Please be patient as the page loads
|
SRBP2_HUMAN
|
||||||
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SRBP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.905839 (rank : 2) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.882959 (rank : 3) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
SRBP2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 133 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
USF1_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 4) | NC score | 0.450678 (rank : 4) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 5) | NC score | 0.447479 (rank : 5) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 6) | NC score | 0.426778 (rank : 6) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
MITF_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 7) | NC score | 0.409881 (rank : 9) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
USF2_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 8) | NC score | 0.424340 (rank : 7) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
MITF_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 9) | NC score | 0.407274 (rank : 10) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
TFE3_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 10) | NC score | 0.415026 (rank : 8) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
TFEB_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 11) | NC score | 0.386725 (rank : 12) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 12) | NC score | 0.387287 (rank : 11) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
MYCN_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 13) | NC score | 0.290005 (rank : 15) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCN_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 14) | NC score | 0.289726 (rank : 16) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYC_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 15) | NC score | 0.282593 (rank : 17) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MAX_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 16) | NC score | 0.381152 (rank : 13) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
CLOCK_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 17) | NC score | 0.145088 (rank : 42) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
CLOCK_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 18) | NC score | 0.146642 (rank : 41) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
MAD4_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 19) | NC score | 0.216620 (rank : 27) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MAX_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 20) | NC score | 0.368871 (rank : 14) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
BHLH2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 21) | NC score | 0.205939 (rank : 30) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
EMSY_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 22) | NC score | 0.069148 (rank : 77) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
MYC_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 23) | NC score | 0.269337 (rank : 19) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
TFAP4_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 24) | NC score | 0.263265 (rank : 20) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
BHLH2_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 25) | NC score | 0.202030 (rank : 31) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
HEY2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 26) | NC score | 0.239406 (rank : 22) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY2_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 27) | NC score | 0.242855 (rank : 21) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
ELF1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 28) | NC score | 0.037412 (rank : 103) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
HAND2_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 29) | NC score | 0.130916 (rank : 50) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P61296, O95300, O95301, P97833 | Gene names | HAND2, DHAND | |||
|
Domain Architecture |
|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND). | |||||
|
HAND2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 30) | NC score | 0.130916 (rank : 51) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61039, Q61100 | Gene names | Hand2, Dhand, Hed, Thing2 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND) (Helix-loop-helix transcription factor expressed in embryo and deciduum) (Thing-2). | |||||
|
SP4_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 31) | NC score | 0.019874 (rank : 126) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62445 | Gene names | Sp4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp4. | |||||
|
HEY1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 32) | NC score | 0.231975 (rank : 23) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
HEY1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 33) | NC score | 0.229242 (rank : 25) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 34) | NC score | 0.050034 (rank : 94) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MYCS_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 35) | NC score | 0.269453 (rank : 18) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
MYF6_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 36) | NC score | 0.117255 (rank : 58) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P23409, Q53X80, Q6FHI9 | Gene names | MYF6 | |||
|
Domain Architecture |
|
|||||
| Description | Myogenic factor 6 (Myf-6). | |||||
|
SP4_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 37) | NC score | 0.019337 (rank : 127) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q02446, O60402 | Gene names | SP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp4 (SPR-1). | |||||
|
HES1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 38) | NC score | 0.187607 (rank : 34) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
HES1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 39) | NC score | 0.187460 (rank : 35) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
R3HD2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 40) | NC score | 0.074696 (rank : 75) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
CAH5B_MOUSE
|
||||||
| θ value | 0.163984 (rank : 41) | NC score | 0.023296 (rank : 120) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZA0, Q8C3J9, Q8K014 | Gene names | Ca5b, Car5b | |||
|
Domain Architecture |
|
|||||
| Description | Carbonic anhydrase 5B, mitochondrial precursor (EC 4.2.1.1) (Carbonic anhydrase VB) (Carbonate dehydratase VB) (CA-VB). | |||||
|
MYCL1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 42) | NC score | 0.229297 (rank : 24) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
SP3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 43) | NC score | 0.018365 (rank : 132) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02447, Q8TD56, Q9BQR1 | Gene names | SP3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp3 (SPR-2). | |||||
|
SP3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 44) | NC score | 0.018508 (rank : 130) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 731 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O70494, Q68FF2, Q8CF64, Q8K378 | Gene names | Sp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp3. | |||||
|
GP152_HUMAN
|
||||||
| θ value | 0.279714 (rank : 45) | NC score | 0.021288 (rank : 122) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
MYF6_MOUSE
|
||||||
| θ value | 0.279714 (rank : 46) | NC score | 0.106140 (rank : 64) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15375 | Gene names | Myf6, Myf-6 | |||
|
Domain Architecture |
|
|||||
| Description | Myogenic factor 6 (Myf-6) (Herculin). | |||||
|
ATF1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 47) | NC score | 0.043632 (rank : 100) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P81269 | Gene names | Atf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TCR-ATF1). | |||||
|
MXI1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 48) | NC score | 0.174325 (rank : 39) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MXI1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 49) | NC score | 0.179617 (rank : 37) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MYCL1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 50) | NC score | 0.228897 (rank : 26) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
NPAS4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 51) | NC score | 0.058405 (rank : 89) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
R3HD1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 52) | NC score | 0.061789 (rank : 87) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15032, Q8IW32 | Gene names | R3HDM1, KIAA0029, R3HDM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R3H domain-containing protein 1. | |||||
|
MYOG_HUMAN
|
||||||
| θ value | 0.62314 (rank : 53) | NC score | 0.114203 (rank : 59) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P15173 | Gene names | MYOG, MYF4 | |||
|
Domain Architecture |
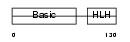
|
|||||
| Description | Myogenin (Myogenic factor 4) (Myf-4). | |||||
|
MYOG_MOUSE
|
||||||
| θ value | 0.62314 (rank : 54) | NC score | 0.112906 (rank : 61) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P12979 | Gene names | Myog | |||
|
Domain Architecture |
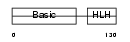
|
|||||
| Description | Myogenin (MYOD1-related protein). | |||||
|
PAPOG_MOUSE
|
||||||
| θ value | 0.62314 (rank : 55) | NC score | 0.025644 (rank : 116) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PCL9, Q8BZC9 | Gene names | Papolg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) polymerase gamma (EC 2.7.7.19) (PAP gamma) (Polynucleotide adenylyltransferase gamma) (SRP RNA 3' adenylating enzyme). | |||||
|
ARNT_HUMAN
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.139568 (rank : 43) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.138344 (rank : 45) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
HAND1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.097796 (rank : 66) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O96004 | Gene names | HAND1, EHAND | |||
|
Domain Architecture |
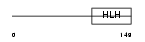
|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND). | |||||
|
HES4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 59) | NC score | 0.186360 (rank : 36) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HCC6 | Gene names | HES4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-4 (Hairy and enhancer of split 4) (bHLH factor Hes4). | |||||
|
BHLH3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.191465 (rank : 33) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
BHLH3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 61) | NC score | 0.192830 (rank : 32) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
CK024_MOUSE
|
||||||
| θ value | 1.06291 (rank : 62) | NC score | 0.041982 (rank : 101) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
HES6_MOUSE
|
||||||
| θ value | 1.06291 (rank : 63) | NC score | 0.161138 (rank : 40) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JHE6, Q3U2D7 | Gene names | Hes6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.039789 (rank : 102) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.047042 (rank : 97) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
TAL1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.101428 (rank : 65) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P17542 | Gene names | TAL1, SCL, TCL5 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein (TAL-1 protein) (Stem cell protein) (T-cell leukemia/lymphoma-5 protein). | |||||
|
AFF2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.027438 (rank : 113) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
ARNT2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.124403 (rank : 54) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.125371 (rank : 53) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
GP156_HUMAN
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.028979 (rank : 111) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NFN8, Q86SN6 | Gene names | GPR156, GABABL, PGR28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor) (G-protein coupled receptor PGR28). | |||||
|
MAD4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.136532 (rank : 47) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
K0319_HUMAN
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.026973 (rank : 114) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5VV43, Q9UJC8, Q9Y4G7 | Gene names | KIAA0319 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0319 precursor. | |||||
|
MAD1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.119221 (rank : 57) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.004060 (rank : 151) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
CEAM7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.011266 (rank : 139) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14002, O15148, O15149, Q13983, Q9UPJ2 | Gene names | CEACAM7, CGM2 | |||
|
Domain Architecture |

|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 7 precursor (Carcinoembryonic antigen CGM2). | |||||
|
HAND1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.080076 (rank : 71) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q64279, Q61099 | Gene names | Hand1, Ehand, Hxt, Thing1 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND) (Helix-loop-helix transcription factor expressed in extraembryonic mesoderm and trophoblast) (Thing-1) (Th1). | |||||
|
MNT_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.136282 (rank : 48) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
NPAS4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.049985 (rank : 95) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
PAX3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.005833 (rank : 148) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23760, Q16448 | Gene names | PAX3, HUP2 | |||
|
Domain Architecture |
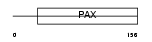
|
|||||
| Description | Paired box protein Pax-3 (HUP2). | |||||
|
PF21B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.037304 (rank : 104) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
RBM25_HUMAN
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.012742 (rank : 138) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
ATF2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.045968 (rank : 98) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
ATF2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.045075 (rank : 99) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
CKLF4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.030223 (rank : 109) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IZR5, Q8IZR4, Q8IZV1 | Gene names | CMTM4, CKLFSF4 | |||
|
Domain Architecture |
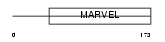
|
|||||
| Description | CKLF-like MARVEL transmembrane domain-containing protein 4 (Chemokine- like factor superfamily member 4). | |||||
|
CKLF4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.030315 (rank : 108) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CJ61, Q8K143 | Gene names | Cmtm4, Cklfsf4 | |||
|
Domain Architecture |
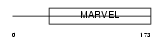
|
|||||
| Description | CKLF-like MARVEL transmembrane domain-containing protein 4 (Chemokine- like factor superfamily member 4). | |||||
|
DCP1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.049961 (rank : 96) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NPI6 | Gene names | DCP1A, SMIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (Smad4-interacting transcriptional co-activator). | |||||
|
LTB1L_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.003487 (rank : 152) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14766 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.013984 (rank : 133) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.007585 (rank : 143) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
NPAS2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.083563 (rank : 69) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
RTN2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.018496 (rank : 131) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70622, O70620 | Gene names | Rtn2, Nspl1 | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-2 (Neuroendocrine-specific protein-like 1) (NSP-like protein 1) (NSPLI). | |||||
|
SYM_MOUSE
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.025570 (rank : 117) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q68FL6 | Gene names | Mars | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
ZN292_HUMAN
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.013589 (rank : 135) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60281, Q9H8G3, Q9H8J4 | Gene names | ZNF292, KIAA0530 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 292. | |||||
|
EI2BE_MOUSE
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.019314 (rank : 128) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CHW4, Q3TU39 | Gene names | Eif2b5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Translation initiation factor eIF-2B subunit epsilon (eIF-2B GDP-GTP exchange factor subunit epsilon). | |||||
|
K1683_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.037161 (rank : 105) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.051912 (rank : 91) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
PPRB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.020504 (rank : 123) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
RFX5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.025147 (rank : 118) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48382 | Gene names | RFX5 | |||
|
Domain Architecture |
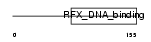
|
|||||
| Description | DNA-binding protein RFX5 (Regulatory factor X subunit 5). | |||||
|
TA2R7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.007008 (rank : 145) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYW3 | Gene names | TAS2R7 | |||
|
Domain Architecture |

|
|||||
| Description | Taste receptor type 2 member 7 (T2R7) (Taste receptor family B member 4) (TRB4). | |||||
|
ATF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.028596 (rank : 112) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P18846, P25168 | Gene names | ATF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TREB36 protein). | |||||
|
HES5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.114189 (rank : 60) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
HES6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.139254 (rank : 44) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96HZ4, Q8N2J2, Q96T93, Q9P2S3 | Gene names | HES6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6) (C- HAIRY1). | |||||
|
MK15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.000745 (rank : 155) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TD08, Q2TCF9, Q8N362 | Gene names | MAPK15, ERK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 15 (EC 2.7.11.24) (Extracellular signal-regulated kinase 8). | |||||
|
NEST_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.005457 (rank : 149) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
PDXL2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.023781 (rank : 119) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
T2R41_MOUSE
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.008377 (rank : 142) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59532, Q7M704 | Gene names | Tas2r41, Tas2r12, Tas2r126, Tas2r26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 2 member 41 (T2R41) (T2R12) (T2R26). | |||||
|
TAL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.091230 (rank : 67) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P22091 | Gene names | Tal1, Scl, Tal-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein homolog (TAL-1 protein) (Stem cell protein). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.034904 (rank : 106) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
WBS14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.075943 (rank : 73) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
ATX2L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.030155 (rank : 110) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
B3GA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.011245 (rank : 140) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NPZ5, Q5JS09, Q8TF38, Q96NK4 | Gene names | B3GAT2, GLCATS, KIAA1963 | |||
|
Domain Architecture |
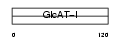
|
|||||
| Description | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 2 (EC 2.4.1.135) (Beta-1,3-glucuronyltransferase 2) (Glucuronosyltransferase-S) (GlcAT-S) (UDP-glucuronosyltransferase-S) (GlcAT-D). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.007198 (rank : 144) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
CAMKV_HUMAN
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.000209 (rank : 156) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
CE290_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.001458 (rank : 154) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CIKS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.033999 (rank : 107) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43734, Q5R3A3, Q9H5W2, Q9H6Y3, Q9NS14, Q9UG72 | Gene names | TRAF3IP2, C6orf4, C6orf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adapter protein CIKS (Connection to IKK and SAPK/JNK) (TRAF3- interacting protein 2) (Nuclear factor NF-kappa-B activator 1) (ACT1). | |||||
|
CT075_MOUSE
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.006649 (rank : 147) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59383 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf75 homolog precursor. | |||||
|
HEN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.062634 (rank : 86) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q02575 | Gene names | NHLH1, HEN1 | |||
|
Domain Architecture |
|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
HEN1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.062873 (rank : 85) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q02576 | Gene names | Nhlh1, Hen1 | |||
|
Domain Architecture |
|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
MAD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.112628 (rank : 62) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q05195 | Gene names | MXD1, MAD | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
MAD3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.177631 (rank : 38) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.026221 (rank : 115) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
NUDT9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.013401 (rank : 137) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BVU5, Q3TZ68, Q8K1J4 | Gene names | Nudt9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribose pyrophosphatase, mitochondrial precursor (EC 3.6.1.13) (ADP-ribose diphosphatase) (Adenosine diphosphoribose pyrophosphatase) (ADPR-PPase) (ADP-ribose phosphohydrolase) (Nucleoside diphosphate- linked moiety X motif 9) (Nudix motif 9). | |||||
|
P80C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.022671 (rank : 121) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P38432 | Gene names | COIL, CLN80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coilin (p80). | |||||
|
CAH5B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.013518 (rank : 136) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2D0 | Gene names | CA5B | |||
|
Domain Architecture |
|
|||||
| Description | Carbonic anhydrase 5B, mitochondrial precursor (EC 4.2.1.1) (Carbonic anhydrase VB) (Carbonate dehydratase VB) (CA-VB). | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.004978 (rank : 150) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
HCFC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.019918 (rank : 125) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
IQEC3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.009949 (rank : 141) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
NFRKB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.050510 (rank : 93) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
SYM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.020410 (rank : 124) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56192, Q14895, Q53H14, Q96A15, Q96BZ0, Q9NSE0 | Gene names | MARS | |||
|
Domain Architecture |

|
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
TMM20_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.013861 (rank : 134) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q2M3R5, Q86YG5, Q8NBA5 | Gene names | TMEM20, C10orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 20. | |||||
|
VASN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.001651 (rank : 153) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CZT5, Q8BJJ0, Q8R2G5 | Gene names | Vasn, Slitl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vasorin precursor (Protein Slit-like 2). | |||||
|
ZEP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.019216 (rank : 129) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
ZHX3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.006832 (rank : 146) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C0Q2, Q80U14 | Gene names | Zhx3, Kiaa0395, Tix1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and homeodomain protein 3 (Triple homeobox protein 1). | |||||
|
BHLH8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.065745 (rank : 82) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7RTS1 | Gene names | BHLHB8, MIST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
BHLH8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.066942 (rank : 81) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QYC3, Q9QYE4 | Gene names | Bhlhb8, Mist1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
BMAL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.124102 (rank : 56) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
BMAL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.124392 (rank : 55) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.063905 (rank : 84) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
FIGLA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.067521 (rank : 79) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6QHK4 | Gene names | FIGLA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
FIGLA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.067719 (rank : 78) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O55208 | Gene names | Figla | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
HES2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.081172 (rank : 70) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y543, Q96EN4, Q9Y542 | Gene names | HES2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
HES2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.087015 (rank : 68) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O54792 | Gene names | Hes2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
HES3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.071277 (rank : 76) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61657, O09083, O09150 | Gene names | Hes3, Hes-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-3 (Hairy and enhancer of split 3). | |||||
|
HES7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.075300 (rank : 74) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BYE0 | Gene names | HES7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
HES7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.067252 (rank : 80) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BKT2, Q99JA6 | Gene names | Hes7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
MAD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.126033 (rank : 52) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
MISS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.050542 (rank : 92) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
MLX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.211956 (rank : 29) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.216353 (rank : 28) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MNT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.131703 (rank : 49) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MYCB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.106965 (rank : 63) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P8Z1, Q9D6E3 | Gene names | Mycb, Bmyc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein B-Myc. | |||||
|
MYCL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.137927 (rank : 46) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
NPAS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.077100 (rank : 72) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
TCFL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.060946 (rank : 88) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
WBS14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.052552 (rank : 90) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
XBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.064610 (rank : 83) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35426, Q8VHM0, Q922G5, Q9ESS3 | Gene names | Xbp1, Treb5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5 homolog). | |||||
|
SRBP2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 133 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
SRBP1_HUMAN
|
||||||
| NC score | 0.905839 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P36956, Q16062 | Gene names | SREBF1, SREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.882959 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
USF1_MOUSE
|
||||||
| NC score | 0.450678 (rank : 4) | θ value | 4.0297e-08 (rank : 4) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q61069 | Gene names | Usf1, Usf | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF1_HUMAN
|
||||||
| NC score | 0.447479 (rank : 5) | θ value | 1.53129e-07 (rank : 5) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P22415 | Gene names | USF1, USF | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 1 (Major late transcription factor 1). | |||||
|
USF2_HUMAN
|
||||||
| NC score | 0.426778 (rank : 6) | θ value | 9.92553e-07 (rank : 6) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q15853, O00671, O00709, Q05750, Q07952, Q15851, Q15852 | Gene names | USF2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (FOS- interacting protein) (FIP) (Major late transcription factor 2). | |||||
|
USF2_MOUSE
|
||||||
| NC score | 0.424340 (rank : 7) | θ value | 1.69304e-06 (rank : 8) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q64705, Q3UIL2, Q8VE59 | Gene names | Usf2 | |||
|
Domain Architecture |

|
|||||
| Description | Upstream stimulatory factor 2 (Upstream transcription factor 2) (Major late transcription factor 2). | |||||
|
TFE3_HUMAN
|
||||||
| NC score | 0.415026 (rank : 8) | θ value | 4.92598e-06 (rank : 10) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P19532, Q92757, Q92758, Q99964 | Gene names | TFE3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E3. | |||||
|
MITF_HUMAN
|
||||||
| NC score | 0.409881 (rank : 9) | θ value | 1.69304e-06 (rank : 7) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O75030, Q14841, Q9P2V0, Q9P2V1, Q9P2V2, Q9P2Y8 | Gene names | MITF | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
MITF_MOUSE
|
||||||
| NC score | 0.407274 (rank : 10) | θ value | 2.88788e-06 (rank : 9) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q08874, O08885, O88203, Q08843, Q60781, Q60782, Q9JIJ0, Q9JIJ1, Q9JIJ2, Q9JIJ3, Q9JIJ4, Q9JIJ5, Q9JIJ6, Q9JKX9 | Gene names | Mitf, Bw, Mi, Vit | |||
|
Domain Architecture |

|
|||||
| Description | Microphthalmia-associated transcription factor. | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.387287 (rank : 11) | θ value | 2.44474e-05 (rank : 12) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
TFEB_HUMAN
|
||||||
| NC score | 0.386725 (rank : 12) | θ value | 2.44474e-05 (rank : 11) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P19484, Q709B3, Q7Z6P9, Q9BRJ5, Q9UJD8 | Gene names | TFEB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
MAX_HUMAN
|
||||||
| NC score | 0.381152 (rank : 13) | θ value | 0.000786445 (rank : 16) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P61244, P25912, P52163 | Gene names | MAX | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X). | |||||
|
MAX_MOUSE
|
||||||
| NC score | 0.368871 (rank : 14) | θ value | 0.00665767 (rank : 20) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P28574 | Gene names | Max, Myn | |||
|
Domain Architecture |

|
|||||
| Description | Protein max (Myc-associated factor X) (Protein myn) (Myc-binding novel HLH/LZ protein). | |||||
|
MYCN_HUMAN
|
||||||
| NC score | 0.290005 (rank : 15) | θ value | 7.1131e-05 (rank : 13) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P04198, Q6LDT9 | Gene names | MYCN, NMYC | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYCN_MOUSE
|
||||||
| NC score | 0.289726 (rank : 16) | θ value | 9.29e-05 (rank : 14) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P03966, Q61978 | Gene names | Mycn, Nmyc, Nmyc1 | |||
|
Domain Architecture |

|
|||||
| Description | N-myc proto-oncogene protein. | |||||
|
MYC_MOUSE
|
||||||
| NC score | 0.282593 (rank : 17) | θ value | 0.00020696 (rank : 15) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MYCS_MOUSE
|
||||||
| NC score | 0.269453 (rank : 18) | θ value | 0.0431538 (rank : 35) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Z304 | Gene names | Mycs | |||
|
Domain Architecture |

|
|||||
| Description | Protein S-myc. | |||||
|
MYC_HUMAN
|
||||||
| NC score | 0.269337 (rank : 19) | θ value | 0.0148317 (rank : 23) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
TFAP4_HUMAN
|
||||||
| NC score | 0.263265 (rank : 20) | θ value | 0.0148317 (rank : 24) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
HEY2_MOUSE
|
||||||
| NC score | 0.242855 (rank : 21) | θ value | 0.0193708 (rank : 27) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QUS4, Q3TZ99, Q8CD44 | Gene names | Hey2, Chf1, Herp, Herp1, Hesr2, Hrt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES-related repressor protein 2). | |||||
|
HEY2_HUMAN
|
||||||
| NC score | 0.239406 (rank : 22) | θ value | 0.0193708 (rank : 26) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UBP5 | Gene names | HEY2, CHF1, GRL, HERP, HERP1, HRT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 2 (Cardiovascular helix-loop-helix factor 1) (Hairy and enhancer of split-related protein 2) (HESR-2) (Hairy-related transcription factor 2) (HES- related repressor protein 2) (Protein gridlock homolog). | |||||
|
HEY1_HUMAN
|
||||||
| NC score | 0.231975 (rank : 23) | θ value | 0.0431538 (rank : 32) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Y5J3, Q5TZS3, Q9NYP4 | Gene names | HEY1, CHF2, HESR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1) (Cardiovascular helix-loop-helix factor 2) (HES-related repressor protein 2) (HERP2). | |||||
|
MYCL1_MOUSE
|
||||||
| NC score | 0.229297 (rank : 24) | θ value | 0.163984 (rank : 42) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P10166, Q5FWI7 | Gene names | Mycl1, Lmyc1, Mycl | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
HEY1_MOUSE
|
||||||
| NC score | 0.229242 (rank : 25) | θ value | 0.0431538 (rank : 33) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9WV93 | Gene names | Hey1, Hesr1 | |||
|
Domain Architecture |

|
|||||
| Description | Hairy/enhancer-of-split related with YRPW motif 1 (Hairy and enhancer of split-related protein 1) (HESR-1). | |||||
|
MYCL1_HUMAN
|
||||||
| NC score | 0.228897 (rank : 26) | θ value | 0.365318 (rank : 50) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P12524, Q14897, Q5QPL0, Q5QPL1, Q9NUE9 | Gene names | MYCL1, LMYC, MYCL | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-1 proto-oncogene protein. | |||||
|
MAD4_HUMAN
|
||||||
| NC score | 0.216620 (rank : 27) | θ value | 0.00665767 (rank : 19) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14582, Q5TZX4 | Gene names | MXD4, MAD4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MLX_MOUSE
|
||||||
| NC score | 0.216353 (rank : 28) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O08609, Q9EQJ7, Q9EQJ8 | Gene names | Mlx, Tcfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
MLX_HUMAN
|
||||||
| NC score | 0.211956 (rank : 29) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UH92, Q53XM6, Q96FL2, Q9H2V0, Q9H2V1, Q9H2V2, Q9NXN3 | Gene names | MLX, TCFL4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-like protein X (Max-like bHLHZip protein) (BigMax protein) (Protein Mlx) (Transcription factor-like protein 4). | |||||
|
BHLH2_HUMAN
|
||||||
| NC score | 0.205939 (rank : 30) | θ value | 0.0113563 (rank : 21) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O14503, Q96TD3 | Gene names | BHLHB2, DEC1, SHARP2, STRA13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Differentially expressed in chondrocytes protein 1) (DEC1) (Enhancer-of-split and hairy-related protein 2) (SHARP-2) (Stimulated with retinoic acid 13). | |||||
|
BHLH2_MOUSE
|
||||||
| NC score | 0.202030 (rank : 31) | θ value | 0.0193708 (rank : 25) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O35185, P97289 | Gene names | Bhlhb2, Clast5, Stra13 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 2 (bHLHB2) (Stimulated with retinoic acid 13) (E47 interaction protein 1) (eipl). | |||||
|
BHLH3_MOUSE
|
||||||
| NC score | 0.192830 (rank : 32) | θ value | 1.06291 (rank : 61) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99PV5 | Gene names | Bhlhb3, Dec2 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (mDEC2). | |||||
|
BHLH3_HUMAN
|
||||||
| NC score | 0.191465 (rank : 33) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9C0J9 | Gene names | BHLHB3, DEC2, SHARP1 | |||
|
Domain Architecture |

|
|||||
| Description | Class B basic helix-loop-helix protein 3 (bHLHB3) (Differentially expressed in chondrocytes protein 2) (hDEC2) (Enhancer-of-split and hairy-related protein 1) (SHARP-1). | |||||
|
HES1_HUMAN
|
||||||
| NC score | 0.187607 (rank : 34) | θ value | 0.125558 (rank : 38) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14469 | Gene names | HES1, HL, HRY | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1) (Hairy- like) (HHL) (Hairy homolog). | |||||
|
HES1_MOUSE
|
||||||
| NC score | 0.187460 (rank : 35) | θ value | 0.125558 (rank : 39) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P35428 | Gene names | Hes1, Hes-1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-1 (Hairy and enhancer of split 1). | |||||
|
HES4_HUMAN
|
||||||
| NC score | 0.186360 (rank : 36) | θ value | 0.813845 (rank : 59) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HCC6 | Gene names | HES4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-4 (Hairy and enhancer of split 4) (bHLH factor Hes4). | |||||
|
MXI1_MOUSE
|
||||||
| NC score | 0.179617 (rank : 37) | θ value | 0.365318 (rank : 49) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P50540 | Gene names | Mxi1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
MAD3_HUMAN
|
||||||
| NC score | 0.177631 (rank : 38) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
MXI1_HUMAN
|
||||||
| NC score | 0.174325 (rank : 39) | θ value | 0.365318 (rank : 48) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P50539, Q15887 | Gene names | MXI1 | |||
|
Domain Architecture |

|
|||||
| Description | MAX-interacting protein 1 (Protein MXI1). | |||||
|
HES6_MOUSE
|
||||||
| NC score | 0.161138 (rank : 40) | θ value | 1.06291 (rank : 63) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JHE6, Q3U2D7 | Gene names | Hes6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6). | |||||
|
CLOCK_MOUSE
|
||||||
| NC score | 0.146642 (rank : 41) | θ value | 0.00665767 (rank : 18) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
CLOCK_HUMAN
|
||||||
| NC score | 0.145088 (rank : 42) | θ value | 0.00390308 (rank : 17) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
ARNT_HUMAN
|
||||||
| NC score | 0.139568 (rank : 43) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P27540 | Gene names | ARNT | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
HES6_HUMAN
|
||||||
| NC score | 0.139254 (rank : 44) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96HZ4, Q8N2J2, Q96T93, Q9P2S3 | Gene names | HES6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor HES-6 (Hairy and enhancer of split 6) (C- HAIRY1). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.138344 (rank : 45) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
MYCL2_HUMAN
|
||||||
| NC score | 0.137927 (rank : 46) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P12525 | Gene names | MYCL2 | |||
|
Domain Architecture |

|
|||||
| Description | L-myc-2 protein. | |||||
|
MAD4_MOUSE
|
||||||
| NC score | 0.136532 (rank : 47) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60948 | Gene names | Mxd4, Mad4 | |||
|
Domain Architecture |

|
|||||
| Description | Max-interacting transcriptional repressor MAD4 (Max-associated protein 4) (MAX dimerization protein 4). | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.136282 (rank : 48) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.131703 (rank : 49) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
HAND2_HUMAN
|
||||||
| NC score | 0.130916 (rank : 50) | θ value | 0.0330416 (rank : 29) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P61296, O95300, O95301, P97833 | Gene names | HAND2, DHAND | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND). | |||||
|
HAND2_MOUSE
|
||||||
| NC score | 0.130916 (rank : 51) | θ value | 0.0330416 (rank : 30) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61039, Q61100 | Gene names | Hand2, Dhand, Hed, Thing2 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 2 (Deciduum, heart, autonomic nervous system and neural crest derivatives-expressed protein 2) (dHAND) (Helix-loop-helix transcription factor expressed in embryo and deciduum) (Thing-2). | |||||
|
MAD3_MOUSE
|
||||||
| NC score | 0.126033 (rank : 52) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
ARNT2_MOUSE
|
||||||
| NC score | 0.125371 (rank : 53) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61324 | Gene names | Arnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
ARNT2_HUMAN
|
||||||
| NC score | 0.124403 (rank : 54) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9HBZ2, O15024 | Gene names | ARNT2, KIAA0307 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2). | |||||
|
BMAL1_MOUSE
|
||||||
| NC score | 0.124392 (rank : 55) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9WTL8, O88295, Q921S4, Q9R0U2, Q9WTL9 | Gene names | Arntl, Bmal1 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Arnt3). | |||||
|
BMAL1_HUMAN
|
||||||
| NC score | 0.124102 (rank : 56) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00327, O00313, O00314, O00315, O00316, O00317, Q99631, Q99649 | Gene names | ARNTL, BMAL1, MOP3 | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator-like protein 1 (Brain and muscle ARNT-like 1) (Member of PAS protein 3) (Basic-helix-loop- helix-PAS orphan MOP3) (bHLH-PAS protein JAP3). | |||||
|
MAD1_MOUSE
|
||||||
| NC score | 0.119221 (rank : 57) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P50538, Q60798, Q61825 | Gene names | Mxd1, Mad, Mad1 | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
MYF6_HUMAN
|
||||||
| NC score | 0.117255 (rank : 58) | θ value | 0.0563607 (rank : 36) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P23409, Q53X80, Q6FHI9 | Gene names | MYF6 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 6 (Myf-6). | |||||
|
MYOG_HUMAN
|
||||||
| NC score | 0.114203 (rank : 59) | θ value | 0.62314 (rank : 53) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P15173 | Gene names | MYOG, MYF4 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenin (Myogenic factor 4) (Myf-4). | |||||
|
HES5_MOUSE
|
||||||
| NC score | 0.114189 (rank : 60) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P70120, Q8BVI1 | Gene names | Hes5, Hes-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-5 (Hairy and enhancer of split 5). | |||||
|
MYOG_MOUSE
|
||||||
| NC score | 0.112906 (rank : 61) | θ value | 0.62314 (rank : 54) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P12979 | Gene names | Myog | |||
|
Domain Architecture |

|
|||||
| Description | Myogenin (MYOD1-related protein). | |||||
|
MAD1_HUMAN
|
||||||
| NC score | 0.112628 (rank : 62) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q05195 | Gene names | MXD1, MAD | |||
|
Domain Architecture |

|
|||||
| Description | MAD protein (MAX dimerizer) (MAX dimerization protein 1). | |||||
|
MYCB_MOUSE
|
||||||
| NC score | 0.106965 (rank : 63) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P8Z1, Q9D6E3 | Gene names | Mycb, Bmyc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein B-Myc. | |||||
|
MYF6_MOUSE
|
||||||
| NC score | 0.106140 (rank : 64) | θ value | 0.279714 (rank : 46) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P15375 | Gene names | Myf6, Myf-6 | |||
|
Domain Architecture |

|
|||||
| Description | Myogenic factor 6 (Myf-6) (Herculin). | |||||
|
TAL1_HUMAN
|
||||||
| NC score | 0.101428 (rank : 65) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P17542 | Gene names | TAL1, SCL, TCL5 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein (TAL-1 protein) (Stem cell protein) (T-cell leukemia/lymphoma-5 protein). | |||||
|
HAND1_HUMAN
|
||||||
| NC score | 0.097796 (rank : 66) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O96004 | Gene names | HAND1, EHAND | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND). | |||||
|
TAL1_MOUSE
|
||||||
| NC score | 0.091230 (rank : 67) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P22091 | Gene names | Tal1, Scl, Tal-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell acute lymphocytic leukemia-1 protein homolog (TAL-1 protein) (Stem cell protein). | |||||
|
HES2_MOUSE
|
||||||
| NC score | 0.087015 (rank : 68) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O54792 | Gene names | Hes2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
NPAS2_MOUSE
|
||||||
| NC score | 0.083563 (rank : 69) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97460 | Gene names | Npas2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2). | |||||
|
HES2_HUMAN
|
||||||
| NC score | 0.081172 (rank : 70) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y543, Q96EN4, Q9Y542 | Gene names | HES2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
HAND1_MOUSE
|
||||||
| NC score | 0.080076 (rank : 71) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q64279, Q61099 | Gene names | Hand1, Ehand, Hxt, Thing1 | |||
|
Domain Architecture |

|
|||||
| Description | Heart- and neural crest derivatives-expressed protein 1 (Extraembryonic tissues, heart, autonomic nervous system and neural crest derivatives-expressed protein 1) (eHAND) (Helix-loop-helix transcription factor expressed in extraembryonic mesoderm and trophoblast) (Thing-1) (Th1). | |||||
|
NPAS2_HUMAN
|
||||||
| NC score | 0.077100 (rank : 72) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
WBS14_HUMAN
|
||||||
| NC score | 0.075943 (rank : 73) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NP71, Q96E48, Q9BY03, Q9BY04, Q9BY05, Q9BY06, Q9Y2P3 | Gene names | MLXIPL, MIO, WBSCR14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein (WS basic-helix- loop-helix leucine zipper protein) (WS-bHLH) (Mlx interactor) (MLX- interacting protein-like). | |||||
|
HES7_HUMAN
|
||||||
| NC score | 0.075300 (rank : 74) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BYE0 | Gene names | HES7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
R3HD2_MOUSE
|
||||||
| NC score | 0.074696 (rank : 75) | θ value | 0.125558 (rank : 40) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
HES3_MOUSE
|
||||||
| NC score | 0.071277 (rank : 76) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61657, O09083, O09150 | Gene names | Hes3, Hes-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-3 (Hairy and enhancer of split 3). | |||||
|
EMSY_MOUSE
|
||||||
| NC score | 0.069148 (rank : 77) | θ value | 0.0148317 (rank : 22) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
FIGLA_MOUSE
|
||||||
| NC score | 0.067719 (rank : 78) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O55208 | Gene names | Figla | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
FIGLA_HUMAN
|
||||||
| NC score | 0.067521 (rank : 79) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6QHK4 | Gene names | FIGLA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Factor in the germline alpha (Transcription factor FIGa) (FIGalpha). | |||||
|
HES7_MOUSE
|
||||||
| NC score | 0.067252 (rank : 80) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BKT2, Q99JA6 | Gene names | Hes7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor HES-7 (Hairy and enhancer of split 7) (bHLH factor Hes7). | |||||
|
BHLH8_MOUSE
|
||||||
| NC score | 0.066942 (rank : 81) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9QYC3, Q9QYE4 | Gene names | Bhlhb8, Mist1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
BHLH8_HUMAN
|
||||||
| NC score | 0.065745 (rank : 82) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7RTS1 | Gene names | BHLHB8, MIST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Class B basic helix-loop-helix protein 8 (bHLHB8) (Muscle, intestine and stomach expression 1) (MIST-1). | |||||
|
XBP1_MOUSE
|
||||||
| NC score | 0.064610 (rank : 83) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35426, Q8VHM0, Q922G5, Q9ESS3 | Gene names | Xbp1, Treb5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X box-binding protein 1 (XBP-1) (Tax-responsive element-binding protein 5 homolog). | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.063905 (rank : 84) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
HEN1_MOUSE
|
||||||
| NC score | 0.062873 (rank : 85) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q02576 | Gene names | Nhlh1, Hen1 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
HEN1_HUMAN
|
||||||
| NC score | 0.062634 (rank : 86) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q02575 | Gene names | NHLH1, HEN1 | |||
|
Domain Architecture |

|
|||||
| Description | Helix-loop-helix protein 1 (HEN1) (Nescient helix loop helix 1) (NSCL- 1). | |||||
|
R3HD1_HUMAN
|
||||||
| NC score | 0.061789 (rank : 87) | θ value | 0.47712 (rank : 52) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15032, Q8IW32 | Gene names | R3HDM1, KIAA0029, R3HDM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R3H domain-containing protein 1. | |||||
|
TCFL5_HUMAN
|
||||||
| NC score | 0.060946 (rank : 88) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UL49, O94771, Q9BYW0 | Gene names | TCFL5, CHA, E2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor-like 5 protein (Cha transcription factor) (HPV-16 E2-binding protein 1) (E2BP-1). | |||||
|
NPAS4_HUMAN
|
||||||
| NC score | 0.058405 (rank : 89) | θ value | 0.365318 (rank : 51) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
WBS14_MOUSE
|
||||||
| NC score | 0.052552 (rank : 90) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.051912 (rank : 91) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.050542 (rank : 92) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
NFRKB_MOUSE
|
||||||
| NC score | 0.050510 (rank : 93) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.050034 (rank : 94) | θ value | 0.0431538 (rank : 34) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
NPAS4_MOUSE
|
||||||
| NC score | 0.049985 (rank : 95) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BGD7, Q3V3U3 | Gene names | Npas4, Nxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF) (Limbic-enhanced PAS protein) (LE-PAS). | |||||
|
DCP1A_HUMAN
|
||||||
| NC score | 0.049961 (rank : 96) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NPI6 | Gene names | DCP1A, SMIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (Smad4-interacting transcriptional co-activator). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.047042 (rank : 97) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
ATF2_HUMAN
|
||||||
| NC score | 0.045968 (rank : 98) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
ATF2_MOUSE
|
||||||
| NC score | 0.045075 (rank : 99) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
ATF1_MOUSE
|
||||||
| NC score | 0.043632 (rank : 100) | θ value | 0.365318 (rank : 47) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P81269 | Gene names | Atf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TCR-ATF1). | |||||
|
CK024_MOUSE
|
||||||
| NC score | 0.041982 (rank : 101) | θ value | 1.06291 (rank : 62) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.039789 (rank : 102) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
ELF1_MOUSE
|
||||||
| NC score | 0.037412 (rank : 103) | θ value | 0.0330416 (rank : 28) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
PF21B_MOUSE
|
||||||
| NC score | 0.037304 (rank : 104) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
K1683_HUMAN
|
||||||
| NC score | 0.037161 (rank : 105) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.034904 (rank : 106) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
CIKS_HUMAN
|
||||||
| NC score | 0.033999 (rank : 107) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43734, Q5R3A3, Q9H5W2, Q9H6Y3, Q9NS14, Q9UG72 | Gene names | TRAF3IP2, C6orf4, C6orf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adapter protein CIKS (Connection to IKK and SAPK/JNK) (TRAF3- interacting protein 2) (Nuclear factor NF-kappa-B activator 1) (ACT1). | |||||
|
CKLF4_MOUSE
|
||||||
| NC score | 0.030315 (rank : 108) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CJ61, Q8K143 | Gene names | Cmtm4, Cklfsf4 | |||
|
Domain Architecture |

|
|||||
| Description | CKLF-like MARVEL transmembrane domain-containing protein 4 (Chemokine- like factor superfamily member 4). | |||||
|
CKLF4_HUMAN
|
||||||
| NC score | 0.030223 (rank : 109) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IZR5, Q8IZR4, Q8IZV1 | Gene names | CMTM4, CKLFSF4 | |||
|
Domain Architecture |

|
|||||
| Description | CKLF-like MARVEL transmembrane domain-containing protein 4 (Chemokine- like factor superfamily member 4). | |||||
|
ATX2L_HUMAN
|
||||||
| NC score | 0.030155 (rank : 110) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
GP156_HUMAN
|
||||||
| NC score | 0.028979 (rank : 111) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NFN8, Q86SN6 | Gene names | GPR156, GABABL, PGR28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 156 (GABAB-related G-protein coupled receptor) (G-protein coupled receptor PGR28). | |||||
|
ATF1_HUMAN
|
||||||
| NC score | 0.028596 (rank : 112) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P18846, P25168 | Gene names | ATF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-1 (Activating transcription factor 1) (TREB36 protein). | |||||
|
AFF2_HUMAN
|
||||||
| NC score | 0.027438 (rank : 113) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
K0319_HUMAN
|
||||||
| NC score | 0.026973 (rank : 114) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5VV43, Q9UJC8, Q9Y4G7 | Gene names | KIAA0319 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0319 precursor. | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.026221 (rank : 115) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
PAPOG_MOUSE
|
||||||
| NC score | 0.025644 (rank : 116) | θ value | 0.62314 (rank : 55) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PCL9, Q8BZC9 | Gene names | Papolg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) polymerase gamma (EC 2.7.7.19) (PAP gamma) (Polynucleotide adenylyltransferase gamma) (SRP RNA 3' adenylating enzyme). | |||||
|
SYM_MOUSE
|
||||||
| NC score | 0.025570 (rank : 117) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q68FL6 | Gene names | Mars | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
RFX5_HUMAN
|
||||||
| NC score | 0.025147 (rank : 118) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48382 | Gene names | RFX5 | |||
|
Domain Architecture |
|
|||||
| Description | DNA-binding protein RFX5 (Regulatory factor X subunit 5). | |||||
|
PDXL2_MOUSE
|
||||||
| NC score | 0.023781 (rank : 119) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CAE9, Q8CFW3 | Gene names | Podxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
CAH5B_MOUSE
|
||||||
| NC score | 0.023296 (rank : 120) | θ value | 0.163984 (rank : 41) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZA0, Q8C3J9, Q8K014 | Gene names | Ca5b, Car5b | |||
|
Domain Architecture |

|
|||||
| Description | Carbonic anhydrase 5B, mitochondrial precursor (EC 4.2.1.1) (Carbonic anhydrase VB) (Carbonate dehydratase VB) (CA-VB). | |||||
|
P80C_HUMAN
|
||||||
| NC score | 0.022671 (rank : 121) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P38432 | Gene names | COIL, CLN80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coilin (p80). | |||||
|
GP152_HUMAN
|
||||||
| NC score | 0.021288 (rank : 122) | θ value | 0.279714 (rank : 45) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
PPRB_HUMAN
|
||||||
| NC score | 0.020504 (rank : 123) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
SYM_HUMAN
|
||||||
| NC score | 0.020410 (rank : 124) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56192, Q14895, Q53H14, Q96A15, Q96BZ0, Q9NSE0 | Gene names | MARS | |||
|
Domain Architecture |

|
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
HCFC1_MOUSE
|
||||||
| NC score | 0.019918 (rank : 125) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
SP4_MOUSE
|
||||||
| NC score | 0.019874 (rank : 126) | θ value | 0.0330416 (rank : 31) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62445 | Gene names | Sp4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp4. | |||||
|
SP4_HUMAN
|
||||||
| NC score | 0.019337 (rank : 127) | θ value | 0.0563607 (rank : 37) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q02446, O60402 | Gene names | SP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp4 (SPR-1). | |||||
|
EI2BE_MOUSE
|
||||||
| NC score | 0.019314 (rank : 128) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CHW4, Q3TU39 | Gene names | Eif2b5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Translation initiation factor eIF-2B subunit epsilon (eIF-2B GDP-GTP exchange factor subunit epsilon). | |||||
|
ZEP1_HUMAN
|
||||||
| NC score | 0.019216 (rank : 129) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
SP3_MOUSE
|
||||||
| NC score | 0.018508 (rank : 130) | θ value | 0.21417 (rank : 44) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 731 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O70494, Q68FF2, Q8CF64, Q8K378 | Gene names | Sp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp3. | |||||
|
RTN2_MOUSE
|
||||||
| NC score | 0.018496 (rank : 131) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70622, O70620 | Gene names | Rtn2, Nspl1 | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-2 (Neuroendocrine-specific protein-like 1) (NSP-like protein 1) (NSPLI). | |||||
|
SP3_HUMAN
|
||||||
| NC score | 0.018365 (rank : 132) | θ value | 0.21417 (rank : 43) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02447, Q8TD56, Q9BQR1 | Gene names | SP3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp3 (SPR-2). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.013984 (rank : 133) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
TMM20_HUMAN
|
||||||
| NC score | 0.013861 (rank : 134) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q2M3R5, Q86YG5, Q8NBA5 | Gene names | TMEM20, C10orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 20. | |||||
|
ZN292_HUMAN
|
||||||
| NC score | 0.013589 (rank : 135) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60281, Q9H8G3, Q9H8J4 | Gene names | ZNF292, KIAA0530 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 292. | |||||
|
CAH5B_HUMAN
|
||||||
| NC score | 0.013518 (rank : 136) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2D0 | Gene names | CA5B | |||
|
Domain Architecture |

|
|||||
| Description | Carbonic anhydrase 5B, mitochondrial precursor (EC 4.2.1.1) (Carbonic anhydrase VB) (Carbonate dehydratase VB) (CA-VB). | |||||
|
NUDT9_MOUSE
|
||||||
| NC score | 0.013401 (rank : 137) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BVU5, Q3TZ68, Q8K1J4 | Gene names | Nudt9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribose pyrophosphatase, mitochondrial precursor (EC 3.6.1.13) (ADP-ribose diphosphatase) (Adenosine diphosphoribose pyrophosphatase) (ADPR-PPase) (ADP-ribose phosphohydrolase) (Nucleoside diphosphate- linked moiety X motif 9) (Nudix motif 9). | |||||
|
RBM25_HUMAN
|
||||||
| NC score | 0.012742 (rank : 138) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
CEAM7_HUMAN
|
||||||
| NC score | 0.011266 (rank : 139) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14002, O15148, O15149, Q13983, Q9UPJ2 | Gene names | CEACAM7, CGM2 | |||
|
Domain Architecture |

|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 7 precursor (Carcinoembryonic antigen CGM2). | |||||
|
B3GA2_HUMAN
|
||||||
| NC score | 0.011245 (rank : 140) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NPZ5, Q5JS09, Q8TF38, Q96NK4 | Gene names | B3GAT2, GLCATS, KIAA1963 | |||
|
Domain Architecture |

|
|||||
| Description | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 2 (EC 2.4.1.135) (Beta-1,3-glucuronyltransferase 2) (Glucuronosyltransferase-S) (GlcAT-S) (UDP-glucuronosyltransferase-S) (GlcAT-D). | |||||
|
IQEC3_MOUSE
|
||||||
| NC score | 0.009949 (rank : 141) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
T2R41_MOUSE
|
||||||
| NC score | 0.008377 (rank : 142) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59532, Q7M704 | Gene names | Tas2r41, Tas2r12, Tas2r126, Tas2r26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 2 member 41 (T2R41) (T2R12) (T2R26). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.007585 (rank : 143) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.007198 (rank : 144) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
TA2R7_HUMAN
|
||||||
| NC score | 0.007008 (rank : 145) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYW3 | Gene names | TAS2R7 | |||
|
Domain Architecture |

|
|||||
| Description | Taste receptor type 2 member 7 (T2R7) (Taste receptor family B member 4) (TRB4). | |||||
|
ZHX3_MOUSE
|
||||||
| NC score | 0.006832 (rank : 146) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C0Q2, Q80U14 | Gene names | Zhx3, Kiaa0395, Tix1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and homeodomain protein 3 (Triple homeobox protein 1). | |||||
|
CT075_MOUSE
|
||||||
| NC score | 0.006649 (rank : 147) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59383 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf75 homolog precursor. | |||||
|
PAX3_HUMAN
|
||||||
| NC score | 0.005833 (rank : 148) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23760, Q16448 | Gene names | PAX3, HUP2 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-3 (HUP2). | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.005457 (rank : 149) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.004978 (rank : 150) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.004060 (rank : 151) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
LTB1L_HUMAN
|
||||||
| NC score | 0.003487 (rank : 152) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14766 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
VASN_MOUSE
|
||||||
| NC score | 0.001651 (rank : 153) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CZT5, Q8BJJ0, Q8R2G5 | Gene names | Vasn, Slitl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vasorin precursor (Protein Slit-like 2). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.001458 (rank : 154) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
MK15_HUMAN
|
||||||
| NC score | 0.000745 (rank : 155) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TD08, Q2TCF9, Q8N362 | Gene names | MAPK15, ERK8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 15 (EC 2.7.11.24) (Extracellular signal-regulated kinase 8). | |||||
|
CAMKV_HUMAN
|
||||||
| NC score | 0.000209 (rank : 156) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 133 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||