Please be patient as the page loads
|
MYC_MOUSE
|
||||||
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MYC_MOUSE
|
||||||
| θ value | 7.71569e-185 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYC_HUMAN
|
||||||
| θ value | 1.19313e-177 (rank : 2) | NC score | 0.977625 (rank : 2) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYCB_MOUSE
|
||||||
| θ value | 1.85029e-45 (rank : 3) | NC score | 0.830056 (rank : 3) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6P8Z1, Q9D6E3 | Gene names | Mycb, Bmyc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein B-Myc. | |||||
|
MYCS_MOUSE
|
||||||
| θ value | 4.72714e-33 (rank : 4) | NC score | 0.809443 (rank : 6) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
MYCN_HUMAN
|
||||||
| θ value | 8.06329e-33 (rank : 5) | NC score | 0.827060 (rank : 5) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCN_MOUSE
|
||||||
| θ value | 3.06405e-32 (rank : 6) | NC score | 0.827098 (rank : 4) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCL1_HUMAN
|
||||||
| θ value | 3.17079e-29 (rank : 7) | NC score | 0.784725 (rank : 7) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCL1_MOUSE
|
||||||
| θ value | 7.0802e-21 (rank : 8) | NC score | 0.750381 (rank : 8) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCL2_HUMAN
|
||||||
| θ value | 1.47631e-18 (rank : 9) | NC score | 0.712192 (rank : 9) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
MAX_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 10) | NC score | 0.622796 (rank : 10) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MAX_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 11) | NC score | 0.612754 (rank : 11) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
MAD3_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 12) | NC score | 0.386049 (rank : 14) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
MAD3_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 13) | NC score | 0.379278 (rank : 15) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
MXI1_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 14) | NC score | 0.394275 (rank : 12) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MXI1_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 15) | NC score | 0.387163 (rank : 13) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
USF1_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 16) | NC score | 0.372626 (rank : 16) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
MNT_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 17) | NC score | 0.350142 (rank : 22) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MNT_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 18) | NC score | 0.359358 (rank : 21) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
USF1_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 19) | NC score | 0.367768 (rank : 17) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
SRBP2_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 20) | NC score | 0.282593 (rank : 29) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
MAD4_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 21) | NC score | 0.359403 (rank : 20) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
PTF1A_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 22) | NC score | 0.242060 (rank : 37) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7RTS3, Q9HC25 | Gene names | PTF1A, PTF1P48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
BHLH8_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 23) | NC score | 0.269632 (rank : 30) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9QYC3, Q9QYE4 | Gene names | Bhlhb8, Mist1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
MAD4_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 24) | NC score | 0.318226 (rank : 26) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
FIGLA_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 25) | NC score | 0.324289 (rank : 24) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O55208 | Gene names | Figla | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
FIGLA_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 26) | NC score | 0.320960 (rank : 25) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6QHK4 | Gene names | FIGLA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
USF2_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 27) | NC score | 0.364873 (rank : 18) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 28) | NC score | 0.362055 (rank : 19) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
MLX_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 29) | NC score | 0.287996 (rank : 28) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 30) | NC score | 0.289930 (rank : 27) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
TFAP4_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 31) | NC score | 0.342529 (rank : 23) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
TWST2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 32) | NC score | 0.223038 (rank : 42) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8WVJ9 | Gene names | TWIST2, DERMO1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
TWST2_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 33) | NC score | 0.223139 (rank : 41) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9D030 | Gene names | Twist2, Dermo1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
BHLH8_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 34) | NC score | 0.260937 (rank : 32) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7RTS1 | Gene names | BHLHB8, MIST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
ATOH1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 35) | NC score | 0.166036 (rank : 65) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q92858 | Gene names | ATOH1, ATH1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein hATH-1). | |||||
|
ATOH1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 36) | NC score | 0.165779 (rank : 66) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P48985 | Gene names | Atoh1, Ath1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein mATH-1) (MATH1). | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 37) | NC score | 0.241253 (rank : 38) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
MYOG_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 38) | NC score | 0.209368 (rank : 49) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P15173 | Gene names | MYOG, MYF4 | |||
|
Domain Architecture |
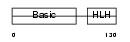
|
|||||
| Description | Myogenin (Myogenic factor 4) (Myf-4). | |||||
|
MYOG_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 39) | NC score | 0.209239 (rank : 51) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P12979 | Gene names | Myog | |||
|
Domain Architecture |
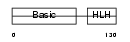
|
|||||
| Description | Myogenin (MYOD1-related protein). | |||||
|
SRBP1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 40) | NC score | 0.249307 (rank : 34) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
WBS14_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 41) | NC score | 0.185908 (rank : 58) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
MYF6_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 42) | NC score | 0.204970 (rank : 54) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P23409, Q53X80, Q6FHI9 | Gene names | MYF6 | |||
|
Domain Architecture |
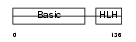
|
|||||
| Description | Myogenic factor 6 (Myf-6). | |||||
|
MYF6_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 43) | NC score | 0.200885 (rank : 55) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P15375 | Gene names | Myf6, Myf-6 | |||
|
Domain Architecture |
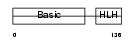
|
|||||
| Description | Myogenic factor 6 (Myf-6) (Herculin). | |||||
|
ASCL2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 44) | NC score | 0.231165 (rank : 39) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O35885, Q9WUJ7 | Gene names | Ascl2, Mash2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash-2). | |||||
|
PTF1A_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 45) | NC score | 0.229715 (rank : 40) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9QX98, Q9QYF5, Q9QYF6 | Gene names | Ptf1a, Ptf1p48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
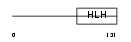
SCX_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 46) | NC score | 0.187368 (rank : 57) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64124 | Gene names | Scx | |||
|
Domain Architecture |
|
|||||
| Description | Basic helix-loop-helix transcription factor scleraxis. | |||||
|
WBS14_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 47) | NC score | 0.153636 (rank : 68) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
ARNT_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 48) | NC score | 0.137753 (rank : 71) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
TWST1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 49) | NC score | 0.222770 (rank : 43) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q15672, Q92487, Q99804 | Gene names | TWIST1, TWIST | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (H-twist). | |||||
|
TWST1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 50) | NC score | 0.222334 (rank : 44) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P26687 | Gene names | Twist1, Twist | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (M-twist). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 51) | NC score | 0.044461 (rank : 128) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 52) | NC score | 0.031624 (rank : 136) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
MITF_MOUSE
|
||||||
| θ value | 0.125558 (rank : 53) | NC score | 0.246798 (rank : 35) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
TFE3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 54) | NC score | 0.266479 (rank : 31) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 0.163984 (rank : 55) | NC score | 0.131563 (rank : 72) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
MITF_HUMAN
|
||||||
| θ value | 0.163984 (rank : 56) | NC score | 0.250261 (rank : 33) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
ARNT2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 57) | NC score | 0.122023 (rank : 78) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 58) | NC score | 0.122717 (rank : 77) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
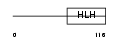
HAND2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 59) | NC score | 0.215401 (rank : 46) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P61296, O95300, O95301, P97833 | Gene names | HAND2, DHAND | |||
|
Domain Architecture |
|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND). | |||||
|
HAND2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 60) | NC score | 0.215401 (rank : 47) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61039, Q61100 | Gene names | Hand2, Dhand, Hed, Thing2 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND) (Helix-loop-helix transcription factor expressed in embryo and deciduum) (Thing-2). | |||||
|
MYOD1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 61) | NC score | 0.173434 (rank : 60) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P10085 | Gene names | Myod1, Myod | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1. | |||||
|
CP250_MOUSE
|
||||||
| θ value | 0.279714 (rank : 62) | NC score | 0.015380 (rank : 168) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
MYOD1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 63) | NC score | 0.170834 (rank : 62) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P15172 | Gene names | MYOD1, MYF3, MYOD | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1 (Myogenic factor 3) (Myf-3). | |||||
|
PZRN3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 64) | NC score | 0.040742 (rank : 129) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UPQ7, Q8N2N7, Q96CC2, Q9NSQ2 | Gene names | PDZRN3, KIAA1095, LNX3, SEMCAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Ligand of Numb-protein X 3) (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
UBP28_MOUSE
|
||||||
| θ value | 0.279714 (rank : 65) | NC score | 0.033233 (rank : 134) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
ASCL1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 66) | NC score | 0.218984 (rank : 45) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P50553, Q9BQ30 | Gene names | ASCL1, ASH1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (HASH1). | |||||
|
ASCL1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 67) | NC score | 0.209748 (rank : 48) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q02067 | Gene names | Ascl1, Ash1, Mash-1, Mash1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (Mash-1). | |||||
|
DHX16_HUMAN
|
||||||
| θ value | 0.365318 (rank : 68) | NC score | 0.022009 (rank : 153) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60231, O60322, Q969X7, Q96QC1 | Gene names | DHX16, DBP2, DDX16, KIAA0577 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 (EC 3.6.1.-) (DEAH-box protein 16) (ATP-dependent RNA helicase #3). | |||||
|
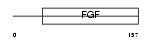
FGF23_HUMAN
|
||||||
| θ value | 0.365318 (rank : 69) | NC score | 0.030673 (rank : 138) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9GZV9, Q4V758 | Gene names | FGF23, HYPF | |||
|
Domain Architecture |
|
|||||
| Description | Fibroblast growth factor 23 precursor (FGF-23) (Tumor-derived hypophosphatemia-inducing factor). | |||||
|
OXR1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 70) | NC score | 0.030608 (rank : 139) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
SNX23_HUMAN
|
||||||
| θ value | 0.365318 (rank : 71) | NC score | 0.015966 (rank : 166) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96L93, Q5HYC0, Q5HYK1, Q5JWW3, Q5TFK5, Q86VL9, Q86YS5, Q8IYU0, Q9BQJ8, Q9BQM0, Q9BQM1, Q9BQM5, Q9H5U0, Q9HCI2, Q9NXN9 | Gene names | C20orf23, KIAA1590, SNX23 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like motor protein C20orf23 (Sorting nexin-23). | |||||
|
DTNA_HUMAN
|
||||||
| θ value | 0.47712 (rank : 72) | NC score | 0.032699 (rank : 135) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y4J8, O15332, O15333, O75697, Q13197, Q13198, Q13199, Q13498, Q13499, Q13500 | Gene names | DTNA, DRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin alpha (Dystrobrevin-alpha) (Dystrophin-related protein 3). | |||||
|
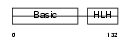
MYF5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 73) | NC score | 0.170629 (rank : 63) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P13349 | Gene names | MYF5 | |||
|
Domain Architecture |
|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
MYF5_MOUSE
|
||||||
| θ value | 0.47712 (rank : 74) | NC score | 0.171269 (rank : 61) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P24699, Q543W7 | Gene names | Myf5, Myf-5 | |||
|
Domain Architecture |
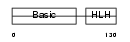
|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
ASXL1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 75) | NC score | 0.036309 (rank : 132) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IXJ9, Q5JWS9, Q8IYY7, Q9H466, Q9NQF8, Q9UFJ0, Q9UFP8, Q9Y2I4 | Gene names | ASXL1, KIAA0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
K0586_HUMAN
|
||||||
| θ value | 0.62314 (rank : 76) | NC score | 0.045939 (rank : 127) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BVV6, O60328 | Gene names | KIAA0586 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0586. | |||||
|
LPIN1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 77) | NC score | 0.021717 (rank : 155) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZP3, Q9CQI2, Q9JLG6 | Gene names | Lpin1, Fld | |||
|
Domain Architecture |
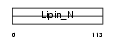
|
|||||
| Description | Lipin-1 (Fatty liver dystrophy protein). | |||||
|
TFEB_HUMAN
|
||||||
| θ value | 0.62314 (rank : 78) | NC score | 0.208625 (rank : 52) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| θ value | 0.62314 (rank : 79) | NC score | 0.209317 (rank : 50) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 80) | NC score | 0.025862 (rank : 148) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
PZRN3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 81) | NC score | 0.034966 (rank : 133) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q69ZS0, Q91Z03, Q9QY54, Q9QY55 | Gene names | Pdzrn3, Kiaa1095, Semcap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 1.38821 (rank : 82) | NC score | 0.024647 (rank : 150) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
DTNA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 83) | NC score | 0.029414 (rank : 142) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D2N4, P97319, Q61498, Q61499, Q9QZZ5, Q9WUL9, Q9WUM0 | Gene names | Dtna, Dtn | |||
|
Domain Architecture |
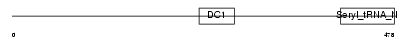
|
|||||
| Description | Dystrobrevin alpha (Alpha-dystrobrevin). | |||||
|
KIF3A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 84) | NC score | 0.018803 (rank : 159) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y496, Q86XE9, Q9Y6V4 | Gene names | KIF3A, KIF3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
CCD91_HUMAN
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | 0.022267 (rank : 152) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
SDC2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.028242 (rank : 144) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43407 | Gene names | Sdc2, Hspg1, Synd2 | |||
|
Domain Architecture |
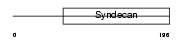
|
|||||
| Description | Syndecan-2 precursor (SYND2) (Fibroglycan) (Heparan sulfate proteoglycan core protein) (HSPG). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 87) | NC score | 0.025919 (rank : 147) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TCF15_MOUSE
|
||||||
| θ value | 1.81305 (rank : 88) | NC score | 0.139190 (rank : 70) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q60756, Q60788 | Gene names | Tcf15, Bhlhec2 | |||
|
Domain Architecture |
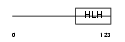
|
|||||
| Description | Transcription factor 15 (bHLH-EC2 protein) (Paraxis) (Meso1). | |||||
|
DACT1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 89) | NC score | 0.020748 (rank : 157) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NYF0, Q86TY0 | Gene names | DACT1, DPR1, HNG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (hDPR1) (Heptacellular carcinoma novel gene 3 protein). | |||||
|
HAND1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.179708 (rank : 59) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O96004 | Gene names | HAND1, EHAND | |||
|
Domain Architecture |
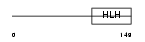
|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND). | |||||
|
HAND1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.191907 (rank : 56) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q64279, Q61099 | Gene names | Hand1, Ehand, Hxt, Thing1 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND) (Helix-loop-helix transcription factor expressed in extraembryonic mesoderm and trophoblast) (Thing-1) (Th1). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.029488 (rank : 141) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
K0319_MOUSE
|
||||||
| θ value | 2.36792 (rank : 93) | NC score | 0.036520 (rank : 130) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SZV5, Q80U39 | Gene names | Kiaa0319 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0319 precursor. | |||||
|
KLRA2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 94) | NC score | 0.013892 (rank : 170) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60660 | Gene names | Klra2, Ly49-b, Ly49b | |||
|
Domain Architecture |
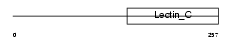
|
|||||
| Description | Killer cell lectin-like receptor 2 (T-cell surface glycoprotein Ly- 49B) (Ly49-B antigen) (Lymphocyte antigen 49B). | |||||
|
SAFB2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 95) | NC score | 0.016467 (rank : 164) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14151, Q8TB13 | Gene names | SAFB2, KIAA0138 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
STX18_HUMAN
|
||||||
| θ value | 2.36792 (rank : 96) | NC score | 0.024782 (rank : 149) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P2W9, Q596L3, Q5TZP5 | Gene names | STX18, GIG9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-18 (Growth-inhibiting gene 9 protein). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 2.36792 (rank : 97) | NC score | 0.022462 (rank : 151) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.046497 (rank : 126) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.015699 (rank : 167) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
CEBPE_HUMAN
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.030426 (rank : 140) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
FBX38_HUMAN
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.026571 (rank : 146) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PIJ6, Q6PK72, Q7Z2U0, Q86VN3, Q9BXY6, Q9H837, Q9HC40 | Gene names | FBXO38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 38 (Modulator of KLF7 activity homolog) (MoKA). | |||||
|
JHD2C_HUMAN
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.014335 (rank : 169) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15652, Q5SQZ8, Q5SQZ9, Q5SR00, Q7Z3E7, Q8N3U0, Q96KB9, Q9P2G7 | Gene names | JMJD1C, JHDM2C, KIAA1380, TRIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C) (Thyroid receptor-interacting protein 8) (TRIP-8). | |||||
|
METL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.036458 (rank : 131) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96IZ6, Q9H9G9, Q9NUI8, Q9P0B5 | Gene names | METTL2 | |||
|
Domain Architecture |

|
|||||
| Description | Methyltransferase-like protein 2 (EC 2.1.1.-). | |||||
|
REN3B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 104) | NC score | 0.021557 (rank : 156) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BZI7, Q9H1J0 | Gene names | UPF3B, RENT3B, UPF3X | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 3B (Nonsense mRNA reducing factor 3B) (Up-frameshift suppressor 3 homolog B) (hUpf3B) (hUpf3p-X). | |||||
|
FA21C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.028903 (rank : 143) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
K0056_HUMAN
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.016648 (rank : 162) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42695 | Gene names | KIAA0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
TCF15_HUMAN
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.128105 (rank : 73) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q12870, Q9NQQ1 | Gene names | TCF15, BHLHEC2 | |||
|
Domain Architecture |
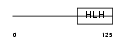
|
|||||
| Description | Transcription factor 15 (bHLH-EC2 protein) (Paraxis). | |||||
|
TTC14_HUMAN
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.027540 (rank : 145) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
ZFYV1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.018547 (rank : 160) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HBF4, Q8WYX7, Q96K57, Q9BXP9, Q9HCI3 | Gene names | ZFYVE1, DFCP1, KIAA1589, TAFF1, ZNFN2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 1 (Double FYVE-containing protein 1) (Tandem FYVE fingers-1) (SR3). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.018350 (rank : 161) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.021731 (rank : 154) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
MTR1L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | -0.003242 (rank : 180) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1180 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13585 | Gene names | GPR50 | |||
|
Domain Architecture |

|
|||||
| Description | Melatonin-related receptor (G protein-coupled receptor 50) (H9). | |||||
|
NEMO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.018948 (rank : 158) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y6K9 | Gene names | IKBKG, FIP3, NEMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (FIP-3). | |||||
|
NUFP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.013107 (rank : 171) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QXX8, Q9CV69 | Gene names | Nufip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 1 (Nuclear FMRP-interacting protein 1). | |||||
|
PTPRD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | -0.000931 (rank : 179) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23468 | Gene names | PTPRD | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
ASXL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.030757 (rank : 137) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59598 | Gene names | Asxl1, Kiaa0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
CEBPA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.016488 (rank : 163) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49715, P78319 | Gene names | CEBPA | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
CEBPA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.016298 (rank : 165) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53566, Q91XB6 | Gene names | Cebpa | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.009427 (rank : 176) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
GOGA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.007526 (rank : 177) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
GP149_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.009945 (rank : 173) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86SP6 | Gene names | GPR149, PGR10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 149 (G-protein coupled receptor PGR10). | |||||
|
INCE_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.009912 (rank : 174) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | -0.000510 (rank : 178) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.009600 (rank : 175) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
NUP88_MOUSE
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.012654 (rank : 172) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CEC0, Q80Z13, Q8K090 | Gene names | Nup88 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear pore complex protein Nup88 (Nucleoporin Nup88) (88 kDa nuclear pore complex protein). | |||||
|
ASCL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.166572 (rank : 64) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99929, Q9UM68 | Gene names | ASCL2 | |||
|
Domain Architecture |
|
|||||
| Description | Achaete-scute homolog 2 (Mash2) (HASH2). | |||||
|
ASCL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.153844 (rank : 67) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NQ33, Q8WYQ6 | Gene names | ASCL3, HASH3, SGN1 | |||
|
Domain Architecture |
|
|||||
| Description | Achaete-scute homolog 3 (bHLH transcriptional regulator Sgn-1). | |||||
|
ASCL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.149600 (rank : 69) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9JJR7 | Gene names | Ascl3, Mash3, Sgn1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 3 (bHLH transcriptional regulator Sgn-1) (Mash- 3). | |||||
|
BHLH2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.055899 (rank : 120) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
BHLH2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.055454 (rank : 121) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
BHLH3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.063686 (rank : 115) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
BHLH3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.058417 (rank : 118) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
BHLH4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.070855 (rank : 106) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NDY6 | Gene names | BHLHB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 4 (bHLHB4). | |||||
|
BHLH4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.068770 (rank : 108) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BGW3, Q71MB7 | Gene names | Bhlhb4, H20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 4 (bHLHB4). | |||||
|
BMAL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.078686 (rank : 100) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.078260 (rank : 101) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
HEN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.106234 (rank : 88) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q02575 | Gene names | NHLH1, HEN1 | |||
|
Domain Architecture |
|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
HEN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.107107 (rank : 86) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q02576 | Gene names | Nhlh1, Hen1 | |||
|
Domain Architecture |
|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
HEN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.103685 (rank : 90) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q02577 | Gene names | NHLH2, HEN2 | |||
|
Domain Architecture |
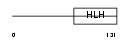
|
|||||
| Description | Helix-loop-helix protein 2 (HEN2) (Nescient helix loop helix 2) (NSCL- 2). | |||||
|
HEN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.103030 (rank : 91) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q64221 | Gene names | Nhlh2, Hen2 | |||
|
Domain Architecture |
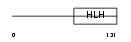
|
|||||
| Description | Helix-loop-helix protein 2 (HEN2) (Nescient helix loop helix 2) (NSCL- 2). | |||||
|
HEY1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.051717 (rank : 123) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HEY1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.051343 (rank : 124) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
HEY2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.054195 (rank : 122) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.056069 (rank : 119) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
ID1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.077927 (rank : 102) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P41134, O00651, O00652, Q16371, Q16377, Q5TE66, Q5TE67, Q969Z7, Q9H0Z5, Q9H109 | Gene names | ID1, ID | |||
|
Domain Architecture |
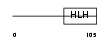
|
|||||
| Description | DNA-binding protein inhibitor ID-1 (Inhibitor of DNA binding 1). | |||||
|
ID1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.072241 (rank : 105) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P20067, Q61101, Q9D897 | Gene names | Id1, Id, Id-1, Idb1 | |||
|
Domain Architecture |
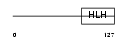
|
|||||
| Description | DNA-binding protein inhibitor ID-1 (Inhibitor of DNA binding 1). | |||||
|
ID4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.050867 (rank : 125) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P47928, Q13005 | Gene names | ID4 | |||
|
Domain Architecture |
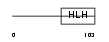
|
|||||
| Description | DNA-binding protein inhibitor ID-4 (Inhibitor of DNA binding 4). | |||||
|
LYL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.113312 (rank : 81) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P12980, O76102 | Gene names | LYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
LYL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.113896 (rank : 80) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P27792 | Gene names | Lyl1, Lyl-1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
MAD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.205753 (rank : 53) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q05195 | Gene names | MXD1, MAD | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
MAD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.246176 (rank : 36) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
MUSC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.091094 (rank : 95) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60682, O75946, Q9BRE7 | Gene names | MSC, ABF1 | |||
|
Domain Architecture |

|
|||||
| Description | Musculin (Activated B-cell factor 1) (ABF-1). | |||||
|
MUSC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.092076 (rank : 94) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O88940 | Gene names | Msc, Myor | |||
|
Domain Architecture |

|
|||||
| Description | Musculin (Myogenic repressor). | |||||
|
NDF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.084301 (rank : 99) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13562, O00343, Q13340, Q5U095, Q96TH0, Q99455, Q9UEC8 | Gene names | NEUROD1, NEUROD | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1) (NeuroD). | |||||
|
NDF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.085424 (rank : 98) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q60867, Q545N9, Q60897 | Gene names | Neurod1, Neurod | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1). | |||||
|
NDF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.106126 (rank : 89) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15784, Q8TBI7, Q9UQC6 | Gene names | NEUROD2, NDRF | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
NDF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.106338 (rank : 87) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q62414, Q61952, Q925V5 | Gene names | Neurod2, Ndrf | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
NDF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.075330 (rank : 104) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HD90 | Gene names | NEUROD4 | |||
|
Domain Architecture |
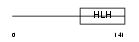
|
|||||
| Description | Neurogenic differentiation factor 4 (NeuroD4). | |||||
|
NDF4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.076220 (rank : 103) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O09105 | Gene names | Neurod4, Ath3, Atoh3 | |||
|
Domain Architecture |
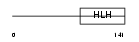
|
|||||
| Description | Neurogenic differentiation factor 4 (NeuroD4) (Atonal protein homolog 3) (Helix-loop-helix protein mATH-3) (MATH3). | |||||
|
NDF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.064284 (rank : 111) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96NK8, Q9H3H6 | Gene names | NEUROD6 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 6 (NeuroD6). | |||||
|
NDF6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.063847 (rank : 114) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P48986 | Gene names | Neurod6, Ath2, Atoh2, Nex1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 6 (NeuroD6) (Atonal protein homolog 2) (Helix-loop-helix protein mATH-2) (MATH2) (NEX-1 protein). | |||||
|
NGN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.098452 (rank : 93) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q92886, Q96HE1 | Gene names | NEUROG1, NEUROD3, NGN, NGN1 | |||
|
Domain Architecture |
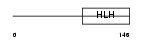
|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein). | |||||
|
NGN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.101286 (rank : 92) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70660 | Gene names | Neurog1, Ath4c, Neurod3, Ngn, Ngn1 | |||
|
Domain Architecture |
|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein) (Helix-loop-helix protein mATH-4C) (MATH4C). | |||||
|
NGN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.109412 (rank : 84) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H2A3, Q8N416 | Gene names | NEUROG2, NGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurogenin 2. | |||||
|
NGN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.111566 (rank : 82) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P70447, P70237 | Gene names | Neurog2, Ath4a, Atoh4, Ngn2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 2 (Atonal protein homolog 4) (Helix-loop-helix protein mATH-4A) (MATH4A). | |||||
|
NGN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.124683 (rank : 76) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Y4Z2, Q9BY24 | Gene names | NEUROG3, NGN3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 3. | |||||
|
NGN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.117041 (rank : 79) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70661 | Gene names | Neurog3, Ath4b, Atoh5, Ngn3 | |||
|
Domain Architecture |
|
|||||
| Description | Neurogenin 3 (Atonal protein homolog 5) (Helix-loop-helix protein mATH-4B) (MATH4B). | |||||
|
OLIG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.065848 (rank : 109) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TAK6, Q7RTS0 | Gene names | OLIG1 | |||
|
Domain Architecture |
|
|||||
| Description | Oligodendrocyte transcription factor 1 (Oligo1). | |||||
|
OLIG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.063995 (rank : 112) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JKN5 | Gene names | Olig1 | |||
|
Domain Architecture |
|
|||||
| Description | Oligodendrocyte transcription factor 1 (Oligo1) (Oligodendrocyte- specific bHLH transcription factor 1). | |||||
|
OLIG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.065021 (rank : 110) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13516, Q86X04, Q9NZ14 | Gene names | OLIG2, BHLHB1, PRKCBP2, RACK17 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 2 (Oligo2) (Basic helix-loop- helix protein class B 1) (Protein kinase C-binding protein RACK17) (Protein kinase C-binding protein 2). | |||||
|
OLIG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.063941 (rank : 113) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9EQW6, Q9JKN4 | Gene names | Olig2 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 2 (Oligo2). | |||||
|
OLIG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.061127 (rank : 117) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7RTU3, Q8N8Q0 | Gene names | OLIG3, BHLHB7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oligodendrocyte transcription factor 3 (Oligo3) (Basic helix-loop- helix domain-containing class B protein 7). | |||||
|
OLIG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.062446 (rank : 116) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6PFG8, Q9EQW5 | Gene names | Olig3, Bhlhb7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oligodendrocyte transcription factor 3 (Oligo3) (Oligodendrocyte- specific bHLH transcription factor 3) (Basic helix-loop-helix domain- containing class B protein 7). | |||||
|
TAL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.125370 (rank : 75) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P17542 | Gene names | TAL1, SCL, TCL5 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein (TAL-1 protein) (Stem cell protein) (T-cell leukemia/lymphoma-5 protein). | |||||
|
TAL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.127236 (rank : 74) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P22091 | Gene names | Tal1, Scl, Tal-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein homolog (TAL-1 protein) (Stem cell protein). | |||||
|
TAL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.107591 (rank : 85) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q16559 | Gene names | TAL2 | |||
|
Domain Architecture |
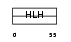
|
|||||
| Description | T-cell acute lymphocytic leukemia-2 protein (TAL-2 protein). | |||||
|
TAL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.109667 (rank : 83) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62282 | Gene names | Tal2 | |||
|
Domain Architecture |
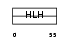
|
|||||
| Description | T-cell acute lymphocytic leukemia-2 protein homolog (TAL-2 protein). | |||||
|
TCF21_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.088338 (rank : 96) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O43680, O43545, Q9BZ14 | Gene names | TCF21, POD1 | |||
|
Domain Architecture |
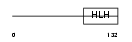
|
|||||
| Description | Transcription factor 21 (Podocyte-expressed 1) (Pod-1) (Epicardin) (Capsulin). | |||||
|
TCF21_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.087753 (rank : 97) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O35437, Q3U023 | Gene names | Tcf21, Pod1 | |||
|
Domain Architecture |
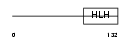
|
|||||
| Description | Transcription factor 21 (Podocyte-expressed 1) (Pod-1) (Epicardin) (Capsulin). | |||||
|
TCFL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.070245 (rank : 107) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
MYC_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 7.71569e-185 (rank : 1) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYC_HUMAN
|
||||||
| NC score | 0.977625 (rank : 2) | θ value | 1.19313e-177 (rank : 2) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYCB_MOUSE
|
||||||
| NC score | 0.830056 (rank : 3) | θ value | 1.85029e-45 (rank : 3) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6P8Z1, Q9D6E3 | Gene names | Mycb, Bmyc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein B-Myc. | |||||
|
MYCN_MOUSE
|
||||||
| NC score | 0.827098 (rank : 4) | θ value | 3.06405e-32 (rank : 6) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCN_HUMAN
|
||||||
| NC score | 0.827060 (rank : 5) | θ value | 8.06329e-33 (rank : 5) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCS_MOUSE
|
||||||
| NC score | 0.809443 (rank : 6) | θ value | 4.72714e-33 (rank : 4) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
MYCL1_HUMAN
|
||||||
| NC score | 0.784725 (rank : 7) | θ value | 3.17079e-29 (rank : 7) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCL1_MOUSE
|
||||||
| NC score | 0.750381 (rank : 8) | θ value | 7.0802e-21 (rank : 8) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MYCL2_HUMAN
|
||||||
| NC score | 0.712192 (rank : 9) | θ value | 1.47631e-18 (rank : 9) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
MAX_HUMAN
|
||||||
| NC score | 0.622796 (rank : 10) | θ value | 1.25267e-09 (rank : 10) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MAX_MOUSE
|
||||||
| NC score | 0.612754 (rank : 11) | θ value | 1.38499e-08 (rank : 11) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
MXI1_MOUSE
|
||||||
| NC score | 0.394275 (rank : 12) | θ value | 1.69304e-06 (rank : 14) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MXI1_HUMAN
|
||||||
| NC score | 0.387163 (rank : 13) | θ value | 2.88788e-06 (rank : 15) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MAD3_HUMAN
|
||||||
| NC score | 0.386049 (rank : 14) | θ value | 5.81887e-07 (rank : 12) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
MAD3_MOUSE
|
||||||
| NC score | 0.379278 (rank : 15) | θ value | 1.29631e-06 (rank : 13) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
USF1_MOUSE
|
||||||
| NC score | 0.372626 (rank : 16) | θ value | 1.09739e-05 (rank : 16) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_HUMAN
|
||||||
| NC score | 0.367768 (rank : 17) | θ value | 3.19293e-05 (rank : 19) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_HUMAN
|
||||||
| NC score | 0.364873 (rank : 18) | θ value | 0.00390308 (rank : 27) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| NC score | 0.362055 (rank : 19) | θ value | 0.00390308 (rank : 28) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
MAD4_HUMAN
|
||||||
| NC score | 0.359403 (rank : 20) | θ value | 0.00035302 (rank : 21) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.359358 (rank : 21) | θ value | 3.19293e-05 (rank : 18) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.350142 (rank : 22) | θ value | 3.19293e-05 (rank : 17) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
TFAP4_HUMAN
|
||||||
| NC score | 0.342529 (rank : 23) | θ value | 0.00665767 (rank : 31) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
FIGLA_MOUSE
|
||||||
| NC score | 0.324289 (rank : 24) | θ value | 0.00298849 (rank : 25) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O55208 | Gene names | Figla | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
FIGLA_HUMAN
|
||||||
| NC score | 0.320960 (rank : 25) | θ value | 0.00390308 (rank : 26) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6QHK4 | Gene names | FIGLA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
MAD4_MOUSE
|
||||||
| NC score | 0.318226 (rank : 26) | θ value | 0.00228821 (rank : 24) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MLX_MOUSE
|
||||||
| NC score | 0.289930 (rank : 27) | θ value | 0.00509761 (rank : 30) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_HUMAN
|
||||||
| NC score | 0.287996 (rank : 28) | θ value | 0.00509761 (rank : 29) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
SRBP2_HUMAN
|
||||||
| NC score | 0.282593 (rank : 29) | θ value | 0.00020696 (rank : 20) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
BHLH8_MOUSE
|
||||||
| NC score | 0.269632 (rank : 30) | θ value | 0.00134147 (rank : 23) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9QYC3, Q9QYE4 | Gene names | Bhlhb8, Mist1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.266479 (rank : 31) | θ value | 0.125558 (rank : 54) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
BHLH8_HUMAN
|
||||||
| NC score | 0.260937 (rank : 32) | θ value | 0.0113563 (rank : 34) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7RTS1 | Gene names | BHLHB8, MIST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.250261 (rank : 33) | θ value | 0.163984 (rank : 56) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
SRBP1_HUMAN
|
||||||
| NC score | 0.249307 (rank : 34) | θ value | 0.0193708 (rank : 40) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.246798 (rank : 35) | θ value | 0.125558 (rank : 53) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MAD1_MOUSE
|
||||||
| NC score | 0.246176 (rank : 36) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
PTF1A_HUMAN
|
||||||
| NC score | 0.242060 (rank : 37) | θ value | 0.000602161 (rank : 22) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7RTS3, Q9HC25 | Gene names | PTF1A, PTF1P48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.241253 (rank : 38) | θ value | 0.0148317 (rank : 37) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
ASCL2_MOUSE
|
||||||
| NC score | 0.231165 (rank : 39) | θ value | 0.0563607 (rank : 44) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O35885, Q9WUJ7 | Gene names | Ascl2, Mash2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash-2). | |||||
|
PTF1A_MOUSE
|
||||||
| NC score | 0.229715 (rank : 40) | θ value | 0.0563607 (rank : 45) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9QX98, Q9QYF5, Q9QYF6 | Gene names | Ptf1a, Ptf1p48 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pancreas transcription factor 1 subunit alpha (Pancreas-specific transcription factor 1a) (bHLH transcription factor p48) (p48 DNA- binding subunit of transcription factor PTF1) (PTF1-p48). | |||||
|
TWST2_MOUSE
|
||||||
| NC score | 0.223139 (rank : 41) | θ value | 0.00869519 (rank : 33) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9D030 | Gene names | Twist2, Dermo1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
TWST2_HUMAN
|
||||||
| NC score | 0.223038 (rank : 42) | θ value | 0.00869519 (rank : 32) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8WVJ9 | Gene names | TWIST2, DERMO1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
TWST1_HUMAN
|
||||||
| NC score | 0.222770 (rank : 43) | θ value | 0.0736092 (rank : 49) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q15672, Q92487, Q99804 | Gene names | TWIST1, TWIST | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (H-twist). | |||||
|
TWST1_MOUSE
|
||||||
| NC score | 0.222334 (rank : 44) | θ value | 0.0736092 (rank : 50) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P26687 | Gene names | Twist1, Twist | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 1 (M-twist). | |||||
|
ASCL1_HUMAN
|
||||||
| NC score | 0.218984 (rank : 45) | θ value | 0.365318 (rank : 66) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P50553, Q9BQ30 | Gene names | ASCL1, ASH1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (HASH1). | |||||
|
HAND2_HUMAN
|
||||||
| NC score | 0.215401 (rank : 46) | θ value | 0.21417 (rank : 59) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P61296, O95300, O95301, P97833 | Gene names | HAND2, DHAND | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND). | |||||
|
HAND2_MOUSE
|
||||||
| NC score | 0.215401 (rank : 47) | θ value | 0.21417 (rank : 60) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61039, Q61100 | Gene names | Hand2, Dhand, Hed, Thing2 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND) (Helix-loop-helix transcription factor expressed in embryo and deciduum) (Thing-2). | |||||
|
ASCL1_MOUSE
|
||||||
| NC score | 0.209748 (rank : 48) | θ value | 0.365318 (rank : 67) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q02067 | Gene names | Ascl1, Ash1, Mash-1, Mash1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (Mash-1). | |||||
|
MYOG_HUMAN
|
||||||
| NC score | 0.209368 (rank : 49) | θ value | 0.0193708 (rank : 38) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P15173 | Gene names | MYOG, MYF4 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenin (Myogenic factor 4) (Myf-4). | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.209317 (rank : 50) | θ value | 0.62314 (rank : 79) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
MYOG_MOUSE
|
||||||
| NC score | 0.209239 (rank : 51) | θ value | 0.0193708 (rank : 39) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P12979 | Gene names | Myog | |||
|
Domain Architecture |

|
|||||
| Description | Myogenin (MYOD1-related protein). | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.208625 (rank : 52) | θ value | 0.62314 (rank : 78) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
MAD1_HUMAN
|
||||||
| NC score | 0.205753 (rank : 53) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q05195 | Gene names | MXD1, MAD | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
MYF6_HUMAN
|
||||||
| NC score | 0.204970 (rank : 54) | θ value | 0.0330416 (rank : 42) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P23409, Q53X80, Q6FHI9 | Gene names | MYF6 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 6 (Myf-6). | |||||
|
MYF6_MOUSE
|
||||||
| NC score | 0.200885 (rank : 55) | θ value | 0.0431538 (rank : 43) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P15375 | Gene names | Myf6, Myf-6 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 6 (Myf-6) (Herculin). | |||||
|
HAND1_MOUSE
|
||||||
| NC score | 0.191907 (rank : 56) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q64279, Q61099 | Gene names | Hand1, Ehand, Hxt, Thing1 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND) (Helix-loop-helix transcription factor expressed in extraembryonic mesoderm and trophoblast) (Thing-1) (Th1). | |||||
|
SCX_MOUSE
|
||||||
| NC score | 0.187368 (rank : 57) | θ value | 0.0563607 (rank : 46) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64124 | Gene names | Scx | |||
|
Domain Architecture |

|
|||||
| Description | Basic helix-loop-helix transcription factor scleraxis. | |||||
|
WBS14_HUMAN
|
||||||
| NC score | 0.185908 (rank : 58) | θ value | 0.0252991 (rank : 41) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
HAND1_HUMAN
|
||||||
| NC score | 0.179708 (rank : 59) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O96004 | Gene names | HAND1, EHAND | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND). | |||||
|
MYOD1_MOUSE
|
||||||
| NC score | 0.173434 (rank : 60) | θ value | 0.21417 (rank : 61) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P10085 | Gene names | Myod1, Myod | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1. | |||||
|
MYF5_MOUSE
|
||||||
| NC score | 0.171269 (rank : 61) | θ value | 0.47712 (rank : 74) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P24699, Q543W7 | Gene names | Myf5, Myf-5 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
MYOD1_HUMAN
|
||||||
| NC score | 0.170834 (rank : 62) | θ value | 0.279714 (rank : 63) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P15172 | Gene names | MYOD1, MYF3, MYOD | |||
|
Domain Architecture |

|
|||||
| Description | Myoblast determination protein 1 (Myogenic factor 3) (Myf-3). | |||||
|
MYF5_HUMAN
|
||||||
| NC score | 0.170629 (rank : 63) | θ value | 0.47712 (rank : 73) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P13349 | Gene names | MYF5 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 5 (Myf-5). | |||||
|
ASCL2_HUMAN
|
||||||
| NC score | 0.166572 (rank : 64) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q99929, Q9UM68 | Gene names | ASCL2 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 2 (Mash2) (HASH2). | |||||
|
ATOH1_HUMAN
|
||||||
| NC score | 0.166036 (rank : 65) | θ value | 0.0148317 (rank : 35) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q92858 | Gene names | ATOH1, ATH1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein hATH-1). | |||||
|
ATOH1_MOUSE
|
||||||
| NC score | 0.165779 (rank : 66) | θ value | 0.0148317 (rank : 36) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P48985 | Gene names | Atoh1, Ath1 | |||
|
Domain Architecture |

|
|||||
| Description | Atonal protein homolog 1 (Helix-loop-helix protein mATH-1) (MATH1). | |||||
|
ASCL3_HUMAN
|
||||||
| NC score | 0.153844 (rank : 67) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NQ33, Q8WYQ6 | Gene names | ASCL3, HASH3, SGN1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 3 (bHLH transcriptional regulator Sgn-1). | |||||
|
WBS14_MOUSE
|
||||||
| NC score | 0.153636 (rank : 68) | θ value | 0.0563607 (rank : 47) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
ASCL3_MOUSE
|
||||||
| NC score | 0.149600 (rank : 69) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9JJR7 | Gene names | Ascl3, Mash3, Sgn1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 3 (bHLH transcriptional regulator Sgn-1) (Mash- 3). | |||||
|
TCF15_MOUSE
|
||||||
| NC score | 0.139190 (rank : 70) | θ value | 1.81305 (rank : 88) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q60756, Q60788 | Gene names | Tcf15, Bhlhec2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 15 (bHLH-EC2 protein) (Paraxis) (Meso1). | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.137753 (rank : 71) | θ value | 0.0736092 (rank : 48) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.131563 (rank : 72) | θ value | 0.163984 (rank : 55) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
TCF15_HUMAN
|
||||||
| NC score | 0.128105 (rank : 73) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q12870, Q9NQQ1 | Gene names | TCF15, BHLHEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 15 (bHLH-EC2 protein) (Paraxis). | |||||
|
TAL1_MOUSE
|
||||||
| NC score | 0.127236 (rank : 74) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P22091 | Gene names | Tal1, Scl, Tal-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein homolog (TAL-1 protein) (Stem cell protein). | |||||
|
TAL1_HUMAN
|
||||||
| NC score | 0.125370 (rank : 75) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P17542 | Gene names | TAL1, SCL, TCL5 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein (TAL-1 protein) (Stem cell protein) (T-cell leukemia/lymphoma-5 protein). | |||||
|
NGN3_HUMAN
|
||||||
| NC score | 0.124683 (rank : 76) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Y4Z2, Q9BY24 | Gene names | NEUROG3, NGN3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 3. | |||||
|
ARNT2_MOUSE
|
||||||
| NC score | 0.122717 (rank : 77) | θ value | 0.21417 (rank : 58) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_HUMAN
|
||||||
| NC score | 0.122023 (rank : 78) | θ value | 0.21417 (rank : 57) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
NGN3_MOUSE
|
||||||
| NC score | 0.117041 (rank : 79) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70661 | Gene names | Neurog3, Ath4b, Atoh5, Ngn3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 3 (Atonal protein homolog 5) (Helix-loop-helix protein mATH-4B) (MATH4B). | |||||
|
LYL1_MOUSE
|
||||||
| NC score | 0.113896 (rank : 80) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P27792 | Gene names | Lyl1, Lyl-1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
LYL1_HUMAN
|
||||||
| NC score | 0.113312 (rank : 81) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P12980, O76102 | Gene names | LYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein lyl-1 (Lymphoblastic leukemia-derived sequence 1). | |||||
|
NGN2_MOUSE
|
||||||
| NC score | 0.111566 (rank : 82) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P70447, P70237 | Gene names | Neurog2, Ath4a, Atoh4, Ngn2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 2 (Atonal protein homolog 4) (Helix-loop-helix protein mATH-4A) (MATH4A). | |||||
|
TAL2_MOUSE
|
||||||
| NC score | 0.109667 (rank : 83) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62282 | Gene names | Tal2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-2 protein homolog (TAL-2 protein). | |||||
|
NGN2_HUMAN
|
||||||
| NC score | 0.109412 (rank : 84) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H2A3, Q8N416 | Gene names | NEUROG2, NGN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurogenin 2. | |||||
|
TAL2_HUMAN
|
||||||
| NC score | 0.107591 (rank : 85) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q16559 | Gene names | TAL2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-2 protein (TAL-2 protein). | |||||
|
HEN1_MOUSE
|
||||||
| NC score | 0.107107 (rank : 86) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q02576 | Gene names | Nhlh1, Hen1 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
NDF2_MOUSE
|
||||||
| NC score | 0.106338 (rank : 87) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q62414, Q61952, Q925V5 | Gene names | Neurod2, Ndrf | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
HEN1_HUMAN
|
||||||
| NC score | 0.106234 (rank : 88) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q02575 | Gene names | NHLH1, HEN1 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
NDF2_HUMAN
|
||||||
| NC score | 0.106126 (rank : 89) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15784, Q8TBI7, Q9UQC6 | Gene names | NEUROD2, NDRF | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 2 (NeuroD2) (NeuroD-related factor) (NDRF). | |||||
|
HEN2_HUMAN
|
||||||
| NC score | 0.103685 (rank : 90) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q02577 | Gene names | NHLH2, HEN2 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 2 (HEN2) (Nescient helix loop helix 2) (NSCL- 2). | |||||
|
HEN2_MOUSE
|
||||||
| NC score | 0.103030 (rank : 91) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q64221 | Gene names | Nhlh2, Hen2 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 2 (HEN2) (Nescient helix loop helix 2) (NSCL- 2). | |||||
|
NGN1_MOUSE
|
||||||
| NC score | 0.101286 (rank : 92) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70660 | Gene names | Neurog1, Ath4c, Neurod3, Ngn, Ngn1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein) (Helix-loop-helix protein mATH-4C) (MATH4C). | |||||
|
NGN1_HUMAN
|
||||||
| NC score | 0.098452 (rank : 93) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q92886, Q96HE1 | Gene names | NEUROG1, NEUROD3, NGN, NGN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 1 (Neurogenic differentiation factor 3) (NeuroD3) (Neurogenic basic-helix-loop-helix protein). | |||||
|
MUSC_MOUSE
|
||||||
| NC score | 0.092076 (rank : 94) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O88940 | Gene names | Msc, Myor | |||
|
Domain Architecture |

|
|||||
| Description | Musculin (Myogenic repressor). | |||||
|
MUSC_HUMAN
|
||||||
| NC score | 0.091094 (rank : 95) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60682, O75946, Q9BRE7 | Gene names | MSC, ABF1 | |||
|
Domain Architecture |

|
|||||
| Description | Musculin (Activated B-cell factor 1) (ABF-1). | |||||
|
TCF21_HUMAN
|
||||||
| NC score | 0.088338 (rank : 96) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O43680, O43545, Q9BZ14 | Gene names | TCF21, POD1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 21 (Podocyte-expressed 1) (Pod-1) (Epicardin) (Capsulin). | |||||
|
TCF21_MOUSE
|
||||||
| NC score | 0.087753 (rank : 97) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O35437, Q3U023 | Gene names | Tcf21, Pod1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 21 (Podocyte-expressed 1) (Pod-1) (Epicardin) (Capsulin). | |||||
|
NDF1_MOUSE
|
||||||
| NC score | 0.085424 (rank : 98) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q60867, Q545N9, Q60897 | Gene names | Neurod1, Neurod | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1). | |||||
|
NDF1_HUMAN
|
||||||
| NC score | 0.084301 (rank : 99) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13562, O00343, Q13340, Q5U095, Q96TH0, Q99455, Q9UEC8 | Gene names | NEUROD1, NEUROD | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1) (NeuroD). | |||||
|
BMAL1_HUMAN
|
||||||
| NC score | 0.078686 (rank : 100) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| NC score | 0.078260 (rank : 101) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
ID1_HUMAN
|
||||||
| NC score | 0.077927 (rank : 102) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P41134, O00651, O00652, Q16371, Q16377, Q5TE66, Q5TE67, Q969Z7, Q9H0Z5, Q9H109 | Gene names | ID1, ID | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein inhibitor ID-1 (Inhibitor of DNA binding 1). | |||||
|
NDF4_MOUSE
|
||||||
| NC score | 0.076220 (rank : 103) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O09105 | Gene names | Neurod4, Ath3, Atoh3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 4 (NeuroD4) (Atonal protein homolog 3) (Helix-loop-helix protein mATH-3) (MATH3). | |||||
|
NDF4_HUMAN
|
||||||
| NC score | 0.075330 (rank : 104) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HD90 | Gene names | NEUROD4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 4 (NeuroD4). | |||||
|
ID1_MOUSE
|
||||||
| NC score | 0.072241 (rank : 105) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P20067, Q61101, Q9D897 | Gene names | Id1, Id, Id-1, Idb1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein inhibitor ID-1 (Inhibitor of DNA binding 1). | |||||
|
BHLH4_HUMAN
|
||||||
| NC score | 0.070855 (rank : 106) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NDY6 | Gene names | BHLHB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 4 (bHLHB4). | |||||
|
TCFL5_HUMAN
|
||||||
| NC score | 0.070245 (rank : 107) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
BHLH4_MOUSE
|
||||||
| NC score | 0.068770 (rank : 108) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BGW3, Q71MB7 | Gene names | Bhlhb4, H20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 4 (bHLHB4). | |||||
|
OLIG1_HUMAN
|
||||||
| NC score | 0.065848 (rank : 109) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TAK6, Q7RTS0 | Gene names | OLIG1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 1 (Oligo1). | |||||
|
OLIG2_HUMAN
|
||||||
| NC score | 0.065021 (rank : 110) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13516, Q86X04, Q9NZ14 | Gene names | OLIG2, BHLHB1, PRKCBP2, RACK17 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 2 (Oligo2) (Basic helix-loop- helix protein class B 1) (Protein kinase C-binding protein RACK17) (Protein kinase C-binding protein 2). | |||||
|
NDF6_HUMAN
|
||||||
| NC score | 0.064284 (rank : 111) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96NK8, Q9H3H6 | Gene names | NEUROD6 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 6 (NeuroD6). | |||||
|
OLIG1_MOUSE
|
||||||
| NC score | 0.063995 (rank : 112) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9JKN5 | Gene names | Olig1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 1 (Oligo1) (Oligodendrocyte- specific bHLH transcription factor 1). | |||||
|
OLIG2_MOUSE
|
||||||
| NC score | 0.063941 (rank : 113) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9EQW6, Q9JKN4 | Gene names | Olig2 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 2 (Oligo2). | |||||
|
NDF6_MOUSE
|
||||||
| NC score | 0.063847 (rank : 114) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P48986 | Gene names | Neurod6, Ath2, Atoh2, Nex1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 6 (NeuroD6) (Atonal protein homolog 2) (Helix-loop-helix protein mATH-2) (MATH2) (NEX-1 protein). | |||||
|
BHLH3_HUMAN
|
||||||
| NC score | 0.063686 (rank : 115) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
OLIG3_MOUSE
|
||||||
| NC score | 0.062446 (rank : 116) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6PFG8, Q9EQW5 | Gene names | Olig3, Bhlhb7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oligodendrocyte transcription factor 3 (Oligo3) (Oligodendrocyte- specific bHLH transcription factor 3) (Basic helix-loop-helix domain- containing class B protein 7). | |||||
|
OLIG3_HUMAN
|
||||||
| NC score | 0.061127 (rank : 117) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7RTU3, Q8N8Q0 | Gene names | OLIG3, BHLHB7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oligodendrocyte transcription factor 3 (Oligo3) (Basic helix-loop- helix domain-containing class B protein 7). | |||||
|
BHLH3_MOUSE
|
||||||
| NC score | 0.058417 (rank : 118) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
HEY2_MOUSE
|
||||||
| NC score | 0.056069 (rank : 119) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
BHLH2_HUMAN
|
||||||
| NC score | 0.055899 (rank : 120) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
BHLH2_MOUSE
|
||||||
| NC score | 0.055454 (rank : 121) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
HEY2_HUMAN
|
||||||
| NC score | 0.054195 (rank : 122) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY1_HUMAN
|
||||||
| NC score | 0.051717 (rank : 123) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HEY1_MOUSE
|
||||||
| NC score | 0.051343 (rank : 124) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
ID4_HUMAN
|
||||||
| NC score | 0.050867 (rank : 125) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P47928, Q13005 | Gene names | ID4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein inhibitor ID-4 (Inhibitor of DNA binding 4). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.046497 (rank : 126) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
K0586_HUMAN
|
||||||
| NC score | 0.045939 (rank : 127) | θ value | 0.62314 (rank : 76) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BVV6, O60328 | Gene names | KIAA0586 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0586. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.044461 (rank : 128) | θ value | 0.0961366 (rank : 51) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
PZRN3_HUMAN
|
||||||
| NC score | 0.040742 (rank : 129) | θ value | 0.279714 (rank : 64) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UPQ7, Q8N2N7, Q96CC2, Q9NSQ2 | Gene names | PDZRN3, KIAA1095, LNX3, SEMCAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Ligand of Numb-protein X 3) (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
K0319_MOUSE
|
||||||
| NC score | 0.036520 (rank : 130) | θ value | 2.36792 (rank : 93) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SZV5, Q80U39 | Gene names | Kiaa0319 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0319 precursor. | |||||
|
METL2_HUMAN
|
||||||
| NC score | 0.036458 (rank : 131) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96IZ6, Q9H9G9, Q9NUI8, Q9P0B5 | Gene names | METTL2 | |||
|
Domain Architecture |

|
|||||
| Description | Methyltransferase-like protein 2 (EC 2.1.1.-). | |||||
|
ASXL1_HUMAN
|
||||||
| NC score | 0.036309 (rank : 132) | θ value | 0.62314 (rank : 75) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IXJ9, Q5JWS9, Q8IYY7, Q9H466, Q9NQF8, Q9UFJ0, Q9UFP8, Q9Y2I4 | Gene names | ASXL1, KIAA0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
PZRN3_MOUSE
|
||||||
| NC score | 0.034966 (rank : 133) | θ value | 1.06291 (rank : 81) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q69ZS0, Q91Z03, Q9QY54, Q9QY55 | Gene names | Pdzrn3, Kiaa1095, Semcap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
UBP28_MOUSE
|
||||||
| NC score | 0.033233 (rank : 134) | θ value | 0.279714 (rank : 65) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5I043, Q6NZP3, Q6ZPP1, Q8BWI1 | Gene names | Usp28, Kiaa1515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 28 (EC 3.1.2.15) (Ubiquitin thioesterase 28) (Ubiquitin-specific-processing protease 28) (Deubiquitinating enzyme 28). | |||||
|
DTNA_HUMAN
|
||||||
| NC score | 0.032699 (rank : 135) | θ value | 0.47712 (rank : 72) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y4J8, O15332, O15333, O75697, Q13197, Q13198, Q13199, Q13498, Q13499, Q13500 | Gene names | DTNA, DRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin alpha (Dystrobrevin-alpha) (Dystrophin-related protein 3). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.031624 (rank : 136) | θ value | 0.0961366 (rank : 52) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
ASXL1_MOUSE
|
||||||
| NC score | 0.030757 (rank : 137) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59598 | Gene names | Asxl1, Kiaa0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
FGF23_HUMAN
|
||||||
| NC score | 0.030673 (rank : 138) | θ value | 0.365318 (rank : 69) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9GZV9, Q4V758 | Gene names | FGF23, HYPF | |||
|
Domain Architecture |

|
|||||
| Description | Fibroblast growth factor 23 precursor (FGF-23) (Tumor-derived hypophosphatemia-inducing factor). | |||||
|
OXR1_MOUSE
|
||||||
| NC score | 0.030608 (rank : 139) | θ value | 0.365318 (rank : 70) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
CEBPE_HUMAN
|
||||||
| NC score | 0.030426 (rank : 140) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.029488 (rank : 141) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
DTNA_MOUSE
|
||||||
| NC score | 0.029414 (rank : 142) | θ value | 1.38821 (rank : 83) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D2N4, P97319, Q61498, Q61499, Q9QZZ5, Q9WUL9, Q9WUM0 | Gene names | Dtna, Dtn | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin alpha (Alpha-dystrobrevin). | |||||
|
FA21C_HUMAN
|
||||||
| NC score | 0.028903 (rank : 143) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
SDC2_MOUSE
|
||||||
| NC score | 0.028242 (rank : 144) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43407 | Gene names | Sdc2, Hspg1, Synd2 | |||
|
Domain Architecture |

|
|||||
| Description | Syndecan-2 precursor (SYND2) (Fibroglycan) (Heparan sulfate proteoglycan core protein) (HSPG). | |||||
|
TTC14_HUMAN
|
||||||
| NC score | 0.027540 (rank : 145) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
FBX38_HUMAN
|
||||||
| NC score | 0.026571 (rank : 146) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PIJ6, Q6PK72, Q7Z2U0, Q86VN3, Q9BXY6, Q9H837, Q9HC40 | Gene names | FBXO38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 38 (Modulator of KLF7 activity homolog) (MoKA). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.025919 (rank : 147) | θ value | 1.81305 (rank : 87) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.025862 (rank : 148) | θ value | 0.813845 (rank : 80) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
STX18_HUMAN
|
||||||
| NC score | 0.024782 (rank : 149) | θ value | 2.36792 (rank : 96) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9P2W9, Q596L3, Q5TZP5 | Gene names | STX18, GIG9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-18 (Growth-inhibiting gene 9 protein). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.024647 (rank : 150) | θ value | 1.38821 (rank : 82) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.022462 (rank : 151) | θ value | 2.36792 (rank : 97) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
CCD91_HUMAN
|
||||||
| NC score | 0.022267 (rank : 152) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
DHX16_HUMAN
|
||||||
| NC score | 0.022009 (rank : 153) | θ value | 0.365318 (rank : 68) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60231, O60322, Q969X7, Q96QC1 | Gene names | DHX16, DBP2, DDX16, KIAA0577 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 (EC 3.6.1.-) (DEAH-box protein 16) (ATP-dependent RNA helicase #3). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.021731 (rank : 154) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
LPIN1_MOUSE
|
||||||
| NC score | 0.021717 (rank : 155) | θ value | 0.62314 (rank : 77) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZP3, Q9CQI2, Q9JLG6 | Gene names | Lpin1, Fld | |||
|
Domain Architecture |

|
|||||
| Description | Lipin-1 (Fatty liver dystrophy protein). | |||||
|
REN3B_HUMAN
|
||||||
| NC score | 0.021557 (rank : 156) | θ value | 3.0926 (rank : 104) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BZI7, Q9H1J0 | Gene names | UPF3B, RENT3B, UPF3X | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 3B (Nonsense mRNA reducing factor 3B) (Up-frameshift suppressor 3 homolog B) (hUpf3B) (hUpf3p-X). | |||||
|
DACT1_HUMAN
|
||||||
| NC score | 0.020748 (rank : 157) | θ value | 2.36792 (rank : 89) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NYF0, Q86TY0 | Gene names | DACT1, DPR1, HNG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (hDPR1) (Heptacellular carcinoma novel gene 3 protein). | |||||
|
NEMO_HUMAN
|
||||||
| NC score | 0.018948 (rank : 158) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y6K9 | Gene names | IKBKG, FIP3, NEMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (FIP-3). | |||||
|
KIF3A_HUMAN
|
||||||
| NC score | 0.018803 (rank : 159) | θ value | 1.38821 (rank : 84) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y496, Q86XE9, Q9Y6V4 | Gene names | KIF3A, KIF3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
ZFYV1_HUMAN
|
||||||
| NC score | 0.018547 (rank : 160) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HBF4, Q8WYX7, Q96K57, Q9BXP9, Q9HCI3 | Gene names | ZFYVE1, DFCP1, KIAA1589, TAFF1, ZNFN2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 1 (Double FYVE-containing protein 1) (Tandem FYVE fingers-1) (SR3). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.018350 (rank : 161) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
K0056_HUMAN
|
||||||
| NC score | 0.016648 (rank : 162) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42695 | Gene names | KIAA0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
CEBPA_HUMAN
|
||||||
| NC score | 0.016488 (rank : 163) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49715, P78319 | Gene names | CEBPA | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
SAFB2_HUMAN
|
||||||
| NC score | 0.016467 (rank : 164) | θ value | 2.36792 (rank : 95) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14151, Q8TB13 | Gene names | SAFB2, KIAA0138 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
CEBPA_MOUSE
|
||||||
| NC score | 0.016298 (rank : 165) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53566, Q91XB6 | Gene names | Cebpa | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
SNX23_HUMAN
|
||||||
| NC score | 0.015966 (rank : 166) | θ value | 0.365318 (rank : 71) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96L93, Q5HYC0, Q5HYK1, Q5JWW3, Q5TFK5, Q86VL9, Q86YS5, Q8IYU0, Q9BQJ8, Q9BQM0, Q9BQM1, Q9BQM5, Q9H5U0, Q9HCI2, Q9NXN9 | Gene names | C20orf23, KIAA1590, SNX23 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like motor protein C20orf23 (Sorting nexin-23). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.015699 (rank : 167) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.015380 (rank : 168) | θ value | 0.279714 (rank : 62) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
JHD2C_HUMAN
|
||||||
| NC score | 0.014335 (rank : 169) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15652, Q5SQZ8, Q5SQZ9, Q5SR00, Q7Z3E7, Q8N3U0, Q96KB9, Q9P2G7 | Gene names | JMJD1C, JHDM2C, KIAA1380, TRIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C) (Thyroid receptor-interacting protein 8) (TRIP-8). | |||||
|
KLRA2_MOUSE
|
||||||
| NC score | 0.013892 (rank : 170) | θ value | 2.36792 (rank : 94) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60660 | Gene names | Klra2, Ly49-b, Ly49b | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell lectin-like receptor 2 (T-cell surface glycoprotein Ly- 49B) (Ly49-B antigen) (Lymphocyte antigen 49B). | |||||
|
NUFP1_MOUSE
|
||||||
| NC score | 0.013107 (rank : 171) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QXX8, Q9CV69 | Gene names | Nufip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 1 (Nuclear FMRP-interacting protein 1). | |||||
|
NUP88_MOUSE
|
||||||
| NC score | 0.012654 (rank : 172) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CEC0, Q80Z13, Q8K090 | Gene names | Nup88 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear pore complex protein Nup88 (Nucleoporin Nup88) (88 kDa nuclear pore complex protein). | |||||
|
GP149_HUMAN
|
||||||
| NC score | 0.009945 (rank : 173) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86SP6 | Gene names | GPR149, PGR10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 149 (G-protein coupled receptor PGR10). | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.009912 (rank : 174) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.009600 (rank : 175) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.009427 (rank : 176) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
GOGA1_MOUSE
|
||||||
| NC score | 0.007526 (rank : 177) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | -0.000510 (rank : 178) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
PTPRD_HUMAN
|
||||||
| NC score | -0.000931 (rank : 179) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23468 | Gene names | PTPRD | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase delta precursor (EC 3.1.3.48) (Protein-tyrosine phosphatase delta) (R-PTP-delta). | |||||
|
MTR1L_HUMAN
|
||||||
| NC score | -0.003242 (rank : 180) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1180 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13585 | Gene names | GPR50 | |||
|
Domain Architecture |

|
|||||
| Description | Melatonin-related receptor (G protein-coupled receptor 50) (H9). | |||||