Please be patient as the page loads
|
NLGN3_MOUSE
|
||||||
| SwissProt Accessions | Q8BYM5, Q8BXR4 | Gene names | Nlgn3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NLGN1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.996875 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N2Q7, Q9UPT2 | Gene names | NLGN1, KIAA1070 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-1 precursor. | |||||
|
NLGN1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.996909 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99K10 | Gene names | Nlgn1, Kiaa1070 | |||
|
Domain Architecture |
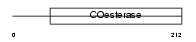
|
|||||
| Description | Neuroligin-1 precursor. | |||||
|
NLGN2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.994902 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NFZ4, Q9P2I1 | Gene names | NLGN2, KIAA1366 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-2 precursor. | |||||
|
NLGN2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.995058 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q69ZK9, Q5F288 | Gene names | Nlgn2, Kiaa1366 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroligin-2 precursor. | |||||
|
NLGN3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.999709 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NZ94, Q8NCD0, Q9NZ95, Q9NZ96, Q9NZ97, Q9P248 | Gene names | NLGN3, KIAA1480, NL3 | |||
|
Domain Architecture |
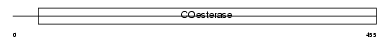
|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
|
NLGN3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BYM5, Q8BXR4 | Gene names | Nlgn3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
|
NLGNX_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.997770 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N0W4, Q6UX10, Q9ULG0 | Gene names | NLGN4X, KIAA1260, NLGN4 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-4, X-linked precursor (Neuroligin X) (HNLX). | |||||
|
NLGNY_HUMAN
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.997654 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8NFZ3, Q7Z3T5, Q9Y2F8 | Gene names | NLGN4Y, KIAA0951 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-4, Y-linked precursor (Neuroligin Y). | |||||
|
ACES_MOUSE
|
||||||
| θ value | 5.32145e-85 (rank : 9) | NC score | 0.948071 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P21836 | Gene names | Ache | |||
|
Domain Architecture |

|
|||||
| Description | Acetylcholinesterase precursor (EC 3.1.1.7) (AChE). | |||||
|
ACES_HUMAN
|
||||||
| θ value | 2.02215e-84 (rank : 10) | NC score | 0.947251 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P22303, Q16169, Q86TM9, Q9BXP7 | Gene names | ACHE | |||
|
Domain Architecture |

|
|||||
| Description | Acetylcholinesterase precursor (EC 3.1.1.7) (AChE). | |||||
|
CHLE_HUMAN
|
||||||
| θ value | 5.69867e-79 (rank : 11) | NC score | 0.945462 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P06276 | Gene names | BCHE, CHE1 | |||
|
Domain Architecture |

|
|||||
| Description | Cholinesterase precursor (EC 3.1.1.8) (Acylcholine acylhydrolase) (Choline esterase II) (Butyrylcholine esterase) (Pseudocholinesterase). | |||||
|
CHLE_MOUSE
|
||||||
| θ value | 2.1655e-78 (rank : 12) | NC score | 0.945043 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q03311 | Gene names | Bche | |||
|
Domain Architecture |

|
|||||
| Description | Cholinesterase precursor (EC 3.1.1.8) (Acylcholine acylhydrolase) (Choline esterase II) (Butyrylcholine esterase) (Pseudocholinesterase). | |||||
|
EST2_HUMAN
|
||||||
| θ value | 8.22888e-78 (rank : 13) | NC score | 0.946899 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O00748, Q16859, Q5MAB8, Q7Z366, Q8IUP4, Q8TCP8 | Gene names | CES2, ICE | |||
|
Domain Architecture |

|
|||||
| Description | Carboxylesterase 2 precursor (EC 3.1.1.1) (CE-2) (hCE-2). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 1.8332e-77 (rank : 14) | NC score | 0.722972 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
EST1_HUMAN
|
||||||
| θ value | 5.33382e-77 (rank : 15) | NC score | 0.931574 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P23141, Q00015, Q13657, Q14062, Q16737, Q16788, Q86UK2, Q96EE8, Q9UK77, Q9ULY2 | Gene names | CES1, CES2, SES1 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 1 precursor (EC 3.1.1.1) (Acyl coenzyme A:cholesterol acyltransferase) (ACAT) (Monocyte/macrophage serine esterase) (HMSE) (Serine esterase 1) (Brain carboxylesterase hBr1) (Triacylglycerol hydrolase) (TGH) (Egasyn). | |||||
|
CEL_MOUSE
|
||||||
| θ value | 1.71583e-75 (rank : 16) | NC score | 0.952184 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q64285 | Gene names | Cel, Lip1 | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CES3_MOUSE
|
||||||
| θ value | 4.99229e-75 (rank : 17) | NC score | 0.929683 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VCT4, Q91ZV9, Q924V8, Q9ULY1 | Gene names | Ces3, Ces1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxylesterase 3 precursor (EC 3.1.1.1) (Triacylglycerol hydrolase) (TGH). | |||||
|
EST31_MOUSE
|
||||||
| θ value | 2.73936e-73 (rank : 18) | NC score | 0.946308 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q63880 | Gene names | Es31 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 31 precursor (EC 3.1.1.1) (ES-Male) (Esterase- 31). | |||||
|
EST22_MOUSE
|
||||||
| θ value | 2.56397e-71 (rank : 19) | NC score | 0.931006 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q64176 | Gene names | Es22 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 22 precursor (EC 3.1.1.1) (Egasyn) (Esterase- 22) (Es-22). | |||||
|
EST1_MOUSE
|
||||||
| θ value | 1.83746e-69 (rank : 20) | NC score | 0.926832 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VCC2, O55136 | Gene names | Ces1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Liver carboxylesterase 1 precursor (EC 3.1.1.1) (Acyl coenzyme A:cholesterol acyltransferase) (ES-x). | |||||
|
ESTN_MOUSE
|
||||||
| θ value | 2.03155e-68 (rank : 21) | NC score | 0.929073 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P23953, O54936, P11374, Q8K125 | Gene names | Es1 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase N precursor (EC 3.1.1.1) (PES-N) (Lung surfactant convertase). | |||||
|
THYG_MOUSE
|
||||||
| θ value | 7.01481e-53 (rank : 22) | NC score | 0.810189 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08710, O88590, Q9QWY7 | Gene names | Tg, Tgn | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
THYG_HUMAN
|
||||||
| θ value | 3.85808e-43 (rank : 23) | NC score | 0.791138 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P01266, O15274, O43899, Q15593, Q15948, Q9NYR1, Q9NYR2, Q9UMZ0, Q9UNY3 | Gene names | TG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.011958 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
SMR3B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.102478 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P02814, Q9UBN0, Q9UCT0 | Gene names | SMR3B, PBII, PROL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog B precursor (Proline-rich protein 3) (Proline-rich peptide P-B) [Contains: Peptide P-A; Peptide D1A]. | |||||
|
RC3H1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.017944 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5TC82, Q5W180, Q5W181, Q8IVE6, Q8N9V1 | Gene names | RC3H1, KIAA2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.021504 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
SMRD2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.011426 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99JR8 | Gene names | Smarcd2, Baf60b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 2 (60 kDa BRG-1/Brm-associated factor subunit B) (BRG1-associated factor 60B). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | -0.000538 (rank : 49) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
IQEC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.010560 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.010506 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
TTBK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.003334 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6IQ55, O94932, Q6ZN52, Q8IVV1 | Gene names | TTBK2, KIAA0847 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2 (EC 2.7.11.1). | |||||
|
ELF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.006664 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.006639 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
OMD_MOUSE
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.003043 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35103 | Gene names | Omd | |||
|
Domain Architecture |
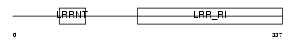
|
|||||
| Description | Osteomodulin precursor (Osteoadherin) (OSAD) (Keratan sulfate proteoglycan osteomodulin) (KSPG osteomodulin). | |||||
|
BUB1B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.006940 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z1S0 | Gene names | Bub1b, Mad3l | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta (EC 2.7.11.1) (MAD3/BUB1-related protein kinase) (Mitotic checkpoint kinase MAD3L). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.006217 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
DNJA3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.005816 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96EY1, O75472, Q8WUJ6, Q8WXJ3, Q96D76, Q96IV1, Q9NYH8 | Gene names | DNAJA3, TID1 | |||
|
Domain Architecture |
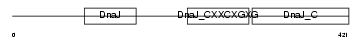
|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (hTid-1). | |||||
|
HGFA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.000117 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q04756, Q14726 | Gene names | HGFAC | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
MYEOV_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.030503 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96EZ4, Q9UGN6, Q9UGN7 | Gene names | MYEOV, OCIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myeloma overexpressed gene protein (Oncogene in multiple myeloma). | |||||
|
TTBK2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.002494 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3UVR3, Q3TSR6, Q3UFW0, Q571D1, Q8BKA4, Q924U8 | Gene names | Ttbk2, Kiaa0847, Ttbk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2. | |||||
|
ANR47_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.003916 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NY19, Q6NZI1, Q6ZQR3, Q8IUV2 | Gene names | ANKRD47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 47. | |||||
|
CASC3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.009168 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
MK04_HUMAN
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | -0.002394 (rank : 50) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P31152 | Gene names | MAPK4, ERK4, PRKM4 | |||
|
Domain Architecture |
|
|||||
| Description | Mitogen-activated protein kinase 4 (EC 2.7.11.24) (Extracellular signal-regulated kinase 4) (ERK-4) (MAP kinase isoform p63) (p63- MAPK). | |||||
|
CCP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.009391 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53805, O00582, O00583, Q96R03, Q9BU69, Q9UF15, Q9UME4 | Gene names | DSCR1, ADAPT78, CSP1, DSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcipressin-1 (Down syndrome critical region protein 1) (Myocyte- enriched calcineurin-interacting protein 1) (MCIP1) (Adapt78). | |||||
|
CCP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.009317 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHG6, Q91WQ4, Q9JK50, Q9JK51, Q9JKK2, Q9JKK3 | Gene names | Dscr1 | |||
|
Domain Architecture |
|
|||||
| Description | Calcipressin-1 (Down syndrome critical region protein 1 homolog) (Myocyte-enriched calcineurin-interacting protein 1) (MCIP1). | |||||
|
DIAP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.017665 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
FA5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.003399 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P12259, Q14285, Q6UPU6 | Gene names | F5 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor V precursor (Activated protein C cofactor) [Contains: Coagulation factor V heavy chain; Coagulation factor V light chain]. | |||||
|
AAAD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.206002 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P22760, Q8N1A9 | Gene names | AADAC, DAC | |||
|
Domain Architecture |

|
|||||
| Description | Arylacetamide deacetylase (EC 3.1.1.-) (AADAC). | |||||
|
AAAD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.115782 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99PG0, Q8VCF2 | Gene names | Aadac, Aada | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylacetamide deacetylase (EC 3.1.1.-) (AADAC). | |||||
|
NLGN3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BYM5, Q8BXR4 | Gene names | Nlgn3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
|
NLGN3_HUMAN
|
||||||
| NC score | 0.999709 (rank : 2) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NZ94, Q8NCD0, Q9NZ95, Q9NZ96, Q9NZ97, Q9P248 | Gene names | NLGN3, KIAA1480, NL3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
|
NLGNX_HUMAN
|
||||||
| NC score | 0.997770 (rank : 3) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N0W4, Q6UX10, Q9ULG0 | Gene names | NLGN4X, KIAA1260, NLGN4 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-4, X-linked precursor (Neuroligin X) (HNLX). | |||||
|
NLGNY_HUMAN
|
||||||
| NC score | 0.997654 (rank : 4) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8NFZ3, Q7Z3T5, Q9Y2F8 | Gene names | NLGN4Y, KIAA0951 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-4, Y-linked precursor (Neuroligin Y). | |||||
|
NLGN1_MOUSE
|
||||||
| NC score | 0.996909 (rank : 5) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99K10 | Gene names | Nlgn1, Kiaa1070 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-1 precursor. | |||||
|
NLGN1_HUMAN
|
||||||
| NC score | 0.996875 (rank : 6) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N2Q7, Q9UPT2 | Gene names | NLGN1, KIAA1070 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-1 precursor. | |||||
|
NLGN2_MOUSE
|
||||||
| NC score | 0.995058 (rank : 7) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q69ZK9, Q5F288 | Gene names | Nlgn2, Kiaa1366 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroligin-2 precursor. | |||||
|
NLGN2_HUMAN
|
||||||
| NC score | 0.994902 (rank : 8) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NFZ4, Q9P2I1 | Gene names | NLGN2, KIAA1366 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-2 precursor. | |||||
|
CEL_MOUSE
|
||||||
| NC score | 0.952184 (rank : 9) | θ value | 1.71583e-75 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q64285 | Gene names | Cel, Lip1 | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
ACES_MOUSE
|
||||||
| NC score | 0.948071 (rank : 10) | θ value | 5.32145e-85 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P21836 | Gene names | Ache | |||
|
Domain Architecture |

|
|||||
| Description | Acetylcholinesterase precursor (EC 3.1.1.7) (AChE). | |||||
|
ACES_HUMAN
|
||||||
| NC score | 0.947251 (rank : 11) | θ value | 2.02215e-84 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P22303, Q16169, Q86TM9, Q9BXP7 | Gene names | ACHE | |||
|
Domain Architecture |

|
|||||
| Description | Acetylcholinesterase precursor (EC 3.1.1.7) (AChE). | |||||
|
EST2_HUMAN
|
||||||
| NC score | 0.946899 (rank : 12) | θ value | 8.22888e-78 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O00748, Q16859, Q5MAB8, Q7Z366, Q8IUP4, Q8TCP8 | Gene names | CES2, ICE | |||
|
Domain Architecture |

|
|||||
| Description | Carboxylesterase 2 precursor (EC 3.1.1.1) (CE-2) (hCE-2). | |||||
|
EST31_MOUSE
|
||||||
| NC score | 0.946308 (rank : 13) | θ value | 2.73936e-73 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q63880 | Gene names | Es31 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 31 precursor (EC 3.1.1.1) (ES-Male) (Esterase- 31). | |||||
|
CHLE_HUMAN
|
||||||
| NC score | 0.945462 (rank : 14) | θ value | 5.69867e-79 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P06276 | Gene names | BCHE, CHE1 | |||
|
Domain Architecture |

|
|||||
| Description | Cholinesterase precursor (EC 3.1.1.8) (Acylcholine acylhydrolase) (Choline esterase II) (Butyrylcholine esterase) (Pseudocholinesterase). | |||||
|
CHLE_MOUSE
|
||||||
| NC score | 0.945043 (rank : 15) | θ value | 2.1655e-78 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q03311 | Gene names | Bche | |||
|
Domain Architecture |

|
|||||
| Description | Cholinesterase precursor (EC 3.1.1.8) (Acylcholine acylhydrolase) (Choline esterase II) (Butyrylcholine esterase) (Pseudocholinesterase). | |||||
|
EST1_HUMAN
|
||||||
| NC score | 0.931574 (rank : 16) | θ value | 5.33382e-77 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P23141, Q00015, Q13657, Q14062, Q16737, Q16788, Q86UK2, Q96EE8, Q9UK77, Q9ULY2 | Gene names | CES1, CES2, SES1 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 1 precursor (EC 3.1.1.1) (Acyl coenzyme A:cholesterol acyltransferase) (ACAT) (Monocyte/macrophage serine esterase) (HMSE) (Serine esterase 1) (Brain carboxylesterase hBr1) (Triacylglycerol hydrolase) (TGH) (Egasyn). | |||||
|
EST22_MOUSE
|
||||||
| NC score | 0.931006 (rank : 17) | θ value | 2.56397e-71 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q64176 | Gene names | Es22 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 22 precursor (EC 3.1.1.1) (Egasyn) (Esterase- 22) (Es-22). | |||||
|
CES3_MOUSE
|
||||||
| NC score | 0.929683 (rank : 18) | θ value | 4.99229e-75 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VCT4, Q91ZV9, Q924V8, Q9ULY1 | Gene names | Ces3, Ces1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxylesterase 3 precursor (EC 3.1.1.1) (Triacylglycerol hydrolase) (TGH). | |||||
|
ESTN_MOUSE
|
||||||
| NC score | 0.929073 (rank : 19) | θ value | 2.03155e-68 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P23953, O54936, P11374, Q8K125 | Gene names | Es1 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase N precursor (EC 3.1.1.1) (PES-N) (Lung surfactant convertase). | |||||
|
EST1_MOUSE
|
||||||
| NC score | 0.926832 (rank : 20) | θ value | 1.83746e-69 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VCC2, O55136 | Gene names | Ces1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Liver carboxylesterase 1 precursor (EC 3.1.1.1) (Acyl coenzyme A:cholesterol acyltransferase) (ES-x). | |||||
|
THYG_MOUSE
|
||||||
| NC score | 0.810189 (rank : 21) | θ value | 7.01481e-53 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08710, O88590, Q9QWY7 | Gene names | Tg, Tgn | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
THYG_HUMAN
|
||||||
| NC score | 0.791138 (rank : 22) | θ value | 3.85808e-43 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P01266, O15274, O43899, Q15593, Q15948, Q9NYR1, Q9NYR2, Q9UMZ0, Q9UNY3 | Gene names | TG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.722972 (rank : 23) | θ value | 1.8332e-77 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
AAAD_HUMAN
|
||||||
| NC score | 0.206002 (rank : 24) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P22760, Q8N1A9 | Gene names | AADAC, DAC | |||
|
Domain Architecture |

|
|||||
| Description | Arylacetamide deacetylase (EC 3.1.1.-) (AADAC). | |||||
|
AAAD_MOUSE
|
||||||
| NC score | 0.115782 (rank : 25) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99PG0, Q8VCF2 | Gene names | Aadac, Aada | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylacetamide deacetylase (EC 3.1.1.-) (AADAC). | |||||
|
SMR3B_HUMAN
|
||||||
| NC score | 0.102478 (rank : 26) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P02814, Q9UBN0, Q9UCT0 | Gene names | SMR3B, PBII, PROL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog B precursor (Proline-rich protein 3) (Proline-rich peptide P-B) [Contains: Peptide P-A; Peptide D1A]. | |||||
|
MYEOV_HUMAN
|
||||||
| NC score | 0.030503 (rank : 27) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96EZ4, Q9UGN6, Q9UGN7 | Gene names | MYEOV, OCIM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myeloma overexpressed gene protein (Oncogene in multiple myeloma). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.021504 (rank : 28) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
RC3H1_HUMAN
|
||||||
| NC score | 0.017944 (rank : 29) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5TC82, Q5W180, Q5W181, Q8IVE6, Q8N9V1 | Gene names | RC3H1, KIAA2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1). | |||||
|
DIAP1_HUMAN
|
||||||
| NC score | 0.017665 (rank : 30) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.011958 (rank : 31) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
SMRD2_MOUSE
|
||||||
| NC score | 0.011426 (rank : 32) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99JR8 | Gene names | Smarcd2, Baf60b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 2 (60 kDa BRG-1/Brm-associated factor subunit B) (BRG1-associated factor 60B). | |||||
|
IQEC2_HUMAN
|
||||||
| NC score | 0.010560 (rank : 33) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_MOUSE
|
||||||
| NC score | 0.010506 (rank : 34) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
CCP1_HUMAN
|
||||||
| NC score | 0.009391 (rank : 35) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53805, O00582, O00583, Q96R03, Q9BU69, Q9UF15, Q9UME4 | Gene names | DSCR1, ADAPT78, CSP1, DSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcipressin-1 (Down syndrome critical region protein 1) (Myocyte- enriched calcineurin-interacting protein 1) (MCIP1) (Adapt78). | |||||
|
CCP1_MOUSE
|
||||||
| NC score | 0.009317 (rank : 36) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHG6, Q91WQ4, Q9JK50, Q9JK51, Q9JKK2, Q9JKK3 | Gene names | Dscr1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcipressin-1 (Down syndrome critical region protein 1 homolog) (Myocyte-enriched calcineurin-interacting protein 1) (MCIP1). | |||||
|
CASC3_MOUSE
|
||||||
| NC score | 0.009168 (rank : 37) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
BUB1B_MOUSE
|
||||||
| NC score | 0.006940 (rank : 38) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z1S0 | Gene names | Bub1b, Mad3l | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta (EC 2.7.11.1) (MAD3/BUB1-related protein kinase) (Mitotic checkpoint kinase MAD3L). | |||||
|
ELF1_HUMAN
|
||||||
| NC score | 0.006664 (rank : 39) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32519, Q9UDE1 | Gene names | ELF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
ELF1_MOUSE
|
||||||
| NC score | 0.006639 (rank : 40) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.006217 (rank : 41) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
DNJA3_HUMAN
|
||||||
| NC score | 0.005816 (rank : 42) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96EY1, O75472, Q8WUJ6, Q8WXJ3, Q96D76, Q96IV1, Q9NYH8 | Gene names | DNAJA3, TID1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (hTid-1). | |||||
|
ANR47_HUMAN
|
||||||
| NC score | 0.003916 (rank : 43) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NY19, Q6NZI1, Q6ZQR3, Q8IUV2 | Gene names | ANKRD47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 47. | |||||
|
FA5_HUMAN
|
||||||
| NC score | 0.003399 (rank : 44) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P12259, Q14285, Q6UPU6 | Gene names | F5 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor V precursor (Activated protein C cofactor) [Contains: Coagulation factor V heavy chain; Coagulation factor V light chain]. | |||||
|
TTBK2_HUMAN
|
||||||
| NC score | 0.003334 (rank : 45) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6IQ55, O94932, Q6ZN52, Q8IVV1 | Gene names | TTBK2, KIAA0847 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2 (EC 2.7.11.1). | |||||
|
OMD_MOUSE
|
||||||
| NC score | 0.003043 (rank : 46) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35103 | Gene names | Omd | |||
|
Domain Architecture |

|
|||||
| Description | Osteomodulin precursor (Osteoadherin) (OSAD) (Keratan sulfate proteoglycan osteomodulin) (KSPG osteomodulin). | |||||
|
TTBK2_MOUSE
|
||||||
| NC score | 0.002494 (rank : 47) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3UVR3, Q3TSR6, Q3UFW0, Q571D1, Q8BKA4, Q924U8 | Gene names | Ttbk2, Kiaa0847, Ttbk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2. | |||||
|
HGFA_HUMAN
|
||||||
| NC score | 0.000117 (rank : 48) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q04756, Q14726 | Gene names | HGFAC | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
TITIN_HUMAN
|
||||||
| NC score | -0.000538 (rank : 49) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
MK04_HUMAN
|
||||||
| NC score | -0.002394 (rank : 50) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P31152 | Gene names | MAPK4, ERK4, PRKM4 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 4 (EC 2.7.11.24) (Extracellular signal-regulated kinase 4) (ERK-4) (MAP kinase isoform p63) (p63- MAPK). | |||||