Please be patient as the page loads
|
ELF1_MOUSE
|
||||||
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ELF1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.983833 (rank : 2) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF4_HUMAN
|
||||||
| θ value | 3.22841e-82 (rank : 3) | NC score | 0.952957 (rank : 3) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q99607, O60435 | Gene names | ELF4, ELFR, MEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELF4_MOUSE
|
||||||
| θ value | 8.22888e-78 (rank : 4) | NC score | 0.934733 (rank : 6) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELF2_HUMAN
|
||||||
| θ value | 8.24801e-70 (rank : 5) | NC score | 0.944643 (rank : 4) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q15723, Q15724, Q15725, Q6P1K5 | Gene names | ELF2, NERF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF2_MOUSE
|
||||||
| θ value | 3.03575e-64 (rank : 6) | NC score | 0.942831 (rank : 5) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELK1_HUMAN
|
||||||
| θ value | 8.36355e-22 (rank : 7) | NC score | 0.853137 (rank : 7) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK1_MOUSE
|
||||||
| θ value | 1.5773e-20 (rank : 8) | NC score | 0.848830 (rank : 10) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P41969 | Gene names | Elk1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ELK4_HUMAN
|
||||||
| θ value | 2.27762e-19 (rank : 9) | NC score | 0.821536 (rank : 19) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
SPDEF_MOUSE
|
||||||
| θ value | 2.97466e-19 (rank : 10) | NC score | 0.850108 (rank : 9) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9WTP3 | Gene names | Spdef, Pdef, Pse | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
SPDEF_HUMAN
|
||||||
| θ value | 3.88503e-19 (rank : 11) | NC score | 0.851675 (rank : 8) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O95238 | Gene names | SPDEF, PDEF, PSE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ELK4_MOUSE
|
||||||
| θ value | 1.13037e-18 (rank : 12) | NC score | 0.829853 (rank : 15) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P41158 | Gene names | Elk4, Sap1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ETV4_HUMAN
|
||||||
| θ value | 4.29542e-18 (rank : 13) | NC score | 0.783031 (rank : 33) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P43268, Q96AW9 | Gene names | ETV4, E1AF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 4 (Adenovirus E1A enhancer-binding protein) (E1A-F). | |||||
|
ETV4_MOUSE
|
||||||
| θ value | 4.29542e-18 (rank : 14) | NC score | 0.781826 (rank : 34) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
ETS2_HUMAN
|
||||||
| θ value | 5.60996e-18 (rank : 15) | NC score | 0.813151 (rank : 22) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15036 | Gene names | ETS2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETS2_MOUSE
|
||||||
| θ value | 5.60996e-18 (rank : 16) | NC score | 0.811496 (rank : 25) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15037 | Gene names | Ets2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETS1_HUMAN
|
||||||
| θ value | 1.63225e-17 (rank : 17) | NC score | 0.810500 (rank : 26) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P14921, Q14278, Q16080 | Gene names | ETS1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS1_MOUSE
|
||||||
| θ value | 1.63225e-17 (rank : 18) | NC score | 0.810354 (rank : 27) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P27577, Q61403 | Gene names | Ets1, Ets-1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETV2_MOUSE
|
||||||
| θ value | 2.7842e-17 (rank : 19) | NC score | 0.837392 (rank : 12) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41163 | Gene names | Etv2, Er71, Etsrp71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ETV6_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 20) | NC score | 0.813063 (rank : 23) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV6_MOUSE
|
||||||
| θ value | 2.7842e-17 (rank : 21) | NC score | 0.812640 (rank : 24) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV7_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 22) | NC score | 0.848054 (rank : 11) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y603, Q9NZ65, Q9NZ66, Q9NZ68, Q9NZR8, Q9UNJ7, Q9Y5K4, Q9Y604 | Gene names | ETV7, TEL2, TELB, TREF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV7 (Transcription factor Tel-2) (ETS-related protein Tel2) (Tel-related Ets factor). | |||||
|
ETV5_HUMAN
|
||||||
| θ value | 4.74913e-17 (rank : 23) | NC score | 0.772808 (rank : 39) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
ETV5_MOUSE
|
||||||
| θ value | 4.74913e-17 (rank : 24) | NC score | 0.756845 (rank : 41) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
ETV1_HUMAN
|
||||||
| θ value | 6.20254e-17 (rank : 25) | NC score | 0.779090 (rank : 36) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV1_MOUSE
|
||||||
| θ value | 6.20254e-17 (rank : 26) | NC score | 0.779866 (rank : 35) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
GABPA_HUMAN
|
||||||
| θ value | 1.058e-16 (rank : 27) | NC score | 0.816534 (rank : 21) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q06546, Q12939 | Gene names | GABPA, E4TF1A | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha) (Transcription factor E4TF1-60) (Nuclear respiratory factor 2 subunit alpha). | |||||
|
GABPA_MOUSE
|
||||||
| θ value | 1.058e-16 (rank : 28) | NC score | 0.817948 (rank : 20) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q00422 | Gene names | Gabpa, E4tf1a | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha). | |||||
|
FLI1_MOUSE
|
||||||
| θ value | 2.35696e-16 (rank : 29) | NC score | 0.794016 (rank : 28) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P26323 | Gene names | Fli1, Fli-1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Retroviral integration site protein Fli-1). | |||||
|
ELK3_HUMAN
|
||||||
| θ value | 3.07829e-16 (rank : 30) | NC score | 0.830196 (rank : 14) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41970, Q6FG57, Q6GU29, Q9UD17 | Gene names | ELK3, NET, SAP2 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP) (SRF accessory protein 2) (SAP-2). | |||||
|
FLI1_HUMAN
|
||||||
| θ value | 3.07829e-16 (rank : 31) | NC score | 0.792582 (rank : 29) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q01543, Q14319, Q92480, Q9UE07 | Gene names | FLI1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Fli-1 proto- oncogene) (ERGB transcription factor). | |||||
|
ELK3_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 32) | NC score | 0.826465 (rank : 16) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41971, P97747, Q62346 | Gene names | Elk3, Erp, Net | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP). | |||||
|
ETV2_HUMAN
|
||||||
| θ value | 5.25075e-16 (rank : 33) | NC score | 0.832919 (rank : 13) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00321, Q9UEA0 | Gene names | ETV2, ER71, ETSRP71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ETV3_HUMAN
|
||||||
| θ value | 6.85773e-16 (rank : 34) | NC score | 0.767076 (rank : 40) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ETV3_MOUSE
|
||||||
| θ value | 6.85773e-16 (rank : 35) | NC score | 0.788824 (rank : 30) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8R4Z4, Q9QZW1 | Gene names | Etv3, Mets, Pe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ERG_HUMAN
|
||||||
| θ value | 1.99529e-15 (rank : 36) | NC score | 0.786126 (rank : 31) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P11308, Q16113 | Gene names | ERG | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG (Transforming protein ERG). | |||||
|
ERG_MOUSE
|
||||||
| θ value | 1.99529e-15 (rank : 37) | NC score | 0.784511 (rank : 32) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P81270, Q8C5L4, Q920K7, Q920K8, Q920K9 | Gene names | Erg, Erg-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG. | |||||
|
ELF5_HUMAN
|
||||||
| θ value | 9.90251e-15 (rank : 38) | NC score | 0.821571 (rank : 18) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKW6, O95175, Q8N2K9, Q96QY3, Q9UKW5 | Gene names | ELF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5) (Epithelium-specific Ets transcription factor 2) (ESE-2) (Epithelium- restricted ESE-1-related Ets factor). | |||||
|
ELF5_MOUSE
|
||||||
| θ value | 9.90251e-15 (rank : 39) | NC score | 0.822654 (rank : 17) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VDK3, Q921H5, Q9Z2K6 | Gene names | Elf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5). | |||||
|
ERF_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 40) | NC score | 0.777109 (rank : 37) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P50548, Q9UPI7 | Gene names | ERF | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF (Ets2 repressor factor). | |||||
|
ERF_MOUSE
|
||||||
| θ value | 1.86753e-13 (rank : 41) | NC score | 0.776611 (rank : 38) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
SPIC_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 42) | NC score | 0.643666 (rank : 42) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8N5J4 | Gene names | SPIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C. | |||||
|
SPIC_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 43) | NC score | 0.612299 (rank : 46) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P3D7, Q9Z0Y0 | Gene names | Spic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C (Pu.1-related factor) (Prf). | |||||
|
SPI1_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 44) | NC score | 0.632085 (rank : 44) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17947 | Gene names | SPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein). | |||||
|
SPI1_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 45) | NC score | 0.634228 (rank : 43) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17433, Q99L57 | Gene names | Spi1, Sfpi-1, Sfpi1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein) (SFFV proviral integration 1 protein). | |||||
|
SPIB_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 46) | NC score | 0.622157 (rank : 45) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q01892, Q15359 | Gene names | SPIB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIB_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 47) | NC score | 0.609260 (rank : 47) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O35906, O35907, O35909, O55199 | Gene names | Spib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-B. | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 48) | NC score | 0.054311 (rank : 58) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
ARID2_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 49) | NC score | 0.078974 (rank : 48) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
HCFC1_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 50) | NC score | 0.077670 (rank : 49) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
RFX1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 51) | NC score | 0.045239 (rank : 64) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P22670 | Gene names | RFX1 | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II regulatory factor RFX1 (RFX) (Enhancer factor C) (EF-C). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 52) | NC score | 0.076218 (rank : 50) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
AAK1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 53) | NC score | 0.011431 (rank : 101) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
TF2AA_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 54) | NC score | 0.048875 (rank : 60) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P52655 | Gene names | GTF2A1, TF2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIA subunit 1 (General transcription factor IIA1) (TFIIA-42) (TFIIAL) [Contains: Transcription initiation factor IIA alpha chain (TFIIA p35 subunit); Transcription initiation factor IIA beta chain (TFIIA p19 subunit)]. | |||||
|
HCFC1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 55) | NC score | 0.076164 (rank : 51) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 56) | NC score | 0.017707 (rank : 87) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SRBP2_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 57) | NC score | 0.037412 (rank : 73) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 58) | NC score | 0.070510 (rank : 53) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 59) | NC score | 0.065690 (rank : 55) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 60) | NC score | 0.014626 (rank : 94) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
SIX4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 61) | NC score | 0.018171 (rank : 86) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UIU6 | Gene names | SIX4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX4 (Sine oculis homeobox homolog 4). | |||||
|
EYA3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 62) | NC score | 0.020747 (rank : 83) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99504, O95463, Q99813 | Gene names | EYA3 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 3 (EC 3.1.3.48). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 63) | NC score | 0.010157 (rank : 108) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
ATF6B_MOUSE
|
||||||
| θ value | 0.21417 (rank : 64) | NC score | 0.038847 (rank : 72) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35451 | Gene names | Crebl1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp). | |||||
|
CN004_MOUSE
|
||||||
| θ value | 0.21417 (rank : 65) | NC score | 0.046099 (rank : 62) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K3X4, Q3USE5, Q8BUS1, Q8C011 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4 homolog. | |||||
|
EMSY_HUMAN
|
||||||
| θ value | 0.21417 (rank : 66) | NC score | 0.072565 (rank : 52) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
ARHGC_MOUSE
|
||||||
| θ value | 0.279714 (rank : 67) | NC score | 0.018517 (rank : 85) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R4H2, Q80U18 | Gene names | Arhgef12, Kiaa0382, Larg | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
ATF7_HUMAN
|
||||||
| θ value | 0.279714 (rank : 68) | NC score | 0.040050 (rank : 71) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
SP4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 69) | NC score | 0.005649 (rank : 123) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62445 | Gene names | Sp4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp4. | |||||
|
AT8B3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 70) | NC score | 0.011718 (rank : 99) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60423, Q8IVB8, Q8N4Y8, Q96M22 | Gene names | ATP8B3, ATP1K, FOS37502_2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IK (EC 3.6.3.1) (ATPase class I type 8B member 3). | |||||
|
ATF6A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 71) | NC score | 0.044387 (rank : 66) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P18850, O15139, Q5VW62, Q6IPB5, Q9UEC9 | Gene names | ATF6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 alpha (Activating transcription factor 6 alpha) (ATF6-alpha). | |||||
|
ATF7_MOUSE
|
||||||
| θ value | 0.365318 (rank : 72) | NC score | 0.036747 (rank : 74) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
SP4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 73) | NC score | 0.003517 (rank : 130) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02446, O60402 | Gene names | SP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp4 (SPR-1). | |||||
|
SUHW3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 74) | NC score | 0.006506 (rank : 120) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 697 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P3Y5, Q6ZPM2, Q8K145 | Gene names | Suhw3, Kiaa1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 3. | |||||
|
CN004_HUMAN
|
||||||
| θ value | 0.47712 (rank : 75) | NC score | 0.043503 (rank : 68) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H1B7, Q8NDQ2, Q96JG2, Q9H3I7 | Gene names | C14orf4, KIAA1865 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4. | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 76) | NC score | 0.054747 (rank : 57) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 77) | NC score | 0.010548 (rank : 106) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
BOC_HUMAN
|
||||||
| θ value | 0.62314 (rank : 78) | NC score | 0.010339 (rank : 107) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BWV1, Q6UXJ5, Q8N2P7, Q8NF26 | Gene names | BOC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brother of CDO precursor (Protein BOC). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 0.813845 (rank : 79) | NC score | 0.024869 (rank : 79) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 0.813845 (rank : 80) | NC score | 0.051314 (rank : 59) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
EMSY_MOUSE
|
||||||
| θ value | 1.06291 (rank : 81) | NC score | 0.069838 (rank : 54) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 82) | NC score | 0.060304 (rank : 56) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
RFX1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 83) | NC score | 0.041025 (rank : 70) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P48377 | Gene names | Rfx1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFX1. | |||||
|
SON_MOUSE
|
||||||
| θ value | 1.06291 (rank : 84) | NC score | 0.042952 (rank : 69) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
TPR_HUMAN
|
||||||
| θ value | 1.06291 (rank : 85) | NC score | 0.001154 (rank : 134) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 86) | NC score | 0.004546 (rank : 125) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 87) | NC score | 0.044062 (rank : 67) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 88) | NC score | 0.045621 (rank : 63) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
RP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 89) | NC score | 0.015046 (rank : 92) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56715 | Gene names | RP1, ORP1 | |||
|
Domain Architecture |
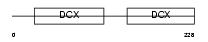
|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein) (Retinitis pigmentosa 1 protein). | |||||
|
ARI4A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 90) | NC score | 0.019035 (rank : 84) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
EWS_HUMAN
|
||||||
| θ value | 1.81305 (rank : 91) | NC score | 0.015309 (rank : 90) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q01844, Q92635 | Gene names | EWSR1, EWS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS (EWS oncogene) (Ewing sarcoma breakpoint region 1 protein). | |||||
|
FGD1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 92) | NC score | 0.012376 (rank : 97) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 93) | NC score | 0.003931 (rank : 126) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
HTF4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 94) | NC score | 0.011342 (rank : 102) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99081 | Gene names | TCF12, HEB, HTF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4). | |||||
|
MCA1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 95) | NC score | 0.028287 (rank : 76) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P31230, Q60659 | Gene names | Scye1, Emap2 | |||
|
Domain Architecture |

|
|||||
| Description | Multisynthetase complex auxiliary component p43 [Contains: Endothelial monocyte-activating polypeptide 2 (EMAP-II) (Small inducible cytokine subfamily E member 1)]. | |||||
|
POGZ_MOUSE
|
||||||
| θ value | 1.81305 (rank : 96) | NC score | 0.006207 (rank : 121) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BZH4, Q4VA94, Q80TZ8, Q8C0K1, Q8K294 | Gene names | Pogz, Kiaa0461 | |||
|
Domain Architecture |

|
|||||
| Description | Pogo transposable element with ZNF domain. | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 97) | NC score | 0.022781 (rank : 81) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SP2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 98) | NC score | 0.001649 (rank : 133) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q02086 | Gene names | SP2, KIAA0048 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp2. | |||||
|
TIMD1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 99) | NC score | 0.044782 (rank : 65) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96D42, O43656 | Gene names | HAVCR1, TIM1, TIMD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatitis A virus cellular receptor 1 precursor (HAVcr-1) (T cell immunoglobulin and mucin domain-containing protein 1) (TIMD-1) (T cell membrane protein 1) (TIM-1) (TIM). | |||||
|
ARVC_MOUSE
|
||||||
| θ value | 2.36792 (rank : 100) | NC score | 0.010897 (rank : 103) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P98203, Q6PGJ6, Q8BQ36, Q8BRF2, Q8C3U7, Q924L2, Q924L3, Q924L4, Q924L5 | Gene names | Arvcf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome homolog. | |||||
|
BOC_MOUSE
|
||||||
| θ value | 2.36792 (rank : 101) | NC score | 0.009851 (rank : 111) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6AZB0, Q6KAM5, Q6P5H3, Q7TMJ3, Q8CE73, Q8CE91, Q8R377, Q923W7 | Gene names | Boc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brother of CDO precursor (Protein BOC). | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.015109 (rank : 91) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
E2AK1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | -0.000693 (rank : 136) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z2R9, Q8C024, Q8K123, Q9CTP5, Q9D601 | Gene names | Eif2ak1, Hri | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2-alpha kinase 1 (EC 2.7.11.1) (Heme-regulated eukaryotic initiation factor eIF-2-alpha kinase) (Heme-regulated inhibitor) (Heme-controlled repressor) (HCR) (Hemin-sensitive initiation factor 2-alpha kinase). | |||||
|
IL1R1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.004930 (rank : 124) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P14778 | Gene names | IL1R1, IL1R, IL1RA, IL1RT1 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-1 receptor type I precursor (IL-1R-1) (IL-1RT1) (IL-1R- alpha) (p80) (CD121a antigen). | |||||
|
MBNL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.010631 (rank : 104) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JKP5 | Gene names | Mbnl1, Exp, Mbnl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like protein (Triplet-expansion RNA-binding protein). | |||||
|
NLGN3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.006639 (rank : 119) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BYM5, Q8BXR4 | Gene names | Nlgn3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
|
SLK_MOUSE
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | -0.002880 (rank : 138) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
SON_HUMAN
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.048725 (rank : 61) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SUHW1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.003804 (rank : 127) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 648 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P59817 | Gene names | SUHW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 1 (3'OY11.1). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.022560 (rank : 82) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
YTHD3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.011898 (rank : 98) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z739, Q63Z37, Q659A3 | Gene names | YTHDF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain family protein 3. | |||||
|
ZYX_HUMAN
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.007974 (rank : 113) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15942 | Gene names | ZYX | |||
|
Domain Architecture |

|
|||||
| Description | Zyxin (Zyxin-2). | |||||
|
ARHGC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.014525 (rank : 95) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZN5, O15086 | Gene names | ARHGEF12, KIAA0382, LARG | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
ARVC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.009906 (rank : 110) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00192 | Gene names | ARVCF | |||
|
Domain Architecture |

|
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome. | |||||
|
GP112_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.031786 (rank : 75) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
GPR98_MOUSE
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.007731 (rank : 115) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VHN7, Q6ZQ69, Q810D2, Q810D3, Q91ZS0, Q91ZS1 | Gene names | Gpr98, Kiaa0686, Mass1, Vlgr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1) (Very large G-protein coupled receptor 1) (Neurepin). | |||||
|
MAST1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | -0.001303 (rank : 137) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
RIOK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.014757 (rank : 93) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BRS2, Q8NDC8, Q96NV9 | Gene names | RIOK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO1 (EC 2.7.11.1) (RIO kinase 1). | |||||
|
SDC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.016041 (rank : 89) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18827, Q96HB7 | Gene names | SDC1, SDC | |||
|
Domain Architecture |
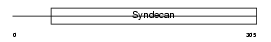
|
|||||
| Description | Syndecan-1 precursor (SYND1) (CD138 antigen). | |||||
|
SSXT_HUMAN
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.025395 (rank : 77) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15532, Q16404, Q9BXC6 | Gene names | SS18, SSXT, SYT | |||
|
Domain Architecture |
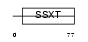
|
|||||
| Description | SSXT protein (Synovial sarcoma, translocated to X chromosome) (SYT protein). | |||||
|
CS016_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.025258 (rank : 78) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
DGKH_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.003638 (rank : 128) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86XP1, Q5VZW0, Q6PI56, Q86XP2, Q8N3N0, Q8N7J9 | Gene names | DGKH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase eta (EC 2.7.1.107) (Diglyceride kinase eta) (DGK-eta) (DAG kinase eta). | |||||
|
DUS16_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.003543 (rank : 129) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BY84, Q9C0G3 | Gene names | DUSP16, KIAA1700, MKP7 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 16 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 7) (MAP kinase phosphatase 7) (MKP-7). | |||||
|
HOMEZ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.010057 (rank : 109) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IX15, Q6P049, Q86XB6, Q9P2A5 | Gene names | HOMEZ, KIAA1443 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox and leucine zipper protein Homez (Homeodomain leucine zipper- containing factor). | |||||
|
MSX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.000634 (rank : 135) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P28360, Q96NY4 | Gene names | MSX1, HOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein MSX-1 (Msh homeobox 1-like protein) (Hox-7). | |||||
|
MSX2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.002364 (rank : 132) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35548 | Gene names | MSX2, HOX8 | |||
|
Domain Architecture |
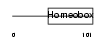
|
|||||
| Description | Homeobox protein MSX-2 (Hox-8). | |||||
|
WDR36_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.002850 (rank : 131) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NI36, Q8N1Q2 | Gene names | WDR36 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 36 (T-cell activation WD repeat-containing protein) (TA-WDRP). | |||||
|
AFF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.012839 (rank : 96) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
EWS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.009804 (rank : 112) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61545 | Gene names | Ewsr1, Ews, Ewsh | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS. | |||||
|
MOR2A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.007971 (rank : 114) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q69ZX6, Q5QNQ7, Q6P547 | Gene names | Morc2a, Kiaa0852, Zcwcc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 2A (Zinc finger CW-type coiled-coil domain protein 1). | |||||
|
NPAS2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.006854 (rank : 118) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
SLK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | -0.003891 (rank : 139) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
SSXT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.023110 (rank : 80) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62280 | Gene names | Ss18, Ssxt, Syt | |||
|
Domain Architecture |
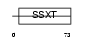
|
|||||
| Description | SSXT protein (SYT protein) (Synovial sarcoma-associated Ss18-alpha). | |||||
|
TAF6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.017573 (rank : 88) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49848 | Gene names | TAF6, TAF2E, TAFII70 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 6 (Transcription initiation factor TFIID 70 kDa subunit) (TAF(II)70) (TAFII-70) (TAFII- 80) (TAFII80). | |||||
|
TSC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.005867 (rank : 122) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61037, P97723, P97724, P97725, P97727, Q61007, Q61008, Q9WUF6 | Gene names | Tsc2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 homolog protein). | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.007559 (rank : 116) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
XRN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.006859 (rank : 117) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IZH2, Q4G0S3, Q68D88, Q6AI24, Q6MZS8, Q86WS7, Q8N8U4, Q9UF39 | Gene names | XRN1, SEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-3' exoribonuclease 1 (EC 3.1.11.-) (Strand-exchange protein 1 homolog). | |||||
|
ZHX1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.010630 (rank : 105) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKY1, Q8IWD8 | Gene names | ZHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
ZHX1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.011611 (rank : 100) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70121, Q8BQ68, Q8C6T4, Q8CJG3 | Gene names | Zhx1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
ELF1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 139 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF1_HUMAN
|
||||||
| NC score | 0.983833 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF4_HUMAN
|
||||||
| NC score | 0.952957 (rank : 3) | θ value | 3.22841e-82 (rank : 3) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q99607, O60435 | Gene names | ELF4, ELFR, MEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELF2_HUMAN
|
||||||
| NC score | 0.944643 (rank : 4) | θ value | 8.24801e-70 (rank : 5) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q15723, Q15724, Q15725, Q6P1K5 | Gene names | ELF2, NERF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF2_MOUSE
|
||||||
| NC score | 0.942831 (rank : 5) | θ value | 3.03575e-64 (rank : 6) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
ELF4_MOUSE
|
||||||
| NC score | 0.934733 (rank : 6) | θ value | 8.22888e-78 (rank : 4) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
ELK1_HUMAN
|
||||||
| NC score | 0.853137 (rank : 7) | θ value | 8.36355e-22 (rank : 7) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P19419, O75606, O95058, Q969X8, Q9UJM4 | Gene names | ELK1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
SPDEF_HUMAN
|
||||||
| NC score | 0.851675 (rank : 8) | θ value | 3.88503e-19 (rank : 11) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O95238 | Gene names | SPDEF, PDEF, PSE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
SPDEF_MOUSE
|
||||||
| NC score | 0.850108 (rank : 9) | θ value | 2.97466e-19 (rank : 10) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9WTP3 | Gene names | Spdef, Pdef, Pse | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAM pointed domain-containing Ets transcription factor (Prostate- derived Ets factor) (Prostate epithelium-specific Ets transcription factor) (Prostate-specific Ets). | |||||
|
ELK1_MOUSE
|
||||||
| NC score | 0.848830 (rank : 10) | θ value | 1.5773e-20 (rank : 8) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P41969 | Gene names | Elk1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-1. | |||||
|
ETV7_HUMAN
|
||||||
| NC score | 0.848054 (rank : 11) | θ value | 2.7842e-17 (rank : 22) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y603, Q9NZ65, Q9NZ66, Q9NZ68, Q9NZR8, Q9UNJ7, Q9Y5K4, Q9Y604 | Gene names | ETV7, TEL2, TELB, TREF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV7 (Transcription factor Tel-2) (ETS-related protein Tel2) (Tel-related Ets factor). | |||||
|
ETV2_MOUSE
|
||||||
| NC score | 0.837392 (rank : 12) | θ value | 2.7842e-17 (rank : 19) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41163 | Gene names | Etv2, Er71, Etsrp71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ETV2_HUMAN
|
||||||
| NC score | 0.832919 (rank : 13) | θ value | 5.25075e-16 (rank : 33) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00321, Q9UEA0 | Gene names | ETV2, ER71, ETSRP71 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 2 (Ets-related protein 71). | |||||
|
ELK3_HUMAN
|
||||||
| NC score | 0.830196 (rank : 14) | θ value | 3.07829e-16 (rank : 30) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P41970, Q6FG57, Q6GU29, Q9UD17 | Gene names | ELK3, NET, SAP2 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP) (SRF accessory protein 2) (SAP-2). | |||||
|
ELK4_MOUSE
|
||||||
| NC score | 0.829853 (rank : 15) | θ value | 1.13037e-18 (rank : 12) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P41158 | Gene names | Elk4, Sap1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
ELK3_MOUSE
|
||||||
| NC score | 0.826465 (rank : 16) | θ value | 4.02038e-16 (rank : 32) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P41971, P97747, Q62346 | Gene names | Elk3, Erp, Net | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-3 (ETS-related protein NET) (ETS- related protein ERP). | |||||
|
ELF5_MOUSE
|
||||||
| NC score | 0.822654 (rank : 17) | θ value | 9.90251e-15 (rank : 39) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VDK3, Q921H5, Q9Z2K6 | Gene names | Elf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5). | |||||
|
ELF5_HUMAN
|
||||||
| NC score | 0.821571 (rank : 18) | θ value | 9.90251e-15 (rank : 38) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKW6, O95175, Q8N2K9, Q96QY3, Q9UKW5 | Gene names | ELF5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-5 (E74-like factor 5) (Epithelium-specific Ets transcription factor 2) (ESE-2) (Epithelium- restricted ESE-1-related Ets factor). | |||||
|
ELK4_HUMAN
|
||||||
| NC score | 0.821536 (rank : 19) | θ value | 2.27762e-19 (rank : 9) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
GABPA_MOUSE
|
||||||
| NC score | 0.817948 (rank : 20) | θ value | 1.058e-16 (rank : 28) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q00422 | Gene names | Gabpa, E4tf1a | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha). | |||||
|
GABPA_HUMAN
|
||||||
| NC score | 0.816534 (rank : 21) | θ value | 1.058e-16 (rank : 27) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q06546, Q12939 | Gene names | GABPA, E4TF1A | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein alpha chain (GABP-subunit alpha) (Transcription factor E4TF1-60) (Nuclear respiratory factor 2 subunit alpha). | |||||
|
ETS2_HUMAN
|
||||||
| NC score | 0.813151 (rank : 22) | θ value | 5.60996e-18 (rank : 15) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15036 | Gene names | ETS2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETV6_HUMAN
|
||||||
| NC score | 0.813063 (rank : 23) | θ value | 2.7842e-17 (rank : 20) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P41212, Q9UMF6 | Gene names | ETV6, TEL, TEL1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETV6_MOUSE
|
||||||
| NC score | 0.812640 (rank : 24) | θ value | 2.7842e-17 (rank : 21) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P97360 | Gene names | Etv6, Tel, Tel1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor ETV6 (ETS-related protein Tel1) (Tel) (ETS translocation variant 6). | |||||
|
ETS2_MOUSE
|
||||||
| NC score | 0.811496 (rank : 25) | θ value | 5.60996e-18 (rank : 16) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P15037 | Gene names | Ets2 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-2 protein. | |||||
|
ETS1_HUMAN
|
||||||
| NC score | 0.810500 (rank : 26) | θ value | 1.63225e-17 (rank : 17) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P14921, Q14278, Q16080 | Gene names | ETS1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
ETS1_MOUSE
|
||||||
| NC score | 0.810354 (rank : 27) | θ value | 1.63225e-17 (rank : 18) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P27577, Q61403 | Gene names | Ets1, Ets-1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
FLI1_MOUSE
|
||||||
| NC score | 0.794016 (rank : 28) | θ value | 2.35696e-16 (rank : 29) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P26323 | Gene names | Fli1, Fli-1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Retroviral integration site protein Fli-1). | |||||
|
FLI1_HUMAN
|
||||||
| NC score | 0.792582 (rank : 29) | θ value | 3.07829e-16 (rank : 31) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q01543, Q14319, Q92480, Q9UE07 | Gene names | FLI1 | |||
|
Domain Architecture |

|
|||||
| Description | Friend leukemia integration 1 transcription factor (Fli-1 proto- oncogene) (ERGB transcription factor). | |||||
|
ETV3_MOUSE
|
||||||
| NC score | 0.788824 (rank : 30) | θ value | 6.85773e-16 (rank : 35) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8R4Z4, Q9QZW1 | Gene names | Etv3, Mets, Pe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ERG_HUMAN
|
||||||
| NC score | 0.786126 (rank : 31) | θ value | 1.99529e-15 (rank : 36) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P11308, Q16113 | Gene names | ERG | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG (Transforming protein ERG). | |||||
|
ERG_MOUSE
|
||||||
| NC score | 0.784511 (rank : 32) | θ value | 1.99529e-15 (rank : 37) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P81270, Q8C5L4, Q920K7, Q920K8, Q920K9 | Gene names | Erg, Erg-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ERG. | |||||
|
ETV4_HUMAN
|
||||||
| NC score | 0.783031 (rank : 33) | θ value | 4.29542e-18 (rank : 13) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P43268, Q96AW9 | Gene names | ETV4, E1AF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 4 (Adenovirus E1A enhancer-binding protein) (E1A-F). | |||||
|
ETV4_MOUSE
|
||||||
| NC score | 0.781826 (rank : 34) | θ value | 4.29542e-18 (rank : 14) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P28322 | Gene names | Etv4, Pea-3, Pea3 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 4 (Polyomavirus enhancer activator 3) (Protein PEA3). | |||||
|
ETV1_MOUSE
|
||||||
| NC score | 0.779866 (rank : 35) | θ value | 6.20254e-17 (rank : 26) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P41164 | Gene names | Etv1, Er81, Etsrp81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ETV1_HUMAN
|
||||||
| NC score | 0.779090 (rank : 36) | θ value | 6.20254e-17 (rank : 25) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P50549, O75849, Q9UQ71, Q9Y636 | Gene names | ETV1, ER81 | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 1 (ER81 protein). | |||||
|
ERF_HUMAN
|
||||||
| NC score | 0.777109 (rank : 37) | θ value | 1.86753e-13 (rank : 40) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P50548, Q9UPI7 | Gene names | ERF | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF (Ets2 repressor factor). | |||||
|
ERF_MOUSE
|
||||||
| NC score | 0.776611 (rank : 38) | θ value | 1.86753e-13 (rank : 41) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
ETV5_HUMAN
|
||||||
| NC score | 0.772808 (rank : 39) | θ value | 4.74913e-17 (rank : 23) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P41161 | Gene names | ETV5, ERM | |||
|
Domain Architecture |

|
|||||
| Description | ETS translocation variant 5 (Ets-related protein ERM). | |||||
|
ETV3_HUMAN
|
||||||
| NC score | 0.767076 (rank : 40) | θ value | 6.85773e-16 (rank : 34) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
ETV5_MOUSE
|
||||||
| NC score | 0.756845 (rank : 41) | θ value | 4.74913e-17 (rank : 24) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
SPIC_HUMAN
|
||||||
| NC score | 0.643666 (rank : 42) | θ value | 3.08544e-08 (rank : 42) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8N5J4 | Gene names | SPIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C. | |||||
|
SPI1_MOUSE
|
||||||
| NC score | 0.634228 (rank : 43) | θ value | 1.09739e-05 (rank : 45) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17433, Q99L57 | Gene names | Spi1, Sfpi-1, Sfpi1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein) (SFFV proviral integration 1 protein). | |||||
|
SPI1_HUMAN
|
||||||
| NC score | 0.632085 (rank : 44) | θ value | 1.09739e-05 (rank : 44) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P17947 | Gene names | SPI1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor PU.1 (31 kDa transforming protein). | |||||
|
SPIB_HUMAN
|
||||||
| NC score | 0.622157 (rank : 45) | θ value | 1.87187e-05 (rank : 46) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q01892, Q15359 | Gene names | SPIB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Spi-B. | |||||
|
SPIC_MOUSE
|
||||||
| NC score | 0.612299 (rank : 46) | θ value | 4.45536e-07 (rank : 43) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q6P3D7, Q9Z0Y0 | Gene names | Spic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-C (Pu.1-related factor) (Prf). | |||||
|
SPIB_MOUSE
|
||||||
| NC score | 0.609260 (rank : 47) | θ value | 9.29e-05 (rank : 47) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O35906, O35907, O35909, O55199 | Gene names | Spib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Spi-B. | |||||
|
ARID2_HUMAN
|
||||||
| NC score | 0.078974 (rank : 48) | θ value | 0.00390308 (rank : 49) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
HCFC1_HUMAN
|
||||||
| NC score | 0.077670 (rank : 49) | θ value | 0.00509761 (rank : 50) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.076218 (rank : 50) | θ value | 0.0113563 (rank : 52) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
HCFC1_MOUSE
|
||||||
| NC score | 0.076164 (rank : 51) | θ value | 0.0193708 (rank : 55) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
EMSY_HUMAN
|
||||||
| NC score | 0.072565 (rank : 52) | θ value | 0.21417 (rank : 66) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.070510 (rank : 53) | θ value | 0.0563607 (rank : 58) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
EMSY_MOUSE
|
||||||
| NC score | 0.069838 (rank : 54) | θ value | 1.06291 (rank : 81) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.065690 (rank : 55) | θ value | 0.0563607 (rank : 59) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.060304 (rank : 56) | θ value | 1.06291 (rank : 82) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.054747 (rank : 57) | θ value | 0.47712 (rank : 76) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.054311 (rank : 58) | θ value | 0.00175202 (rank : 48) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.051314 (rank : 59) | θ value | 0.813845 (rank : 80) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
TF2AA_HUMAN
|
||||||
| NC score | 0.048875 (rank : 60) | θ value | 0.0148317 (rank : 54) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P52655 | Gene names | GTF2A1, TF2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIA subunit 1 (General transcription factor IIA1) (TFIIA-42) (TFIIAL) [Contains: Transcription initiation factor IIA alpha chain (TFIIA p35 subunit); Transcription initiation factor IIA beta chain (TFIIA p19 subunit)]. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.048725 (rank : 61) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CN004_MOUSE
|
||||||
| NC score | 0.046099 (rank : 62) | θ value | 0.21417 (rank : 65) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K3X4, Q3USE5, Q8BUS1, Q8C011 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4 homolog. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.045621 (rank : 63) | θ value | 1.38821 (rank : 88) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
RFX1_HUMAN
|
||||||
| NC score | 0.045239 (rank : 64) | θ value | 0.00869519 (rank : 51) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P22670 | Gene names | RFX1 | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II regulatory factor RFX1 (RFX) (Enhancer factor C) (EF-C). | |||||
|
TIMD1_HUMAN
|
||||||
| NC score | 0.044782 (rank : 65) | θ value | 1.81305 (rank : 99) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96D42, O43656 | Gene names | HAVCR1, TIM1, TIMD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatitis A virus cellular receptor 1 precursor (HAVcr-1) (T cell immunoglobulin and mucin domain-containing protein 1) (TIMD-1) (T cell membrane protein 1) (TIM-1) (TIM). | |||||
|
ATF6A_HUMAN
|
||||||
| NC score | 0.044387 (rank : 66) | θ value | 0.365318 (rank : 71) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P18850, O15139, Q5VW62, Q6IPB5, Q9UEC9 | Gene names | ATF6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 alpha (Activating transcription factor 6 alpha) (ATF6-alpha). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.044062 (rank : 67) | θ value | 1.38821 (rank : 87) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
CN004_HUMAN
|
||||||
| NC score | 0.043503 (rank : 68) | θ value | 0.47712 (rank : 75) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H1B7, Q8NDQ2, Q96JG2, Q9H3I7 | Gene names | C14orf4, KIAA1865 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4. | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.042952 (rank : 69) | θ value | 1.06291 (rank : 84) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
RFX1_MOUSE
|
||||||
| NC score | 0.041025 (rank : 70) | θ value | 1.06291 (rank : 83) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P48377 | Gene names | Rfx1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFX1. | |||||
|
ATF7_HUMAN
|
||||||
| NC score | 0.040050 (rank : 71) | θ value | 0.279714 (rank : 68) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
ATF6B_MOUSE
|
||||||
| NC score | 0.038847 (rank : 72) | θ value | 0.21417 (rank : 64) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35451 | Gene names | Crebl1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp). | |||||
|
SRBP2_HUMAN
|
||||||
| NC score | 0.037412 (rank : 73) | θ value | 0.0330416 (rank : 57) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q12772, Q9UH04 | Gene names | SREBF2, SREBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 2 (SREBP-2) (Sterol regulatory element-binding transcription factor 2). | |||||
|
ATF7_MOUSE
|
||||||
| NC score | 0.036747 (rank : 74) | θ value | 0.365318 (rank : 72) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8R0S1 | Gene names | Atf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
GP112_HUMAN
|
||||||
| NC score | 0.031786 (rank : 75) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
MCA1_MOUSE
|
||||||
| NC score | 0.028287 (rank : 76) | θ value | 1.81305 (rank : 95) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P31230, Q60659 | Gene names | Scye1, Emap2 | |||
|
Domain Architecture |

|
|||||
| Description | Multisynthetase complex auxiliary component p43 [Contains: Endothelial monocyte-activating polypeptide 2 (EMAP-II) (Small inducible cytokine subfamily E member 1)]. | |||||
|
SSXT_HUMAN
|
||||||
| NC score | 0.025395 (rank : 77) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15532, Q16404, Q9BXC6 | Gene names | SS18, SSXT, SYT | |||
|
Domain Architecture |

|
|||||
| Description | SSXT protein (Synovial sarcoma, translocated to X chromosome) (SYT protein). | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.025258 (rank : 78) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.024869 (rank : 79) | θ value | 0.813845 (rank : 79) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SSXT_MOUSE
|
||||||
| NC score | 0.023110 (rank : 80) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62280 | Gene names | Ss18, Ssxt, Syt | |||
|
Domain Architecture |

|
|||||
| Description | SSXT protein (SYT protein) (Synovial sarcoma-associated Ss18-alpha). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.022781 (rank : 81) | θ value | 1.81305 (rank : 97) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.022560 (rank : 82) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
EYA3_HUMAN
|
||||||
| NC score | 0.020747 (rank : 83) | θ value | 0.125558 (rank : 62) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99504, O95463, Q99813 | Gene names | EYA3 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 3 (EC 3.1.3.48). | |||||
|
ARI4A_HUMAN
|
||||||
| NC score | 0.019035 (rank : 84) | θ value | 1.81305 (rank : 90) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
ARHGC_MOUSE
|
||||||
| NC score | 0.018517 (rank : 85) | θ value | 0.279714 (rank : 67) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R4H2, Q80U18 | Gene names | Arhgef12, Kiaa0382, Larg | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
SIX4_HUMAN
|
||||||
| NC score | 0.018171 (rank : 86) | θ value | 0.0736092 (rank : 61) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UIU6 | Gene names | SIX4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX4 (Sine oculis homeobox homolog 4). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.017707 (rank : 87) | θ value | 0.0252991 (rank : 56) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TAF6_HUMAN
|
||||||
| NC score | 0.017573 (rank : 88) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49848 | Gene names | TAF6, TAF2E, TAFII70 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 6 (Transcription initiation factor TFIID 70 kDa subunit) (TAF(II)70) (TAFII-70) (TAFII- 80) (TAFII80). | |||||
|
SDC1_HUMAN
|
||||||
| NC score | 0.016041 (rank : 89) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18827, Q96HB7 | Gene names | SDC1, SDC | |||
|
Domain Architecture |

|
|||||
| Description | Syndecan-1 precursor (SYND1) (CD138 antigen). | |||||
|
EWS_HUMAN
|
||||||
| NC score | 0.015309 (rank : 90) | θ value | 1.81305 (rank : 91) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q01844, Q92635 | Gene names | EWSR1, EWS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS (EWS oncogene) (Ewing sarcoma breakpoint region 1 protein). | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.015109 (rank : 91) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
RP1_HUMAN
|
||||||
| NC score | 0.015046 (rank : 92) | θ value | 1.38821 (rank : 89) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56715 | Gene names | RP1, ORP1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein) (Retinitis pigmentosa 1 protein). | |||||
|
RIOK1_HUMAN
|
||||||
| NC score | 0.014757 (rank : 93) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BRS2, Q8NDC8, Q96NV9 | Gene names | RIOK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO1 (EC 2.7.11.1) (RIO kinase 1). | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.014626 (rank : 94) | θ value | 0.0736092 (rank : 60) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
ARHGC_HUMAN
|
||||||
| NC score | 0.014525 (rank : 95) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZN5, O15086 | Gene names | ARHGEF12, KIAA0382, LARG | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
AFF2_HUMAN
|
||||||
| NC score | 0.012839 (rank : 96) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P51816, O43786, O60215, P78407, Q13521, Q14323, Q9UNA5 | Gene names | AFF2, FMR2, OX19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation 2 protein) (Protein FMR-2) (FMR2P) (Protein Ox19) (Fragile X E mental retardation syndrome protein). | |||||
|
FGD1_HUMAN
|
||||||
| NC score | 0.012376 (rank : 97) | θ value | 1.81305 (rank : 92) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
YTHD3_HUMAN
|
||||||
| NC score | 0.011898 (rank : 98) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z739, Q63Z37, Q659A3 | Gene names | YTHDF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain family protein 3. | |||||
|
AT8B3_HUMAN
|
||||||
| NC score | 0.011718 (rank : 99) | θ value | 0.365318 (rank : 70) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60423, Q8IVB8, Q8N4Y8, Q96M22 | Gene names | ATP8B3, ATP1K, FOS37502_2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase IK (EC 3.6.3.1) (ATPase class I type 8B member 3). | |||||
|
ZHX1_MOUSE
|
||||||
| NC score | 0.011611 (rank : 100) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P70121, Q8BQ68, Q8C6T4, Q8CJG3 | Gene names | Zhx1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
AAK1_MOUSE
|
||||||
| NC score | 0.011431 (rank : 101) | θ value | 0.0148317 (rank : 53) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
HTF4_HUMAN
|
||||||
| NC score | 0.011342 (rank : 102) | θ value | 1.81305 (rank : 94) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99081 | Gene names | TCF12, HEB, HTF4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 12 (Transcription factor HTF-4) (E-box-binding protein) (DNA-binding protein HTF4). | |||||
|
ARVC_MOUSE
|
||||||
| NC score | 0.010897 (rank : 103) | θ value | 2.36792 (rank : 100) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P98203, Q6PGJ6, Q8BQ36, Q8BRF2, Q8C3U7, Q924L2, Q924L3, Q924L4, Q924L5 | Gene names | Arvcf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome homolog. | |||||
|
MBNL_MOUSE
|
||||||
| NC score | 0.010631 (rank : 104) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JKP5 | Gene names | Mbnl1, Exp, Mbnl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscleblind-like protein (Triplet-expansion RNA-binding protein). | |||||
|
ZHX1_HUMAN
|
||||||
| NC score | 0.010630 (rank : 105) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKY1, Q8IWD8 | Gene names | ZHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.010548 (rank : 106) | θ value | 0.47712 (rank : 77) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
BOC_HUMAN
|
||||||
| NC score | 0.010339 (rank : 107) | θ value | 0.62314 (rank : 78) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BWV1, Q6UXJ5, Q8N2P7, Q8NF26 | Gene names | BOC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brother of CDO precursor (Protein BOC). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.010157 (rank : 108) | θ value | 0.21417 (rank : 63) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
HOMEZ_HUMAN
|
||||||
| NC score | 0.010057 (rank : 109) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IX15, Q6P049, Q86XB6, Q9P2A5 | Gene names | HOMEZ, KIAA1443 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox and leucine zipper protein Homez (Homeodomain leucine zipper- containing factor). | |||||
|
ARVC_HUMAN
|
||||||
| NC score | 0.009906 (rank : 110) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00192 | Gene names | ARVCF | |||
|
Domain Architecture |

|
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome. | |||||
|
BOC_MOUSE
|
||||||
| NC score | 0.009851 (rank : 111) | θ value | 2.36792 (rank : 101) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6AZB0, Q6KAM5, Q6P5H3, Q7TMJ3, Q8CE73, Q8CE91, Q8R377, Q923W7 | Gene names | Boc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brother of CDO precursor (Protein BOC). | |||||
|
EWS_MOUSE
|
||||||
| NC score | 0.009804 (rank : 112) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61545 | Gene names | Ewsr1, Ews, Ewsh | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS. | |||||
|
ZYX_HUMAN
|
||||||
| NC score | 0.007974 (rank : 113) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15942 | Gene names | ZYX | |||
|
Domain Architecture |

|
|||||
| Description | Zyxin (Zyxin-2). | |||||
|
MOR2A_MOUSE
|
||||||
| NC score | 0.007971 (rank : 114) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q69ZX6, Q5QNQ7, Q6P547 | Gene names | Morc2a, Kiaa0852, Zcwcc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 2A (Zinc finger CW-type coiled-coil domain protein 1). | |||||
|
GPR98_MOUSE
|
||||||
| NC score | 0.007731 (rank : 115) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VHN7, Q6ZQ69, Q810D2, Q810D3, Q91ZS0, Q91ZS1 | Gene names | Gpr98, Kiaa0686, Mass1, Vlgr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1) (Very large G-protein coupled receptor 1) (Neurepin). | |||||
|
WNK1_HUMAN
|
||||||
| NC score | 0.007559 (rank : 116) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
XRN1_HUMAN
|
||||||
| NC score | 0.006859 (rank : 117) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IZH2, Q4G0S3, Q68D88, Q6AI24, Q6MZS8, Q86WS7, Q8N8U4, Q9UF39 | Gene names | XRN1, SEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-3' exoribonuclease 1 (EC 3.1.11.-) (Strand-exchange protein 1 homolog). | |||||
|
NPAS2_HUMAN
|
||||||
| NC score | 0.006854 (rank : 118) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99743, Q99629 | Gene names | NPAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 2 (Neuronal PAS2) (Member of PAS protein 4) (MOP4). | |||||
|
NLGN3_MOUSE
|
||||||
| NC score | 0.006639 (rank : 119) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BYM5, Q8BXR4 | Gene names | Nlgn3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
|
SUHW3_MOUSE
|
||||||
| NC score | 0.006506 (rank : 120) | θ value | 0.365318 (rank : 74) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 697 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P3Y5, Q6ZPM2, Q8K145 | Gene names | Suhw3, Kiaa1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 3. | |||||
|
POGZ_MOUSE
|
||||||
| NC score | 0.006207 (rank : 121) | θ value | 1.81305 (rank : 96) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BZH4, Q4VA94, Q80TZ8, Q8C0K1, Q8K294 | Gene names | Pogz, Kiaa0461 | |||
|
Domain Architecture |

|
|||||
| Description | Pogo transposable element with ZNF domain. | |||||
|
TSC2_MOUSE
|
||||||
| NC score | 0.005867 (rank : 122) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61037, P97723, P97724, P97725, P97727, Q61007, Q61008, Q9WUF6 | Gene names | Tsc2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 homolog protein). | |||||
|
SP4_MOUSE
|
||||||
| NC score | 0.005649 (rank : 123) | θ value | 0.279714 (rank : 69) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62445 | Gene names | Sp4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp4. | |||||
|
IL1R1_HUMAN
|
||||||
| NC score | 0.004930 (rank : 124) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P14778 | Gene names | IL1R1, IL1R, IL1RA, IL1RT1 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-1 receptor type I precursor (IL-1R-1) (IL-1RT1) (IL-1R- alpha) (p80) (CD121a antigen). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.004546 (rank : 125) | θ value | 1.38821 (rank : 86) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.003931 (rank : 126) | θ value | 1.81305 (rank : 93) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
SUHW1_HUMAN
|
||||||
| NC score | 0.003804 (rank : 127) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 648 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P59817 | Gene names | SUHW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 1 (3'OY11.1). | |||||
|
DGKH_HUMAN
|
||||||
| NC score | 0.003638 (rank : 128) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86XP1, Q5VZW0, Q6PI56, Q86XP2, Q8N3N0, Q8N7J9 | Gene names | DGKH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Diacylglycerol kinase eta (EC 2.7.1.107) (Diglyceride kinase eta) (DGK-eta) (DAG kinase eta). | |||||
|
DUS16_HUMAN
|
||||||
| NC score | 0.003543 (rank : 129) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BY84, Q9C0G3 | Gene names | DUSP16, KIAA1700, MKP7 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 16 (EC 3.1.3.48) (EC 3.1.3.16) (Mitogen-activated protein kinase phosphatase 7) (MAP kinase phosphatase 7) (MKP-7). | |||||
|
SP4_HUMAN
|
||||||
| NC score | 0.003517 (rank : 130) | θ value | 0.365318 (rank : 73) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02446, O60402 | Gene names | SP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp4 (SPR-1). | |||||
|
WDR36_HUMAN
|
||||||
| NC score | 0.002850 (rank : 131) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NI36, Q8N1Q2 | Gene names | WDR36 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 36 (T-cell activation WD repeat-containing protein) (TA-WDRP). | |||||
|
MSX2_HUMAN
|
||||||
| NC score | 0.002364 (rank : 132) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35548 | Gene names | MSX2, HOX8 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein MSX-2 (Hox-8). | |||||
|
SP2_HUMAN
|
||||||
| NC score | 0.001649 (rank : 133) | θ value | 1.81305 (rank : 98) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q02086 | Gene names | SP2, KIAA0048 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp2. | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.001154 (rank : 134) | θ value | 1.06291 (rank : 85) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
MSX1_HUMAN
|
||||||
| NC score | 0.000634 (rank : 135) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P28360, Q96NY4 | Gene names | MSX1, HOX7 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein MSX-1 (Msh homeobox 1-like protein) (Hox-7). | |||||
|
E2AK1_MOUSE
|
||||||
| NC score | -0.000693 (rank : 136) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z2R9, Q8C024, Q8K123, Q9CTP5, Q9D601 | Gene names | Eif2ak1, Hri | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2-alpha kinase 1 (EC 2.7.11.1) (Heme-regulated eukaryotic initiation factor eIF-2-alpha kinase) (Heme-regulated inhibitor) (Heme-controlled repressor) (HCR) (Hemin-sensitive initiation factor 2-alpha kinase). | |||||
|
MAST1_HUMAN
|
||||||
| NC score | -0.001303 (rank : 137) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
SLK_MOUSE
|
||||||
| NC score | -0.002880 (rank : 138) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
SLK_HUMAN
|
||||||
| NC score | -0.003891 (rank : 139) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 139 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||