HUMAN
|
||||||
| NC score | 0.024600 (rank : 87) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q03692 | Gene names | COL10A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(X) chain precursor. | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.024461 (rank : 88) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
TMAP1_HUMAN
|
||||||
| NC score | 0.024001 (rank : 89) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P84157 | Gene names | TMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane anchor protein 1. | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.023945 (rank : 90) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
TAF6L_MOUSE
|
||||||
| NC score | 0.023529 (rank : 91) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R2K4, Q8C5T2 | Gene names | Taf6l, Paf65a | |||
|
Domain Architecture |

|
|||||
| Description | TAF6-like RNA polymerase II p300/CBP-associated factor-associated factor 65 kDa subunit 6L (PCAF-associated factor 65 alpha) (PAF65- alpha). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.023401 (rank : 92) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
CNOT3_MOUSE
|
||||||
| NC score | 0.023193 (rank : 93) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
FYB_MOUSE
|
||||||
| NC score | 0.023135 (rank : 94) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35601, Q9Z2H3 | Gene names | Fyb | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.023115 (rank : 95) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.022393 (rank : 96) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.022010 (rank : 97) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
CBS_HUMAN
|
||||||
| NC score | 0.021491 (rank : 98) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35520, Q99425, Q9BWC5 | Gene names | CBS | |||
|
Domain Architecture |

|
|||||
| Description | Cystathionine beta-synthase (EC 4.2.1.22) (Serine sulfhydrase) (Beta- thionase). | |||||
|
CO5A1_MOUSE
|
||||||
| NC score | 0.021391 (rank : 99) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O88207 | Gene names | Col5a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
CO5A1_HUMAN
|
||||||
| NC score | 0.021063 (rank : 100) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P20908, Q15094, Q5SUX4 | Gene names | COL5A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(V) chain precursor. | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.018791 (rank : 101) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
FOSL2_MOUSE
|
||||||
| NC score | 0.018591 (rank : 102) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P47930 | Gene names | Fosl2, Fra-2, Fra2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fos-related antigen 2. | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.018557 (rank : 103) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
GASP2_MOUSE
|
||||||
| NC score | 0.016680 (rank : 104) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
SF3B4_HUMAN
|
||||||
| NC score | 0.015354 (rank : 105) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
RFIP3_HUMAN
|
||||||
| NC score | 0.013682 (rank : 106) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.013667 (rank : 107) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
H6ST3_MOUSE
|
||||||
| NC score | 0.013039 (rank : 108) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QYK4 | Gene names | Hs6st3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3) (mHS6ST- 3). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.012981 (rank : 109) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
FOXD4_MOUSE
|
||||||
| NC score | 0.012073 (rank : 110) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60688, Q61573 | Gene names | Foxd4, Fkh2, Fkhl9, Freac5 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D4 (Forkhead-related protein FKHL9) (Forkhead- related transcription factor 5) (FREAC-5) (Transcription factor FKH- 2). | |||||
|
EGR4_HUMAN
|
||||||
| NC score | 0.009550 (rank : 111) | θ value | 0.0736092 (rank : 23) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
MYBPH_MOUSE
|
||||||
| NC score | 0.008212 (rank : 112) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70402 | Gene names | Mybph | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-binding protein H (MyBP-H) (H-protein). | |||||
|
PCGF4_HUMAN
|
||||||
| NC score | 0.007488 (rank : 113) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35226, Q16030, Q96F37 | Gene names | PCGF4, BMI1, RNF51 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb group RING finger protein 4 (Polycomb complex protein BMI-1) (RING finger protein 51). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.006463 (rank : 114) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
SYT3_HUMAN
|
||||||
| NC score | 0.004815 (rank : 115) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQG1, Q8N5Z1, Q8N640 | Gene names | SYT3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
SUHW3_HUMAN
|
||||||
| NC score | 0.002962 (rank : 116) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8ND82, Q9NXR3 | Gene names | SUHW3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 3. | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.002578 (rank : 117) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
TMPS9_HUMAN
|
||||||
| NC score | 0.002127 (rank : 118) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z410, Q6ZND6, Q7Z411 | Gene names | TMPRSS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 9 (EC 3.4.21.-) (Polyserase-1) (Polyserase-I) (Polyserine protease 1) [Contains: Serase-1; Serase-2; Serase-3]. | |||||