Please be patient as the page loads
|
THYG_MOUSE
|
||||||
| SwissProt Accessions | O08710, O88590, Q9QWY7 | Gene names | Tg, Tgn | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
THYG_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.990719 (rank : 2) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | P01266, O15274, O43899, Q15593, Q15948, Q9NYR1, Q9NYR2, Q9UMZ0, Q9UNY3 | Gene names | TG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
THYG_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 161 | |
| SwissProt Accessions | O08710, O88590, Q9QWY7 | Gene names | Tg, Tgn | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
ACES_MOUSE
|
||||||
| θ value | 6.32992e-62 (rank : 3) | NC score | 0.843846 (rank : 4) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P21836 | Gene names | Ache | |||
|
Domain Architecture |

|
|||||
| Description | Acetylcholinesterase precursor (EC 3.1.1.7) (AChE). | |||||
|
ACES_HUMAN
|
||||||
| θ value | 1.55911e-60 (rank : 4) | NC score | 0.844364 (rank : 3) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P22303, Q16169, Q86TM9, Q9BXP7 | Gene names | ACHE | |||
|
Domain Architecture |

|
|||||
| Description | Acetylcholinesterase precursor (EC 3.1.1.7) (AChE). | |||||
|
CHLE_MOUSE
|
||||||
| θ value | 2.94036e-59 (rank : 5) | NC score | 0.843386 (rank : 5) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q03311 | Gene names | Bche | |||
|
Domain Architecture |

|
|||||
| Description | Cholinesterase precursor (EC 3.1.1.8) (Acylcholine acylhydrolase) (Choline esterase II) (Butyrylcholine esterase) (Pseudocholinesterase). | |||||
|
CHLE_HUMAN
|
||||||
| θ value | 1.45929e-58 (rank : 6) | NC score | 0.840715 (rank : 6) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P06276 | Gene names | BCHE, CHE1 | |||
|
Domain Architecture |

|
|||||
| Description | Cholinesterase precursor (EC 3.1.1.8) (Acylcholine acylhydrolase) (Choline esterase II) (Butyrylcholine esterase) (Pseudocholinesterase). | |||||
|
NLGN3_MOUSE
|
||||||
| θ value | 7.01481e-53 (rank : 7) | NC score | 0.810189 (rank : 17) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BYM5, Q8BXR4 | Gene names | Nlgn3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
|
NLGNX_HUMAN
|
||||||
| θ value | 9.16162e-53 (rank : 8) | NC score | 0.809915 (rank : 19) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N0W4, Q6UX10, Q9ULG0 | Gene names | NLGN4X, KIAA1260, NLGN4 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-4, X-linked precursor (Neuroligin X) (HNLX). | |||||
|
NLGNY_HUMAN
|
||||||
| θ value | 9.16162e-53 (rank : 9) | NC score | 0.810148 (rank : 18) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NFZ3, Q7Z3T5, Q9Y2F8 | Gene names | NLGN4Y, KIAA0951 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-4, Y-linked precursor (Neuroligin Y). | |||||
|
NLGN2_HUMAN
|
||||||
| θ value | 2.041e-52 (rank : 10) | NC score | 0.812449 (rank : 12) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NFZ4, Q9P2I1 | Gene names | NLGN2, KIAA1366 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-2 precursor. | |||||
|
NLGN2_MOUSE
|
||||||
| θ value | 5.93839e-52 (rank : 11) | NC score | 0.812058 (rank : 13) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q69ZK9, Q5F288 | Gene names | Nlgn2, Kiaa1366 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroligin-2 precursor. | |||||
|
NLGN3_HUMAN
|
||||||
| θ value | 2.49495e-50 (rank : 12) | NC score | 0.808596 (rank : 20) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZ94, Q8NCD0, Q9NZ95, Q9NZ96, Q9NZ97, Q9P248 | Gene names | NLGN3, KIAA1480, NL3 | |||
|
Domain Architecture |
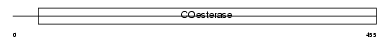
|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
|
EST2_HUMAN
|
||||||
| θ value | 1.15895e-47 (rank : 13) | NC score | 0.829779 (rank : 7) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O00748, Q16859, Q5MAB8, Q7Z366, Q8IUP4, Q8TCP8 | Gene names | CES2, ICE | |||
|
Domain Architecture |

|
|||||
| Description | Carboxylesterase 2 precursor (EC 3.1.1.1) (CE-2) (hCE-2). | |||||
|
CES3_MOUSE
|
||||||
| θ value | 6.35935e-46 (rank : 14) | NC score | 0.813010 (rank : 11) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VCT4, Q91ZV9, Q924V8, Q9ULY1 | Gene names | Ces3, Ces1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxylesterase 3 precursor (EC 3.1.1.1) (Triacylglycerol hydrolase) (TGH). | |||||
|
NLGN1_MOUSE
|
||||||
| θ value | 1.08474e-45 (rank : 15) | NC score | 0.804528 (rank : 21) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99K10 | Gene names | Nlgn1, Kiaa1070 | |||
|
Domain Architecture |
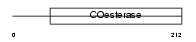
|
|||||
| Description | Neuroligin-1 precursor. | |||||
|
NLGN1_HUMAN
|
||||||
| θ value | 4.122e-45 (rank : 16) | NC score | 0.804342 (rank : 22) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N2Q7, Q9UPT2 | Gene names | NLGN1, KIAA1070 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-1 precursor. | |||||
|
EST22_MOUSE
|
||||||
| θ value | 4.55743e-44 (rank : 17) | NC score | 0.813374 (rank : 10) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q64176 | Gene names | Es22 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 22 precursor (EC 3.1.1.1) (Egasyn) (Esterase- 22) (Es-22). | |||||
|
EST1_HUMAN
|
||||||
| θ value | 1.01529e-43 (rank : 18) | NC score | 0.811279 (rank : 15) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P23141, Q00015, Q13657, Q14062, Q16737, Q16788, Q86UK2, Q96EE8, Q9UK77, Q9ULY2 | Gene names | CES1, CES2, SES1 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 1 precursor (EC 3.1.1.1) (Acyl coenzyme A:cholesterol acyltransferase) (ACAT) (Monocyte/macrophage serine esterase) (HMSE) (Serine esterase 1) (Brain carboxylesterase hBr1) (Triacylglycerol hydrolase) (TGH) (Egasyn). | |||||
|
CEL_MOUSE
|
||||||
| θ value | 6.58091e-43 (rank : 19) | NC score | 0.826710 (rank : 8) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q64285 | Gene names | Cel, Lip1 | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
ESTN_MOUSE
|
||||||
| θ value | 3.61106e-41 (rank : 20) | NC score | 0.811455 (rank : 14) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P23953, O54936, P11374, Q8K125 | Gene names | Es1 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase N precursor (EC 3.1.1.1) (PES-N) (Lung surfactant convertase). | |||||
|
EST1_MOUSE
|
||||||
| θ value | 1.05066e-40 (rank : 21) | NC score | 0.810760 (rank : 16) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VCC2, O55136 | Gene names | Ces1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Liver carboxylesterase 1 precursor (EC 3.1.1.1) (Acyl coenzyme A:cholesterol acyltransferase) (ES-x). | |||||
|
EST31_MOUSE
|
||||||
| θ value | 6.37413e-38 (rank : 22) | NC score | 0.823114 (rank : 9) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q63880 | Gene names | Es31 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 31 precursor (EC 3.1.1.1) (ES-Male) (Esterase- 31). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 1.85459e-37 (rank : 23) | NC score | 0.623517 (rank : 23) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
NID2_HUMAN
|
||||||
| θ value | 5.99374e-20 (rank : 24) | NC score | 0.200004 (rank : 46) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
NID2_MOUSE
|
||||||
| θ value | 6.62687e-19 (rank : 25) | NC score | 0.199085 (rank : 47) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O88322 | Gene names | Nid2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Entactin-2). | |||||
|
SMOC2_HUMAN
|
||||||
| θ value | 1.09485e-13 (rank : 26) | NC score | 0.351411 (rank : 25) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H3U7, Q5TAT7, Q5TAT8, Q86VV9, Q96SF3, Q9H1L3, Q9H1L4, Q9H3U0, Q9H4F7, Q9HCV2 | Gene names | SMOC2, SMAP2 | |||
|
Domain Architecture |
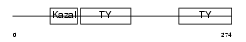
|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2) (Smooth muscle-associated protein 2) (SMAP-2). | |||||
|
SMOC2_MOUSE
|
||||||
| θ value | 1.09485e-13 (rank : 27) | NC score | 0.352299 (rank : 24) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8CD91, Q8VCM1, Q9D9K2, Q9ER95 | Gene names | Smoc2 | |||
|
Domain Architecture |
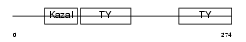
|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2). | |||||
|
SMOC1_HUMAN
|
||||||
| θ value | 2.98157e-11 (rank : 28) | NC score | 0.338295 (rank : 26) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9H4F8, Q96F78 | Gene names | SMOC1 | |||
|
Domain Architecture |
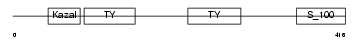
|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1). | |||||
|
SMOC1_MOUSE
|
||||||
| θ value | 2.98157e-11 (rank : 29) | NC score | 0.330585 (rank : 27) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BLY1, Q9WVN9 | Gene names | Smoc1, Srg | |||
|
Domain Architecture |
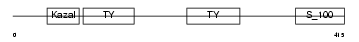
|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1) (SPARC-related gene protein). | |||||
|
NID1_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 30) | NC score | 0.172764 (rank : 52) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P14543, Q14942, Q59FL2, Q5TAF2, Q5TAF3, Q86XD7 | Gene names | NID1, NID | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
TICN3_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 31) | NC score | 0.280434 (rank : 34) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BQ16, O75705, Q6UW53, Q96Q26 | Gene names | SPOCK3, TICN3 | |||
|
Domain Architecture |
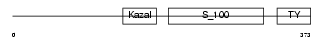
|
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
TICN3_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 32) | NC score | 0.277570 (rank : 35) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BKV0, Q9ER59 | Gene names | Spock3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
TICN1_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 33) | NC score | 0.284321 (rank : 31) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q62288 | Gene names | Spock1, Spock, Ticn1 | |||
|
Domain Architecture |
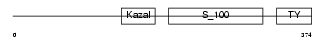
|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
SCUB3_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 34) | NC score | 0.105243 (rank : 56) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8IX30, Q5CZB3, Q86UZ9, Q8NAU9 | Gene names | SCUBE3, CEGF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
TICN1_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 35) | NC score | 0.285384 (rank : 30) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q08629 | Gene names | SPOCK1, SPOCK, TIC1, TICN1 | |||
|
Domain Architecture |
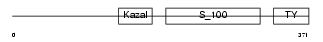
|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
SCUB3_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 36) | NC score | 0.105436 (rank : 55) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q66PY1, Q68FG9 | Gene names | Scube3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
TICN2_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 37) | NC score | 0.281854 (rank : 33) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ER58 | Gene names | Spock2, Ticn2 | |||
|
Domain Architecture |
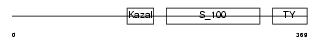
|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
TICN2_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 38) | NC score | 0.283740 (rank : 32) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q92563, Q6UW87 | Gene names | SPOCK2, KIAA0275, TICN2 | |||
|
Domain Architecture |
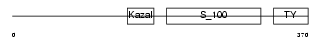
|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
NID1_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 39) | NC score | 0.169264 (rank : 53) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P10493, Q8BQI3, Q8C3U8, Q8C9P6 | Gene names | Nid1, Ent | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
HG2A_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 40) | NC score | 0.289598 (rank : 29) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P04233, Q14597, Q29832, Q5U0J8, Q8WLP6 | Gene names | CD74, DHLAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HLA class II histocompatibility antigen gamma chain (HLA-DR antigens- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (p33) (CD74 antigen). | |||||
|
HG2A_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 41) | NC score | 0.302982 (rank : 28) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P04441, O19452 | Gene names | Cd74, Ii | |||
|
Domain Architecture |
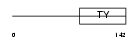
|
|||||
| Description | H-2 class II histocompatibility antigen gamma chain (MHC class II- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (CD74 antigen). | |||||
|
IBP1_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 42) | NC score | 0.213767 (rank : 40) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
|
IBP6_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 43) | NC score | 0.248541 (rank : 36) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P24592, Q14492 | Gene names | IGFBP6, IBP6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
IBP1_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 44) | NC score | 0.225619 (rank : 37) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P08833, Q8IYP5 | Gene names | IGFBP1, IBP1 | |||
|
Domain Architecture |
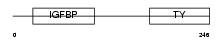
|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1) (Placental protein 12) (PP12). | |||||
|
IBP5_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 45) | NC score | 0.207607 (rank : 42) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q07079 | Gene names | Igfbp5, Igfbp-5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP5_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 46) | NC score | 0.207449 (rank : 43) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P24593 | Gene names | IGFBP5, IBP5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP3_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 47) | NC score | 0.219414 (rank : 39) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P17936, Q2V509, Q6P1M6 | Gene names | IGFBP3, IBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
EPHB3_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 48) | NC score | 0.036275 (rank : 136) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 996 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P54753, Q7Z740 | Gene names | EPHB3, ETK2, HEK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 3 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor HEK-2). | |||||
|
EPHB3_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 49) | NC score | 0.034509 (rank : 140) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 996 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P54754, Q62214 | Gene names | Ephb3, Etk2, Mdk5, Sek4 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 3 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor MDK-5) (Developmental kinase 5) (SEK-4). | |||||
|
IBP6_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 50) | NC score | 0.219533 (rank : 38) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P47880 | Gene names | Igfbp6, Igfbp-6 | |||
|
Domain Architecture |
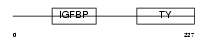
|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
TACD1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 51) | NC score | 0.193159 (rank : 48) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P16422, P18180, Q96C47 | Gene names | TACSTD1, GA733-2, M1S2, TROP1 | |||
|
Domain Architecture |
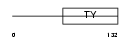
|
|||||
| Description | Tumor-associated calcium signal transducer 1 precursor (Major gastrointestinal tumor-associated protein GA733-2) (Epithelial cell surface antigen) (Epithelial glycoprotein) (EGP) (Adenocarcinoma- associated antigen) (KSA) (KS 1/4 antigen) (Cell surface glycoprotein Trop-1) (CD326 antigen). | |||||
|
IBP3_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 52) | NC score | 0.208775 (rank : 41) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P47878 | Gene names | Igfbp3, Igfbp-3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
MEGF9_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 53) | NC score | 0.076525 (rank : 60) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
LTBP2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 54) | NC score | 0.061338 (rank : 68) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
PKHG4_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 55) | NC score | 0.030390 (rank : 157) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q58EX7, Q4G0J8, Q4H485, Q56A69, Q9H7K4, Q9UFW0 | Gene names | PLEKHG4, PRTPHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Puratrophin-1 (Pleckstrin homology domain-containing family G member 4) (Purkinje cell atrophy-associated protein 1). | |||||
|
IBP4_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 56) | NC score | 0.204977 (rank : 45) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P22692 | Gene names | IGFBP4, IBP4 | |||
|
Domain Architecture |
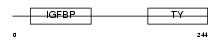
|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
IBP4_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 57) | NC score | 0.206249 (rank : 44) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P47879, O35666 | Gene names | Igfbp4, Igfbp-4 | |||
|
Domain Architecture |
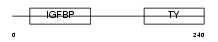
|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
CREL1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 58) | NC score | 0.080653 (rank : 59) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q91XD7, Q8BGJ8 | Gene names | Creld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
STAB2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 59) | NC score | 0.068565 (rank : 62) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
STAB2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 60) | NC score | 0.075581 (rank : 61) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
EPHB2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 61) | NC score | 0.032955 (rank : 145) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 983 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P29323, O43477, Q5T0U6, Q5T0U7, Q5T0U8 | Gene names | EPHB2, DRT, EPTH3, ERK, HEK5 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 2 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EPH-3) (DRT) (Receptor protein-tyrosine kinase HEK5) (ERK) (NY-REN-47 antigen). | |||||
|
EPHB2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 62) | NC score | 0.032953 (rank : 146) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P54763, Q62213, Q9QVY4 | Gene names | Ephb2, Epth3, Nuk, Sek3 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 2 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EPH-3) (Neural kinase) (Nuk receptor tyrosine kinase) (SEK-3). | |||||
|
EPHB6_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 63) | NC score | 0.041549 (rank : 127) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O15197 | Gene names | EPHB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 6 precursor (Tyrosine-protein kinase-defective receptor EPH-6) (HEP). | |||||
|
TACD2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 64) | NC score | 0.156324 (rank : 54) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P09758, Q15658, Q96QD2 | Gene names | TACSTD2, GA733-1, M1S1, TROP2 | |||
|
Domain Architecture |
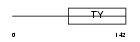
|
|||||
| Description | Tumor-associated calcium signal transducer 2 precursor (Pancreatic carcinoma marker protein GA733-1) (Cell surface glycoprotein Trop-2). | |||||
|
CREL1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 65) | NC score | 0.085896 (rank : 58) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q96HD1, Q6I9X5, Q8NFT4, Q9Y409 | Gene names | CRELD1, CIRRIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
EPHA2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 66) | NC score | 0.030967 (rank : 153) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q03145, Q60633, Q62212 | Gene names | Epha2, Eck, Sek2 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 2 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor ECK) (Epithelial cell kinase) (MPK-5) (SEK-2). | |||||
|
EPHB4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 67) | NC score | 0.035556 (rank : 137) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P54760, Q9BTA5, Q9BXP0 | Gene names | EPHB4, HTK | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 4 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor HTK). | |||||
|
IBP2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 68) | NC score | 0.190387 (rank : 49) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P47877, Q61722 | Gene names | Igfbp2, Igfbp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
FRAS1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 69) | NC score | 0.039524 (rank : 131) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
EPHB4_MOUSE
|
||||||
| θ value | 0.21417 (rank : 70) | NC score | 0.033182 (rank : 142) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1013 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P54761, Q60627 | Gene names | Ephb4, Htk, Mdk2, Myk1 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 4 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor MDK-2) (Developmental kinase 2) (Tyrosine kinase MYK- 1). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 71) | NC score | 0.009688 (rank : 182) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
IBP2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 72) | NC score | 0.179774 (rank : 51) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P18065, Q14619, Q9UCL3 | Gene names | IGFBP2, BP2 | |||
|
Domain Architecture |
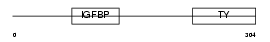
|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
TNR9_HUMAN
|
||||||
| θ value | 0.279714 (rank : 73) | NC score | 0.067512 (rank : 63) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q07011 | Gene names | TNFRSF9, CD137, ILA | |||
|
Domain Architecture |
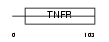
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 9 precursor (4-1BB ligand receptor) (T-cell antigen 4-1BB homolog) (T-cell antigen ILA) (CD137 antigen). | |||||
|
EPHA4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 74) | NC score | 0.030791 (rank : 155) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q03137 | Gene names | Epha4, Sek, Sek1 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 4 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor SEK) (MPK-3). | |||||
|
EPHB6_MOUSE
|
||||||
| θ value | 0.365318 (rank : 75) | NC score | 0.039121 (rank : 132) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O08644, Q8BN76, Q8K0A9 | Gene names | Ephb6, Cekl | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 6 precursor (Tyrosine-protein kinase-defective receptor EPH-6) (MEP). | |||||
|
WBP1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 76) | NC score | 0.030914 (rank : 154) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96G27, O95637 | Gene names | WBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 1 (WBP-1). | |||||
|
WIF1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 77) | NC score | 0.047516 (rank : 114) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9WUA1 | Gene names | Wif1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
EPHA6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 78) | NC score | 0.032402 (rank : 149) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UF33 | Gene names | EPHA6, EHK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephrin type-A receptor 6 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EHK-2) (EPH homology kinase 2). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 0.47712 (rank : 79) | NC score | 0.064383 (rank : 66) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
TNR16_HUMAN
|
||||||
| θ value | 0.47712 (rank : 80) | NC score | 0.051787 (rank : 91) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P08138 | Gene names | NGFR, TNFRSF16 | |||
|
Domain Architecture |
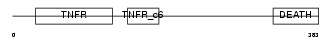
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Gp80-LNGFR) (p75 ICD) (Low affinity neurotrophin receptor p75NTR) (CD271 antigen). | |||||
|
TNR21_MOUSE
|
||||||
| θ value | 0.47712 (rank : 81) | NC score | 0.042559 (rank : 122) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9EPU5, Q91W77, Q91XH9 | Gene names | Tnfrsf21, Dr6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 21 precursor (TNFR- related death receptor 6) (Death receptor 6). | |||||
|
CJ047_MOUSE
|
||||||
| θ value | 0.62314 (rank : 82) | NC score | 0.027839 (rank : 162) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C5R2, Q8R017, Q8R2Z9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47 homolog. | |||||
|
EPHA6_MOUSE
|
||||||
| θ value | 0.62314 (rank : 83) | NC score | 0.032853 (rank : 147) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q62413, Q8CCN2 | Gene names | Epha6, Ehk-2, Ehk2 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 6 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EHK-2) (EPH homology kinase 2). | |||||
|
NLTP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 84) | NC score | 0.027102 (rank : 164) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22307, Q15432, Q16622, Q99430 | Gene names | SCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Nonspecific lipid-transfer protein (EC 2.3.1.176) (Propanoyl-CoA C- acyltransferase) (NSL-TP) (Sterol carrier protein 2) (SCP-2) (Sterol carrier protein X) (SCP-X) (SCP-chi) (SCPX). | |||||
|
NLTP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 85) | NC score | 0.026957 (rank : 166) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32020, Q9DBM7 | Gene names | Scp2, Scp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Nonspecific lipid-transfer protein (EC 2.3.1.176) (Propanoyl-CoA C- acyltransferase) (NSL-TP) (Sterol carrier protein 2) (SCP-2) (Sterol carrier protein X) (SCP-X) (SCP-chi) (SCPX). | |||||
|
TNR21_HUMAN
|
||||||
| θ value | 0.813845 (rank : 86) | NC score | 0.040427 (rank : 130) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75509, Q96D86 | Gene names | TNFRSF21, DR6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 21 precursor (TNFR- related death receptor 6) (Death receptor 6). | |||||
|
AASS_HUMAN
|
||||||
| θ value | 1.06291 (rank : 87) | NC score | 0.020346 (rank : 171) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UDR5, O95462 | Gene names | AASS | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-aminoadipic semialdehyde synthase, mitochondrial precursor (LKR/SDH) [Includes: Lysine ketoglutarate reductase (EC 1.5.1.8) (LOR) (LKR); Saccharopine dehydrogenase (EC 1.5.1.9) (SDH)]. | |||||
|
ATRN_MOUSE
|
||||||
| θ value | 1.06291 (rank : 88) | NC score | 0.037562 (rank : 135) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9WU60, Q9R263, Q9WU77 | Gene names | Atrn, mg, Mgca | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany protein). | |||||
|
FRAS1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 89) | NC score | 0.040912 (rank : 128) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
FUT1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 90) | NC score | 0.016497 (rank : 173) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09160 | Gene names | Fut1 | |||
|
Domain Architecture |

|
|||||
| Description | Galactoside 2-alpha-L-fucosyltransferase 1 (EC 2.4.1.69) (GDP-L- fucose:beta-D-galactoside 2-alpha-L-fucosyltransferase 1) (Alpha(1,2)FT 1) (Fucosyltransferase 1). | |||||
|
LAMC2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 91) | NC score | 0.054407 (rank : 81) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q13753, Q02536, Q02537, Q13752, Q14941, Q5VYE8 | Gene names | LAMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain) (Cell- scattering factor 140 kDa subunit) (CSF 140 kDa subunit) (Large adhesive scatter factor 140 kDa subunit) (Ladsin 140 kDa subunit). | |||||
|
LAMC2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 92) | NC score | 0.059503 (rank : 72) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q61092 | Gene names | Lamc2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain). | |||||
|
LTBP3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 93) | NC score | 0.055247 (rank : 79) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q61810, Q8BNQ6 | Gene names | Ltbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
ATRN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 94) | NC score | 0.038002 (rank : 134) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
PCSK5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 95) | NC score | 0.042786 (rank : 121) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q92824, Q13527 | Gene names | PCSK5, PC5, PC6 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (hPC6). | |||||
|
TNR9_MOUSE
|
||||||
| θ value | 1.38821 (rank : 96) | NC score | 0.052077 (rank : 89) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P20334 | Gene names | Tnfrsf9, Cd137, Cd157, Ila, Ly63 | |||
|
Domain Architecture |
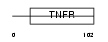
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 9 precursor (4-1BB ligand receptor) (T-cell antigen 4-1BB) (CD137 antigen). | |||||
|
EPHA4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 97) | NC score | 0.029116 (rank : 161) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P54764 | Gene names | EPHA4, HEK8, SEK | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 4 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor SEK) (Receptor protein-tyrosine kinase HEK8). | |||||
|
LAMA3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 98) | NC score | 0.038088 (rank : 133) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
SREC_HUMAN
|
||||||
| θ value | 1.81305 (rank : 99) | NC score | 0.049340 (rank : 113) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
TEP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 100) | NC score | 0.004658 (rank : 191) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99973 | Gene names | TEP1, TLP1, TP1 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase protein component 1 (Telomerase-associated protein 1) (Telomerase protein 1) (p240) (p80 telomerase homolog). | |||||
|
CSTN2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 101) | NC score | 0.007137 (rank : 187) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H4D0, Q9BSS0 | Gene names | CLSTN2, CS2 | |||
|
Domain Architecture |
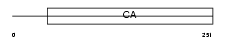
|
|||||
| Description | Calsyntenin-2 precursor. | |||||
|
EPHA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 102) | NC score | 0.029742 (rank : 159) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P21709, Q15405 | Gene names | EPHA1, EPH, EPHT, EPHT1 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 1 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EPH). | |||||
|
EPHAA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 103) | NC score | 0.033178 (rank : 143) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5JZY3, Q6NW42 | Gene names | EPHA10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephrin type-A receptor 10 precursor (EC 2.7.10.1). | |||||
|
LAMA1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 104) | NC score | 0.047143 (rank : 115) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P19137 | Gene names | Lama1, Lama, Lama-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
LAMA3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 105) | NC score | 0.043934 (rank : 117) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q61789, O08751, Q61788, Q61966, Q9JHQ7 | Gene names | Lama3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Nicein subunit alpha). | |||||
|
NOTC3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 106) | NC score | 0.041629 (rank : 126) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
TENR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 107) | NC score | 0.031018 (rank : 152) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q92752, Q15568, Q5R3G0 | Gene names | TNR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin). | |||||
|
TNR16_MOUSE
|
||||||
| θ value | 2.36792 (rank : 108) | NC score | 0.049885 (rank : 112) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Z0W1 | Gene names | Ngfr, Tnfrsf16 | |||
|
Domain Architecture |
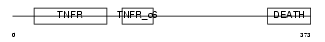
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Low affinity neurotrophin receptor p75NTR). | |||||
|
EPHA7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | 0.032964 (rank : 144) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1053 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q15375, Q59G40, Q5VTU0, Q8N368, Q9H124 | Gene names | EPHA7, EHK3, HEK11 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 7 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EHK-3) (EPH homology kinase 3) (Receptor protein- tyrosine kinase HEK11). | |||||
|
KR261_HUMAN
|
||||||
| θ value | 3.0926 (rank : 110) | NC score | 0.013561 (rank : 177) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PEX3 | Gene names | KRTAP26-1, KAP26.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 26-1. | |||||
|
MEGF6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 111) | NC score | 0.061171 (rank : 69) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
MEGF8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 112) | NC score | 0.062155 (rank : 67) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
NOTC3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 113) | NC score | 0.044913 (rank : 116) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
PCSK5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 114) | NC score | 0.050252 (rank : 108) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q04592, Q62040 | Gene names | Pcsk5 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (Subtilisin-like proprotein convertase 6) (SPC6). | |||||
|
TR10B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 115) | NC score | 0.025891 (rank : 169) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O14763, O14720, O15508, O15517, O15531, Q9BVE0 | Gene names | TNFRSF10B, DR5, KILLER, TRAILR2, TRICK2, ZTNFR9 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10B precursor (Death receptor 5) (TNF-related apoptosis-inducing ligand receptor 2) (TRAIL receptor 2) (TRAIL-R2) (CD262 antigen). | |||||
|
TR19L_HUMAN
|
||||||
| θ value | 3.0926 (rank : 116) | NC score | 0.030657 (rank : 156) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q969Z4, Q86V34, Q96JU1, Q9BUX7 | Gene names | TNFRSF19L, RELT | |||
|
Domain Architecture |
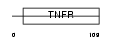
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 19L precursor (Receptor expressed in lymphoid tissues). | |||||
|
USH2A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 117) | NC score | 0.042919 (rank : 120) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | O75445, Q5VVM9, Q6S362, Q9NS27 | Gene names | USH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein) (Usher syndrome type IIa protein). | |||||
|
WIF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 118) | NC score | 0.042282 (rank : 124) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y5W5, Q6UXI1, Q8WVG4 | Gene names | WIF1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
APBB2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 119) | NC score | 0.012860 (rank : 178) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
APBB2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | 0.011557 (rank : 179) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBR4, Q6DFX8 | Gene names | Apbb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2. | |||||
|
EPHA1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.029313 (rank : 160) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q60750, Q8CED9, Q9ESJ2 | Gene names | Epha1, Esk | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 1 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor ESK). | |||||
|
FBLN1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 122) | NC score | 0.057683 (rank : 76) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q08879, Q08878, Q8C3B1, Q91ZC9, Q922K8 | Gene names | Fbln1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-1 precursor (Basement-membrane protein 90) (BM-90). | |||||
|
KR102_HUMAN
|
||||||
| θ value | 4.03905 (rank : 123) | NC score | 0.025992 (rank : 168) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
MKS3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 124) | NC score | 0.026275 (rank : 167) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BR76, Q78U07 | Gene names | Tmem67, Mks3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Meckelin (Meckel syndrome type 3 protein homolog) (Transmembrane protein 67). | |||||
|
TENR_MOUSE
|
||||||
| θ value | 4.03905 (rank : 125) | NC score | 0.030295 (rank : 158) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8BYI9, O88717 | Gene names | Tnr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin) (Neural recognition molecule J1-160/180). | |||||
|
ARI4A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.006795 (rank : 189) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
CASR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.007091 (rank : 188) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P41180, Q13912, Q16108, Q16109, Q16110, Q16379, Q4PJ19 | Gene names | CASR, GPRC2A, PCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular calcium-sensing receptor precursor (CaSR) (Parathyroid Cell calcium-sensing receptor). | |||||
|
LAMA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.043833 (rank : 118) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 804 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P25391 | Gene names | LAMA1, LAMA | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
LAMA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.040553 (rank : 129) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
MEGF8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.060768 (rank : 71) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P60882, Q80TR3, Q80V41, Q8BMN9, Q8JZW7, Q8K0J3 | Gene names | Megf8, Egfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
NDUB9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.014169 (rank : 175) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y6M9, Q9UQE8 | Gene names | NDUFB9, UQOR22 | |||
|
Domain Architecture |
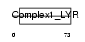
|
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9 (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase B22 subunit) (Complex I-B22) (CI-B22). | |||||
|
TNR5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.031945 (rank : 151) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P27512, Q99NE0, Q99NE1, Q99NE2, Q99NE3 | Gene names | Cd40, Tnfrsf5 | |||
|
Domain Architecture |
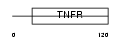
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 5 precursor (CD40L receptor) (B-cell surface antigen CD40) (BP50) (CDw40). | |||||
|
TR10C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.032334 (rank : 150) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14798, O14755, Q6UXM5 | Gene names | TNFRSF10C, DCR1, LIT, TRAILR3, TRID | |||
|
Domain Architecture |
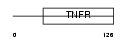
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10C precursor (Decoy receptor 1) (DcR1) (Decoy TRAIL receptor without death domain) (TNF- related apoptosis-inducing ligand receptor 3) (TRAIL receptor 3) (TRAIL-R3) (Trail receptor without an intracellular domain) (Lymphocyte inhibitor of TRAIL) (Antagonist decoy receptor for TRAIL/Apo-2L) (CD263 antigen). | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.043050 (rank : 119) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
CELR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.015192 (rank : 174) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NYQ6, O95722, Q5TH47, Q9BWQ5, Q9Y506, Q9Y526 | Gene names | CELSR1, CDHF9, FMI2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor (Flamingo homolog 2) (hFmi2). | |||||
|
EPHA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.027020 (rank : 165) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P29317 | Gene names | EPHA2, ECK | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 2 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor ECK) (Epithelial cell kinase). | |||||
|
GAK8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.003838 (rank : 192) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HDB9 | Gene names | ERVK5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q12.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(II) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
LRP12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.009461 (rank : 183) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y561 | Gene names | LRP12, ST7 | |||
|
Domain Architecture |
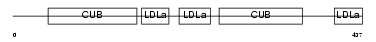
|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor (Suppressor of tumorigenicity protein 7). | |||||
|
MKS3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.033197 (rank : 141) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5HYA8, Q3ZCX3, Q7Z5T8, Q8IZ06 | Gene names | TMEM67, MKS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Meckelin (Meckel syndrome type 3 protein) (Transmembrane protein 67). | |||||
|
RABX5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.008273 (rank : 186) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJ41 | Gene names | RABGEF1, RABEX5 | |||
|
Domain Architecture |

|
|||||
| Description | Rab5 GDP/GTP exchange factor (Rabex-5) (Rabaptin-5-associated exchange factor for Rab5). | |||||
|
RSPO1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.017820 (rank : 172) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q2MKA7, Q5T0F2, Q8N7L5 | Gene names | RSPO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-1 precursor (Roof plate-specific spondin-1) (hRspo1). | |||||
|
SNX6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.009045 (rank : 184) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UNH7, Q9Y449 | Gene names | SNX6 | |||
|
Domain Architecture |
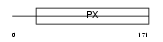
|
|||||
| Description | Sorting nexin-6 (TRAF4-associated factor 2). | |||||
|
SNX6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.009036 (rank : 185) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P8X1 | Gene names | Snx6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-6. | |||||
|
TNR3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.034874 (rank : 138) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P50284 | Gene names | Ltbr, Tnfcr, Tnfrsf3 | |||
|
Domain Architecture |
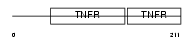
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 3 precursor (Lymphotoxin-beta receptor). | |||||
|
AN32A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.002572 (rank : 193) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P39687 | Gene names | ANP32A, C15orf1, LANP, MAPM, PHAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein) (Lanp) (Putative HLA-DR-associated protein I) (PHAPI) (Mapmodulin). | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | -0.004241 (rank : 197) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CRIS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.005049 (rank : 190) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q03401 | Gene names | Crisp1, Aeg-1, Aeg1 | |||
|
Domain Architecture |
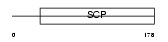
|
|||||
| Description | Cysteine-rich secretory protein 1 precursor (CRISP-1) (Acidic epididymal glycoprotein 1) (Sperm-coating glycoprotein 1) (SCP 1). | |||||
|
DIAP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.011052 (rank : 180) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60879, O60878, Q8WX06, Q8WX48, Q9UJL2 | Gene names | DIAPH2, DIA | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2). | |||||
|
DLL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.042064 (rank : 125) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
EPHB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.027271 (rank : 163) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 988 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P54762, O43569, O95142, O95143 | Gene names | EPHB1, EPHT2, NET | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 1 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EPH-2) (NET) (HEK6) (ELK). | |||||
|
FBN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.051230 (rank : 97) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
LAMA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.034585 (rank : 139) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
LAMC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.042384 (rank : 123) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9Y6N6 | Gene names | LAMC3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
LTBP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.055116 (rank : 80) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
MMP17_MOUSE
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.001133 (rank : 194) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0S3 | Gene names | Mmp17, Mt4mmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-17 precursor (EC 3.4.24.-) (MMP-17) (Membrane-type matrix metalloproteinase 4) (MT-MMP 4) (Membrane-type-4 matrix metalloproteinase) (MT4-MMP). | |||||
|
PCDG4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | -0.000202 (rank : 195) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y5G9, Q9Y5D3 | Gene names | PCDHGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A4 precursor (PCDH-gamma-A4). | |||||
|
PCDG5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | -0.000218 (rank : 196) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y5G8, Q9Y5D2 | Gene names | PCDHGA5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A5 precursor (PCDH-gamma-A5). | |||||
|
PUS7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.010666 (rank : 181) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96PZ0, Q75MG4, Q9NX19 | Gene names | PUS7, KIAA1897 | |||
|
Domain Architecture |
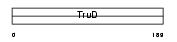
|
|||||
| Description | Pseudouridylate synthase 7 homolog (EC 5.4.99.-). | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.022168 (rank : 170) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
TNR25_HUMAN
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.032779 (rank : 148) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q93038, O00275, O00276, O00277, O00278, O00279, O00280, O14865, O14866, P78507, P78515, Q92983, Q93036, Q93037, Q99722, Q99830, Q99831, Q9BY86, Q9UME0, Q9UME1, Q9UME5 | Gene names | TNFRSF25, APO3, DDR3, DR3, TNFRSF12, WSL, WSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 25 precursor (WSL-1 protein) (Apoptosis-mediating receptor DR3) (Apoptosis-mediating receptor TRAMP) (Death domain receptor 3) (WSL protein) (Apoptosis- inducing receptor AIR) (Apo-3) (Lymphocyte-associated receptor of death) (LARD). | |||||
|
TNR27_MOUSE
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.013917 (rank : 176) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BX35, Q8BM50 | Gene names | Eda2r, Tnfrsf27, Xedar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 27 (X-linked ectodysplasin-A2 receptor) (EDA-A2 receptor). | |||||
|
AAAD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.180217 (rank : 50) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P22760, Q8N1A9 | Gene names | AADAC, DAC | |||
|
Domain Architecture |

|
|||||
| Description | Arylacetamide deacetylase (EC 3.1.1.-) (AADAC). | |||||
|
AAAD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.100211 (rank : 57) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99PG0, Q8VCF2 | Gene names | Aadac, Aada | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylacetamide deacetylase (EC 3.1.1.-) (AADAC). | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.059318 (rank : 74) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
C1QR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.052054 (rank : 90) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NPY3, O00274 | Gene names | CD93, C1QR1, MXRA4 | |||
|
Domain Architecture |
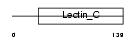
|
|||||
| Description | Complement component C1q receptor precursor (Complement component 1 q subcomponent receptor 1) (C1qR) (C1qRp) (C1qR(p)) (C1q/MBL/SPA receptor) (CD93 antigen) (CDw93). | |||||
|
C1QR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.051457 (rank : 95) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O89103, Q3U2X0 | Gene names | Cd93, Aa4, C1qr1, C1qrp, Ly68 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C1q receptor precursor (Complement component 1 q subcomponent receptor 1) (C1qRp) (C1qR(p)) (C1q/MBL/SPA receptor) (CD93 antigen) (Cell surface antigen AA4) (Lymphocyte antigen 68). | |||||
|
EGF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.053505 (rank : 83) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P01133 | Gene names | EGF | |||
|
Domain Architecture |

|
|||||
| Description | Pro-epidermal growth factor precursor (EGF) [Contains: Epidermal growth factor (Urogastrone)]. | |||||
|
EGF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.053846 (rank : 82) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P01132, Q6P9J2 | Gene names | Egf | |||
|
Domain Architecture |

|
|||||
| Description | Pro-epidermal growth factor precursor (EGF) [Contains: Epidermal growth factor]. | |||||
|
FBLN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.057196 (rank : 77) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P23142, P23143, P23144, P37888, Q5TIC4, Q8TBH8, Q9HBQ5, Q9UC21, Q9UGR4, Q9UH41 | Gene names | FBLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-1 precursor. | |||||
|
FBLN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.051660 (rank : 92) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P98095 | Gene names | FBLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
FBLN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.050902 (rank : 100) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
FBLN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.051382 (rank : 96) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q12805 | Gene names | EFEMP1, FBLN3, FBNL | |||
|
Domain Architecture |
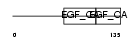
|
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 1 precursor (Fibulin-3) (FIBL-3) (Fibrillin-like protein) (Extracellular protein S1-5). | |||||
|
FBLN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.050757 (rank : 102) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BPB5, Q6Y3N6 | Gene names | Efemp1, Fbln3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 1 precursor (Fibulin-3) (FIBL-3). | |||||
|
FBLN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.050214 (rank : 109) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O95967, O75967 | Gene names | EFEMP2, FBLN4 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 2 precursor (Fibulin-4) (FIBL-4) (UPH1 protein). | |||||
|
FBLN5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.050498 (rank : 103) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UBX5, O75966, Q6UWA3 | Gene names | FBLN5, DANCE | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-5 precursor (FIBL-5) (Developmental arteries and neural crest EGF-like protein) (Dance) (Urine p50 protein) (UP50). | |||||
|
FBLN5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.050768 (rank : 101) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9WVH9, Q541Z7 | Gene names | Fbln5, Dance | |||
|
Domain Architecture |
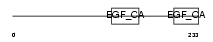
|
|||||
| Description | Fibulin-5 precursor (FIBL-5) (Developmental arteries and neural crest EGF-like protein) (Dance). | |||||
|
FBN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.050293 (rank : 106) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
FBN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.050139 (rank : 111) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q61555, Q63957 | Gene names | Fbn2, Fbn-2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
FBN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.051220 (rank : 98) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q75N90, Q75N91, Q75N92, Q75N93, Q86SJ5, Q96JP8 | Gene names | FBN3, KIAA1776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrillin-3 precursor. | |||||
|
FST_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.050470 (rank : 104) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P19883, Q9BTH0 | Gene names | FST | |||
|
Domain Architecture |
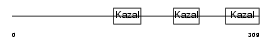
|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
FST_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.050319 (rank : 105) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P47931 | Gene names | Fst | |||
|
Domain Architecture |
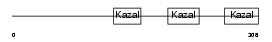
|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
LTBP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.050978 (rank : 99) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9NS15, O15107, Q96HB9, Q9H7K2, Q9UFN4 | Gene names | LTBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
LTBP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.052745 (rank : 86) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
NELL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.052383 (rank : 88) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q92832, Q9Y472 | Gene names | NELL1, NRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL1 precursor (NEL-like protein 1) (Nel-related protein 1). | |||||
|
NELL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.051600 (rank : 93) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q99435 | Gene names | NELL2, NRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (Nel-related protein 2). | |||||
|
NELL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.051556 (rank : 94) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q61220 | Gene names | Nell2, Mel91 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (MEL91 protein). | |||||
|
SMR3B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.058997 (rank : 75) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02814, Q9UBN0, Q9UCT0 | Gene names | SMR3B, PBII, PROL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog B precursor (Proline-rich protein 3) (Proline-rich peptide P-B) [Contains: Peptide P-A; Peptide D1A]. | |||||
|
SPRC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.066152 (rank : 64) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P09486 | Gene names | SPARC, ON | |||
|
Domain Architecture |
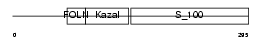
|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
SPRC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.064825 (rank : 65) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P07214 | Gene names | Sparc | |||
|
Domain Architecture |
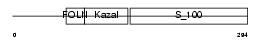
|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
SPRL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.055375 (rank : 78) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
SPRL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.050190 (rank : 110) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
STAB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.060917 (rank : 70) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
STAB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.059483 (rank : 73) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
TRBM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.050284 (rank : 107) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P07204, Q9UC32 | Gene names | THBD, THRM | |||
|
Domain Architecture |
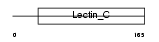
|
|||||
| Description | Thrombomodulin precursor (TM) (Fetomodulin) (CD141 antigen). | |||||
|
TRBM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.052903 (rank : 84) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P15306 | Gene names | Thbd | |||
|
Domain Architecture |
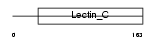
|
|||||
| Description | Thrombomodulin precursor (TM) (Fetomodulin) (CD141 antigen). | |||||
|
UROM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 196) | NC score | 0.052557 (rank : 87) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P07911, Q540J6, Q6ZS84, Q8IYG0 | Gene names | UMOD | |||
|
Domain Architecture |

|
|||||
| Description | Uromodulin precursor (Tamm-Horsfall urinary glycoprotein) (THP). | |||||
|
UROM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 197) | NC score | 0.052879 (rank : 85) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q91X17, Q3TN64, Q3TP60, Q62285 | Gene names | Umod | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uromodulin precursor (Tamm-Horsfall urinary glycoprotein) (THP). | |||||
|
THYG_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 161 | |
| SwissProt Accessions | O08710, O88590, Q9QWY7 | Gene names | Tg, Tgn | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
THYG_HUMAN
|
||||||
| NC score | 0.990719 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | P01266, O15274, O43899, Q15593, Q15948, Q9NYR1, Q9NYR2, Q9UMZ0, Q9UNY3 | Gene names | TG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
ACES_HUMAN
|
||||||
| NC score | 0.844364 (rank : 3) | θ value | 1.55911e-60 (rank : 4) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P22303, Q16169, Q86TM9, Q9BXP7 | Gene names | ACHE | |||
|
Domain Architecture |

|
|||||
| Description | Acetylcholinesterase precursor (EC 3.1.1.7) (AChE). | |||||
|
ACES_MOUSE
|
||||||
| NC score | 0.843846 (rank : 4) | θ value | 6.32992e-62 (rank : 3) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P21836 | Gene names | Ache | |||
|
Domain Architecture |

|
|||||
| Description | Acetylcholinesterase precursor (EC 3.1.1.7) (AChE). | |||||
|
CHLE_MOUSE
|
||||||
| NC score | 0.843386 (rank : 5) | θ value | 2.94036e-59 (rank : 5) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q03311 | Gene names | Bche | |||
|
Domain Architecture |

|
|||||
| Description | Cholinesterase precursor (EC 3.1.1.8) (Acylcholine acylhydrolase) (Choline esterase II) (Butyrylcholine esterase) (Pseudocholinesterase). | |||||
|
CHLE_HUMAN
|
||||||
| NC score | 0.840715 (rank : 6) | θ value | 1.45929e-58 (rank : 6) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P06276 | Gene names | BCHE, CHE1 | |||
|
Domain Architecture |

|
|||||
| Description | Cholinesterase precursor (EC 3.1.1.8) (Acylcholine acylhydrolase) (Choline esterase II) (Butyrylcholine esterase) (Pseudocholinesterase). | |||||
|
EST2_HUMAN
|
||||||
| NC score | 0.829779 (rank : 7) | θ value | 1.15895e-47 (rank : 13) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O00748, Q16859, Q5MAB8, Q7Z366, Q8IUP4, Q8TCP8 | Gene names | CES2, ICE | |||
|
Domain Architecture |

|
|||||
| Description | Carboxylesterase 2 precursor (EC 3.1.1.1) (CE-2) (hCE-2). | |||||
|
CEL_MOUSE
|
||||||
| NC score | 0.826710 (rank : 8) | θ value | 6.58091e-43 (rank : 19) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q64285 | Gene names | Cel, Lip1 | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
EST31_MOUSE
|
||||||
| NC score | 0.823114 (rank : 9) | θ value | 6.37413e-38 (rank : 22) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q63880 | Gene names | Es31 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 31 precursor (EC 3.1.1.1) (ES-Male) (Esterase- 31). | |||||
|
EST22_MOUSE
|
||||||
| NC score | 0.813374 (rank : 10) | θ value | 4.55743e-44 (rank : 17) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q64176 | Gene names | Es22 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 22 precursor (EC 3.1.1.1) (Egasyn) (Esterase- 22) (Es-22). | |||||
|
CES3_MOUSE
|
||||||
| NC score | 0.813010 (rank : 11) | θ value | 6.35935e-46 (rank : 14) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VCT4, Q91ZV9, Q924V8, Q9ULY1 | Gene names | Ces3, Ces1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxylesterase 3 precursor (EC 3.1.1.1) (Triacylglycerol hydrolase) (TGH). | |||||
|
NLGN2_HUMAN
|
||||||
| NC score | 0.812449 (rank : 12) | θ value | 2.041e-52 (rank : 10) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NFZ4, Q9P2I1 | Gene names | NLGN2, KIAA1366 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-2 precursor. | |||||
|
NLGN2_MOUSE
|
||||||
| NC score | 0.812058 (rank : 13) | θ value | 5.93839e-52 (rank : 11) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q69ZK9, Q5F288 | Gene names | Nlgn2, Kiaa1366 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroligin-2 precursor. | |||||
|
ESTN_MOUSE
|
||||||
| NC score | 0.811455 (rank : 14) | θ value | 3.61106e-41 (rank : 20) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P23953, O54936, P11374, Q8K125 | Gene names | Es1 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase N precursor (EC 3.1.1.1) (PES-N) (Lung surfactant convertase). | |||||
|
EST1_HUMAN
|
||||||
| NC score | 0.811279 (rank : 15) | θ value | 1.01529e-43 (rank : 18) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P23141, Q00015, Q13657, Q14062, Q16737, Q16788, Q86UK2, Q96EE8, Q9UK77, Q9ULY2 | Gene names | CES1, CES2, SES1 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 1 precursor (EC 3.1.1.1) (Acyl coenzyme A:cholesterol acyltransferase) (ACAT) (Monocyte/macrophage serine esterase) (HMSE) (Serine esterase 1) (Brain carboxylesterase hBr1) (Triacylglycerol hydrolase) (TGH) (Egasyn). | |||||
|
EST1_MOUSE
|
||||||
| NC score | 0.810760 (rank : 16) | θ value | 1.05066e-40 (rank : 21) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VCC2, O55136 | Gene names | Ces1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Liver carboxylesterase 1 precursor (EC 3.1.1.1) (Acyl coenzyme A:cholesterol acyltransferase) (ES-x). | |||||
|
NLGN3_MOUSE
|
||||||
| NC score | 0.810189 (rank : 17) | θ value | 7.01481e-53 (rank : 7) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BYM5, Q8BXR4 | Gene names | Nlgn3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
|
NLGNY_HUMAN
|
||||||
| NC score | 0.810148 (rank : 18) | θ value | 9.16162e-53 (rank : 9) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NFZ3, Q7Z3T5, Q9Y2F8 | Gene names | NLGN4Y, KIAA0951 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-4, Y-linked precursor (Neuroligin Y). | |||||
|
NLGNX_HUMAN
|
||||||
| NC score | 0.809915 (rank : 19) | θ value | 9.16162e-53 (rank : 8) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N0W4, Q6UX10, Q9ULG0 | Gene names | NLGN4X, KIAA1260, NLGN4 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-4, X-linked precursor (Neuroligin X) (HNLX). | |||||
|
NLGN3_HUMAN
|
||||||
| NC score | 0.808596 (rank : 20) | θ value | 2.49495e-50 (rank : 12) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZ94, Q8NCD0, Q9NZ95, Q9NZ96, Q9NZ97, Q9P248 | Gene names | NLGN3, KIAA1480, NL3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
|
NLGN1_MOUSE
|
||||||
| NC score | 0.804528 (rank : 21) | θ value | 1.08474e-45 (rank : 15) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99K10 | Gene names | Nlgn1, Kiaa1070 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-1 precursor. | |||||
|
NLGN1_HUMAN
|
||||||
| NC score | 0.804342 (rank : 22) | θ value | 4.122e-45 (rank : 16) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N2Q7, Q9UPT2 | Gene names | NLGN1, KIAA1070 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-1 precursor. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.623517 (rank : 23) | θ value | 1.85459e-37 (rank : 23) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
SMOC2_MOUSE
|
||||||
| NC score | 0.352299 (rank : 24) | θ value | 1.09485e-13 (rank : 27) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8CD91, Q8VCM1, Q9D9K2, Q9ER95 | Gene names | Smoc2 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2). | |||||
|
SMOC2_HUMAN
|
||||||
| NC score | 0.351411 (rank : 25) | θ value | 1.09485e-13 (rank : 26) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H3U7, Q5TAT7, Q5TAT8, Q86VV9, Q96SF3, Q9H1L3, Q9H1L4, Q9H3U0, Q9H4F7, Q9HCV2 | Gene names | SMOC2, SMAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2) (Smooth muscle-associated protein 2) (SMAP-2). | |||||
|
SMOC1_HUMAN
|
||||||
| NC score | 0.338295 (rank : 26) | θ value | 2.98157e-11 (rank : 28) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9H4F8, Q96F78 | Gene names | SMOC1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1). | |||||
|
SMOC1_MOUSE
|
||||||
| NC score | 0.330585 (rank : 27) | θ value | 2.98157e-11 (rank : 29) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BLY1, Q9WVN9 | Gene names | Smoc1, Srg | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1) (SPARC-related gene protein). | |||||
|
HG2A_MOUSE
|
||||||
| NC score | 0.302982 (rank : 28) | θ value | 9.29e-05 (rank : 41) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P04441, O19452 | Gene names | Cd74, Ii | |||
|
Domain Architecture |

|
|||||
| Description | H-2 class II histocompatibility antigen gamma chain (MHC class II- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (CD74 antigen). | |||||
|
HG2A_HUMAN
|
||||||
| NC score | 0.289598 (rank : 29) | θ value | 2.44474e-05 (rank : 40) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P04233, Q14597, Q29832, Q5U0J8, Q8WLP6 | Gene names | CD74, DHLAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HLA class II histocompatibility antigen gamma chain (HLA-DR antigens- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (p33) (CD74 antigen). | |||||
|
TICN1_HUMAN
|
||||||
| NC score | 0.285384 (rank : 30) | θ value | 8.97725e-08 (rank : 35) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q08629 | Gene names | SPOCK1, SPOCK, TIC1, TICN1 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
TICN1_MOUSE
|
||||||
| NC score | 0.284321 (rank : 31) | θ value | 3.08544e-08 (rank : 33) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q62288 | Gene names | Spock1, Spock, Ticn1 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
TICN2_HUMAN
|
||||||
| NC score | 0.283740 (rank : 32) | θ value | 1.99992e-07 (rank : 38) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q92563, Q6UW87 | Gene names | SPOCK2, KIAA0275, TICN2 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
TICN2_MOUSE
|
||||||
| NC score | 0.281854 (rank : 33) | θ value | 1.53129e-07 (rank : 37) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ER58 | Gene names | Spock2, Ticn2 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
TICN3_HUMAN
|
||||||
| NC score | 0.280434 (rank : 34) | θ value | 1.38499e-08 (rank : 31) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BQ16, O75705, Q6UW53, Q96Q26 | Gene names | SPOCK3, TICN3 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
TICN3_MOUSE
|
||||||
| NC score | 0.277570 (rank : 35) | θ value | 1.38499e-08 (rank : 32) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BKV0, Q9ER59 | Gene names | Spock3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
IBP6_HUMAN
|
||||||
| NC score | 0.248541 (rank : 36) | θ value | 0.000158464 (rank : 43) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P24592, Q14492 | Gene names | IGFBP6, IBP6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
IBP1_HUMAN
|
||||||
| NC score | 0.225619 (rank : 37) | θ value | 0.00035302 (rank : 44) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P08833, Q8IYP5 | Gene names | IGFBP1, IBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1) (Placental protein 12) (PP12). | |||||
|
IBP6_MOUSE
|
||||||
| NC score | 0.219533 (rank : 38) | θ value | 0.00869519 (rank : 50) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P47880 | Gene names | Igfbp6, Igfbp-6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
IBP3_HUMAN
|
||||||
| NC score | 0.219414 (rank : 39) | θ value | 0.00175202 (rank : 47) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P17936, Q2V509, Q6P1M6 | Gene names | IGFBP3, IBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
IBP1_MOUSE
|
||||||
| NC score | 0.213767 (rank : 40) | θ value | 9.29e-05 (rank : 42) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
|
IBP3_MOUSE
|
||||||
| NC score | 0.208775 (rank : 41) | θ value | 0.0148317 (rank : 52) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P47878 | Gene names | Igfbp3, Igfbp-3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
IBP5_MOUSE
|
||||||
| NC score | 0.207607 (rank : 42) | θ value | 0.00035302 (rank : 45) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q07079 | Gene names | Igfbp5, Igfbp-5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP5_HUMAN
|
||||||
| NC score | 0.207449 (rank : 43) | θ value | 0.000786445 (rank : 46) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P24593 | Gene names | IGFBP5, IBP5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP4_MOUSE
|
||||||
| NC score | 0.206249 (rank : 44) | θ value | 0.0431538 (rank : 57) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P47879, O35666 | Gene names | Igfbp4, Igfbp-4 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
IBP4_HUMAN
|
||||||
| NC score | 0.204977 (rank : 45) | θ value | 0.0431538 (rank : 56) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P22692 | Gene names | IGFBP4, IBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
NID2_HUMAN
|
||||||
| NC score | 0.200004 (rank : 46) | θ value | 5.99374e-20 (rank : 24) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
NID2_MOUSE
|
||||||
| NC score | 0.199085 (rank : 47) | θ value | 6.62687e-19 (rank : 25) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O88322 | Gene names | Nid2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Entactin-2). | |||||
|
TACD1_HUMAN
|
||||||
| NC score | 0.193159 (rank : 48) | θ value | 0.00869519 (rank : 51) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P16422, P18180, Q96C47 | Gene names | TACSTD1, GA733-2, M1S2, TROP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor-associated calcium signal transducer 1 precursor (Major gastrointestinal tumor-associated protein GA733-2) (Epithelial cell surface antigen) (Epithelial glycoprotein) (EGP) (Adenocarcinoma- associated antigen) (KSA) (KS 1/4 antigen) (Cell surface glycoprotein Trop-1) (CD326 antigen). | |||||
|
IBP2_MOUSE
|
||||||
| NC score | 0.190387 (rank : 49) | θ value | 0.125558 (rank : 68) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P47877, Q61722 | Gene names | Igfbp2, Igfbp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
AAAD_HUMAN
|
||||||
| NC score | 0.180217 (rank : 50) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P22760, Q8N1A9 | Gene names | AADAC, DAC | |||
|
Domain Architecture |

|
|||||
| Description | Arylacetamide deacetylase (EC 3.1.1.-) (AADAC). | |||||
|
IBP2_HUMAN
|
||||||
| NC score | 0.179774 (rank : 51) | θ value | 0.279714 (rank : 72) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P18065, Q14619, Q9UCL3 | Gene names | IGFBP2, BP2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
NID1_HUMAN
|
||||||
| NC score | 0.172764 (rank : 52) | θ value | 1.38499e-08 (rank : 30) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P14543, Q14942, Q59FL2, Q5TAF2, Q5TAF3, Q86XD7 | Gene names | NID1, NID | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
NID1_MOUSE
|
||||||
| NC score | 0.169264 (rank : 53) | θ value | 2.61198e-07 (rank : 39) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P10493, Q8BQI3, Q8C3U8, Q8C9P6 | Gene names | Nid1, Ent | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
TACD2_HUMAN
|
||||||
| NC score | 0.156324 (rank : 54) | θ value | 0.0961366 (rank : 64) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P09758, Q15658, Q96QD2 | Gene names | TACSTD2, GA733-1, M1S1, TROP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor-associated calcium signal transducer 2 precursor (Pancreatic carcinoma marker protein GA733-1) (Cell surface glycoprotein Trop-2). | |||||
|
SCUB3_MOUSE
|
||||||
| NC score | 0.105436 (rank : 55) | θ value | 1.17247e-07 (rank : 36) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q66PY1, Q68FG9 | Gene names | Scube3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
SCUB3_HUMAN
|
||||||
| NC score | 0.105243 (rank : 56) | θ value | 6.87365e-08 (rank : 34) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8IX30, Q5CZB3, Q86UZ9, Q8NAU9 | Gene names | SCUBE3, CEGF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
AAAD_MOUSE
|
||||||
| NC score | 0.100211 (rank : 57) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99PG0, Q8VCF2 | Gene names | Aadac, Aada | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylacetamide deacetylase (EC 3.1.1.-) (AADAC). | |||||
|
CREL1_HUMAN
|
||||||
| NC score | 0.085896 (rank : 58) | θ value | 0.125558 (rank : 65) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q96HD1, Q6I9X5, Q8NFT4, Q9Y409 | Gene names | CRELD1, CIRRIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
CREL1_MOUSE
|
||||||
| NC score | 0.080653 (rank : 59) | θ value | 0.0563607 (rank : 58) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q91XD7, Q8BGJ8 | Gene names | Creld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
MEGF9_HUMAN
|
||||||
| NC score | 0.076525 (rank : 60) | θ value | 0.0193708 (rank : 53) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
STAB2_MOUSE
|
||||||
| NC score | 0.075581 (rank : 61) | θ value | 0.0736092 (rank : 60) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
STAB2_HUMAN
|
||||||
| NC score | 0.068565 (rank : 62) | θ value | 0.0563607 (rank : 59) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
TNR9_HUMAN
|
||||||
| NC score | 0.067512 (rank : 63) | θ value | 0.279714 (rank : 73) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q07011 | Gene names | TNFRSF9, CD137, ILA | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 9 precursor (4-1BB ligand receptor) (T-cell antigen 4-1BB homolog) (T-cell antigen ILA) (CD137 antigen). | |||||
|
SPRC_HUMAN
|
||||||
| NC score | 0.066152 (rank : 64) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P09486 | Gene names | SPARC, ON | |||
|
Domain Architecture |

|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
SPRC_MOUSE
|
||||||
| NC score | 0.064825 (rank : 65) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P07214 | Gene names | Sparc | |||
|
Domain Architecture |

|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.064383 (rank : 66) | θ value | 0.47712 (rank : 79) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MEGF8_HUMAN
|
||||||
| NC score | 0.062155 (rank : 67) | θ value | 3.0926 (rank : 112) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
LTBP2_MOUSE
|
||||||
| NC score | 0.061338 (rank : 68) | θ value | 0.0252991 (rank : 54) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
MEGF6_HUMAN
|
||||||
| NC score | 0.061171 (rank : 69) | θ value | 3.0926 (rank : 111) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
STAB1_HUMAN
|
||||||
| NC score | 0.060917 (rank : 70) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
MEGF8_MOUSE
|
||||||
| NC score | 0.060768 (rank : 71) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P60882, Q80TR3, Q80V41, Q8BMN9, Q8JZW7, Q8K0J3 | Gene names | Megf8, Egfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
LAMC2_MOUSE
|
||||||
| NC score | 0.059503 (rank : 72) | θ value | 1.06291 (rank : 92) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q61092 | Gene names | Lamc2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain). | |||||
|
STAB1_MOUSE
|
||||||
| NC score | 0.059483 (rank : 73) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.059318 (rank : 74) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
SMR3B_HUMAN
|
||||||
| NC score | 0.058997 (rank : 75) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02814, Q9UBN0, Q9UCT0 | Gene names | SMR3B, PBII, PROL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog B precursor (Proline-rich protein 3) (Proline-rich peptide P-B) [Contains: Peptide P-A; Peptide D1A]. | |||||
|
FBLN1_MOUSE
|
||||||
| NC score | 0.057683 (rank : 76) | θ value | 4.03905 (rank : 122) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q08879, Q08878, Q8C3B1, Q91ZC9, Q922K8 | Gene names | Fbln1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-1 precursor (Basement-membrane protein 90) (BM-90). | |||||
|
FBLN1_HUMAN
|
||||||
| NC score | 0.057196 (rank : 77) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P23142, P23143, P23144, P37888, Q5TIC4, Q8TBH8, Q9HBQ5, Q9UC21, Q9UGR4, Q9UH41 | Gene names | FBLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-1 precursor. | |||||
|
SPRL1_HUMAN
|
||||||
| NC score | 0.055375 (rank : 78) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
LTBP3_MOUSE
|
||||||
| NC score | 0.055247 (rank : 79) | θ value | 1.06291 (rank : 93) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q61810, Q8BNQ6 | Gene names | Ltbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
LTBP2_HUMAN
|
||||||
| NC score | 0.055116 (rank : 80) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
LAMC2_HUMAN
|
||||||
| NC score | 0.054407 (rank : 81) | θ value | 1.06291 (rank : 91) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q13753, Q02536, Q02537, Q13752, Q14941, Q5VYE8 | Gene names | LAMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain) (Cell- scattering factor 140 kDa subunit) (CSF 140 kDa subunit) (Large adhesive scatter factor 140 kDa subunit) (Ladsin 140 kDa subunit). | |||||
|
EGF_MOUSE
|
||||||
| NC score | 0.053846 (rank : 82) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P01132, Q6P9J2 | Gene names | Egf | |||
|
Domain Architecture |

|
|||||
| Description | Pro-epidermal growth factor precursor (EGF) [Contains: Epidermal growth factor]. | |||||
|
EGF_HUMAN
|
||||||
| NC score | 0.053505 (rank : 83) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P01133 | Gene names | EGF | |||
|
Domain Architecture |

|
|||||
| Description | Pro-epidermal growth factor precursor (EGF) [Contains: Epidermal growth factor (Urogastrone)]. | |||||
|
TRBM_MOUSE
|
||||||
| NC score | 0.052903 (rank : 84) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P15306 | Gene names | Thbd | |||
|
Domain Architecture |

|
|||||
| Description | Thrombomodulin precursor (TM) (Fetomodulin) (CD141 antigen). | |||||
|
UROM_MOUSE
|
||||||
| NC score | 0.052879 (rank : 85) | θ value | θ > 10 (rank : 197) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q91X17, Q3TN64, Q3TP60, Q62285 | Gene names | Umod | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uromodulin precursor (Tamm-Horsfall urinary glycoprotein) (THP). | |||||
|
LTBP4_MOUSE
|
||||||
| NC score | 0.052745 (rank : 86) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
UROM_HUMAN
|
||||||
| NC score | 0.052557 (rank : 87) | θ value | θ > 10 (rank : 196) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P07911, Q540J6, Q6ZS84, Q8IYG0 | Gene names | UMOD | |||
|
Domain Architecture |

|
|||||
| Description | Uromodulin precursor (Tamm-Horsfall urinary glycoprotein) (THP). | |||||
|
NELL1_HUMAN
|
||||||
| NC score | 0.052383 (rank : 88) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q92832, Q9Y472 | Gene names | NELL1, NRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL1 precursor (NEL-like protein 1) (Nel-related protein 1). | |||||
|
TNR9_MOUSE
|
||||||
| NC score | 0.052077 (rank : 89) | θ value | 1.38821 (rank : 96) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P20334 | Gene names | Tnfrsf9, Cd137, Cd157, Ila, Ly63 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 9 precursor (4-1BB ligand receptor) (T-cell antigen 4-1BB) (CD137 antigen). | |||||
|
C1QR1_HUMAN
|
||||||
| NC score | 0.052054 (rank : 90) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NPY3, O00274 | Gene names | CD93, C1QR1, MXRA4 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C1q receptor precursor (Complement component 1 q subcomponent receptor 1) (C1qR) (C1qRp) (C1qR(p)) (C1q/MBL/SPA receptor) (CD93 antigen) (CDw93). | |||||
|
TNR16_HUMAN
|
||||||
| NC score | 0.051787 (rank : 91) | θ value | 0.47712 (rank : 80) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P08138 | Gene names | NGFR, TNFRSF16 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Gp80-LNGFR) (p75 ICD) (Low affinity neurotrophin receptor p75NTR) (CD271 antigen). | |||||
|
FBLN2_HUMAN
|
||||||
| NC score | 0.051660 (rank : 92) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P98095 | Gene names | FBLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
NELL2_HUMAN
|
||||||
| NC score | 0.051600 (rank : 93) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q99435 | Gene names | NELL2, NRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (Nel-related protein 2). | |||||
|
NELL2_MOUSE
|
||||||
| NC score | 0.051556 (rank : 94) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q61220 | Gene names | Nell2, Mel91 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (MEL91 protein). | |||||
|
C1QR1_MOUSE
|
||||||
| NC score | 0.051457 (rank : 95) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O89103, Q3U2X0 | Gene names | Cd93, Aa4, C1qr1, C1qrp, Ly68 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C1q receptor precursor (Complement component 1 q subcomponent receptor 1) (C1qRp) (C1qR(p)) (C1q/MBL/SPA receptor) (CD93 antigen) (Cell surface antigen AA4) (Lymphocyte antigen 68). | |||||
|
FBLN3_HUMAN
|
||||||
| NC score | 0.051382 (rank : 96) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q12805 | Gene names | EFEMP1, FBLN3, FBNL | |||
|
Domain Architecture |

|
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 1 precursor (Fibulin-3) (FIBL-3) (Fibrillin-like protein) (Extracellular protein S1-5). | |||||
|
FBN2_HUMAN
|
||||||
| NC score | 0.051230 (rank : 97) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
FBN3_HUMAN
|
||||||
| NC score | 0.051220 (rank : 98) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q75N90, Q75N91, Q75N92, Q75N93, Q86SJ5, Q96JP8 | Gene names | FBN3, KIAA1776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrillin-3 precursor. | |||||
|
LTBP3_HUMAN
|
||||||
| NC score | 0.050978 (rank : 99) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9NS15, O15107, Q96HB9, Q9H7K2, Q9UFN4 | Gene names | LTBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
FBLN2_MOUSE
|
||||||
| NC score | 0.050902 (rank : 100) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
FBLN5_MOUSE
|
||||||
| NC score | 0.050768 (rank : 101) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9WVH9, Q541Z7 | Gene names | Fbln5, Dance | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-5 precursor (FIBL-5) (Developmental arteries and neural crest EGF-like protein) (Dance). | |||||
|
FBLN3_MOUSE
|
||||||
| NC score | 0.050757 (rank : 102) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BPB5, Q6Y3N6 | Gene names | Efemp1, Fbln3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 1 precursor (Fibulin-3) (FIBL-3). | |||||
|
FBLN5_HUMAN
|
||||||
| NC score | 0.050498 (rank : 103) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UBX5, O75966, Q6UWA3 | Gene names | FBLN5, DANCE | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-5 precursor (FIBL-5) (Developmental arteries and neural crest EGF-like protein) (Dance) (Urine p50 protein) (UP50). | |||||
|
FST_HUMAN
|
||||||
| NC score | 0.050470 (rank : 104) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P19883, Q9BTH0 | Gene names | FST | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
FST_MOUSE
|
||||||
| NC score | 0.050319 (rank : 105) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P47931 | Gene names | Fst | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
FBN1_HUMAN
|
||||||
| NC score | 0.050293 (rank : 106) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
TRBM_HUMAN
|
||||||
| NC score | 0.050284 (rank : 107) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P07204, Q9UC32 | Gene names | THBD, THRM | |||
|
Domain Architecture |

|
|||||
| Description | Thrombomodulin precursor (TM) (Fetomodulin) (CD141 antigen). | |||||
|
PCSK5_MOUSE
|
||||||
| NC score | 0.050252 (rank : 108) | θ value | 3.0926 (rank : 114) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q04592, Q62040 | Gene names | Pcsk5 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (Subtilisin-like proprotein convertase 6) (SPC6). | |||||
|
FBLN4_HUMAN
|
||||||
| NC score | 0.050214 (rank : 109) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O95967, O75967 | Gene names | EFEMP2, FBLN4 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 2 precursor (Fibulin-4) (FIBL-4) (UPH1 protein). | |||||
|
SPRL1_MOUSE
|
||||||
| NC score | 0.050190 (rank : 110) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
FBN2_MOUSE
|
||||||
| NC score | 0.050139 (rank : 111) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q61555, Q63957 | Gene names | Fbn2, Fbn-2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
TNR16_MOUSE
|
||||||
| NC score | 0.049885 (rank : 112) | θ value | 2.36792 (rank : 108) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Z0W1 | Gene names | Ngfr, Tnfrsf16 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Low affinity neurotrophin receptor p75NTR). | |||||
|
SREC_HUMAN
|
||||||
| NC score | 0.049340 (rank : 113) | θ value | 1.81305 (rank : 99) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
WIF1_MOUSE
|
||||||
| NC score | 0.047516 (rank : 114) | θ value | 0.365318 (rank : 77) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9WUA1 | Gene names | Wif1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
LAMA1_MOUSE
|
||||||
| NC score | 0.047143 (rank : 115) | θ value | 2.36792 (rank : 104) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P19137 | Gene names | Lama1, Lama, Lama-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
NOTC3_HUMAN
|
||||||
| NC score | 0.044913 (rank : 116) | θ value | 3.0926 (rank : 113) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
LAMA3_MOUSE
|
||||||
| NC score | 0.043934 (rank : 117) | θ value | 2.36792 (rank : 105) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q61789, O08751, Q61788, Q61966, Q9JHQ7 | Gene names | Lama3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Nicein subunit alpha). | |||||
|
LAMA1_HUMAN
|
||||||
| NC score | 0.043833 (rank : 118) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 804 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P25391 | Gene names | LAMA1, LAMA | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.043050 (rank : 119) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
USH2A_HUMAN
|
||||||
| NC score | 0.042919 (rank : 120) | θ value | 3.0926 (rank : 117) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | O75445, Q5VVM9, Q6S362, Q9NS27 | Gene names | USH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein) (Usher syndrome type IIa protein). | |||||
|
PCSK5_HUMAN
|
||||||
| NC score | 0.042786 (rank : 121) | θ value | 1.38821 (rank : 95) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q92824, Q13527 | Gene names | PCSK5, PC5, PC6 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (hPC6). | |||||
|
TNR21_MOUSE
|
||||||
| NC score | 0.042559 (rank : 122) | θ value | 0.47712 (rank : 81) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9EPU5, Q91W77, Q91XH9 | Gene names | Tnfrsf21, Dr6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 21 precursor (TNFR- related death receptor 6) (Death receptor 6). | |||||
|
LAMC3_HUMAN
|
||||||
| NC score | 0.042384 (rank : 123) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9Y6N6 | Gene names | LAMC3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
WIF1_HUMAN
|
||||||
| NC score | 0.042282 (rank : 124) | θ value | 3.0926 (rank : 118) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Y5W5, Q6UXI1, Q8WVG4 | Gene names | WIF1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
DLL1_HUMAN
|
||||||
| NC score | 0.042064 (rank : 125) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
NOTC3_MOUSE
|
||||||
| NC score | 0.041629 (rank : 126) | θ value | 2.36792 (rank : 106) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
EPHB6_HUMAN
|
||||||
| NC score | 0.041549 (rank : 127) | θ value | 0.0961366 (rank : 63) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O15197 | Gene names | EPHB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 6 precursor (Tyrosine-protein kinase-defective receptor EPH-6) (HEP). | |||||
|
FRAS1_HUMAN
|
||||||
| NC score | 0.040912 (rank : 128) | θ value | 1.06291 (rank : 89) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
LAMA2_HUMAN
|
||||||
| NC score | 0.040553 (rank : 129) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
TNR21_HUMAN
|
||||||
| NC score | 0.040427 (rank : 130) | θ value | 0.813845 (rank : 86) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75509, Q96D86 | Gene names | TNFRSF21, DR6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 21 precursor (TNFR- related death receptor 6) (Death receptor 6). | |||||
|
FRAS1_MOUSE
|
||||||
| NC score | 0.039524 (rank : 131) | θ value | 0.163984 (rank : 69) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
EPHB6_MOUSE
|
||||||
| NC score | 0.039121 (rank : 132) | θ value | 0.365318 (rank : 75) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O08644, Q8BN76, Q8K0A9 | Gene names | Ephb6, Cekl | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 6 precursor (Tyrosine-protein kinase-defective receptor EPH-6) (MEP). | |||||
|
LAMA3_HUMAN
|
||||||
| NC score | 0.038088 (rank : 133) | θ value | 1.81305 (rank : 98) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
ATRN_HUMAN
|
||||||
| NC score | 0.038002 (rank : 134) | θ value | 1.38821 (rank : 94) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
ATRN_MOUSE
|
||||||
| NC score | 0.037562 (rank : 135) | θ value | 1.06291 (rank : 88) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9WU60, Q9R263, Q9WU77 | Gene names | Atrn, mg, Mgca | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany protein). | |||||
|
EPHB3_HUMAN
|
||||||
| NC score | 0.036275 (rank : 136) | θ value | 0.00228821 (rank : 48) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 996 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P54753, Q7Z740 | Gene names | EPHB3, ETK2, HEK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 3 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor HEK-2). | |||||
|
EPHB4_HUMAN
|
||||||
| NC score | 0.035556 (rank : 137) | θ value | 0.125558 (rank : 67) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P54760, Q9BTA5, Q9BXP0 | Gene names | EPHB4, HTK | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 4 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor HTK). | |||||
|
TNR3_MOUSE
|
||||||
| NC score | 0.034874 (rank : 138) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P50284 | Gene names | Ltbr, Tnfcr, Tnfrsf3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 3 precursor (Lymphotoxin-beta receptor). | |||||
|
LAMA2_MOUSE
|
||||||
| NC score | 0.034585 (rank : 139) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
EPHB3_MOUSE
|
||||||
| NC score | 0.034509 (rank : 140) | θ value | 0.00869519 (rank : 49) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 996 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P54754, Q62214 | Gene names | Ephb3, Etk2, Mdk5, Sek4 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 3 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor MDK-5) (Developmental kinase 5) (SEK-4). | |||||
|
MKS3_HUMAN
|
||||||
| NC score | 0.033197 (rank : 141) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5HYA8, Q3ZCX3, Q7Z5T8, Q8IZ06 | Gene names | TMEM67, MKS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Meckelin (Meckel syndrome type 3 protein) (Transmembrane protein 67). | |||||
|
EPHB4_MOUSE
|
||||||
| NC score | 0.033182 (rank : 142) | θ value | 0.21417 (rank : 70) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1013 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P54761, Q60627 | Gene names | Ephb4, Htk, Mdk2, Myk1 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 4 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor MDK-2) (Developmental kinase 2) (Tyrosine kinase MYK- 1). | |||||
|
EPHAA_HUMAN
|
||||||
| NC score | 0.033178 (rank : 143) | θ value | 2.36792 (rank : 103) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5JZY3, Q6NW42 | Gene names | EPHA10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephrin type-A receptor 10 precursor (EC 2.7.10.1). | |||||
|
EPHA7_HUMAN
|
||||||
| NC score | 0.032964 (rank : 144) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1053 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q15375, Q59G40, Q5VTU0, Q8N368, Q9H124 | Gene names | EPHA7, EHK3, HEK11 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 7 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EHK-3) (EPH homology kinase 3) (Receptor protein- tyrosine kinase HEK11). | |||||
|
EPHB2_HUMAN
|
||||||
| NC score | 0.032955 (rank : 145) | θ value | 0.0961366 (rank : 61) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 983 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P29323, O43477, Q5T0U6, Q5T0U7, Q5T0U8 | Gene names | EPHB2, DRT, EPTH3, ERK, HEK5 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 2 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EPH-3) (DRT) (Receptor protein-tyrosine kinase HEK5) (ERK) (NY-REN-47 antigen). | |||||
|
EPHB2_MOUSE
|
||||||
| NC score | 0.032953 (rank : 146) | θ value | 0.0961366 (rank : 62) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P54763, Q62213, Q9QVY4 | Gene names | Ephb2, Epth3, Nuk, Sek3 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 2 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EPH-3) (Neural kinase) (Nuk receptor tyrosine kinase) (SEK-3). | |||||
|
EPHA6_MOUSE
|
||||||
| NC score | 0.032853 (rank : 147) | θ value | 0.62314 (rank : 83) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q62413, Q8CCN2 | Gene names | Epha6, Ehk-2, Ehk2 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 6 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EHK-2) (EPH homology kinase 2). | |||||
|
TNR25_HUMAN
|
||||||
| NC score | 0.032779 (rank : 148) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q93038, O00275, O00276, O00277, O00278, O00279, O00280, O14865, O14866, P78507, P78515, Q92983, Q93036, Q93037, Q99722, Q99830, Q99831, Q9BY86, Q9UME0, Q9UME1, Q9UME5 | Gene names | TNFRSF25, APO3, DDR3, DR3, TNFRSF12, WSL, WSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 25 precursor (WSL-1 protein) (Apoptosis-mediating receptor DR3) (Apoptosis-mediating receptor TRAMP) (Death domain receptor 3) (WSL protein) (Apoptosis- inducing receptor AIR) (Apo-3) (Lymphocyte-associated receptor of death) (LARD). | |||||
|
EPHA6_HUMAN
|
||||||
| NC score | 0.032402 (rank : 149) | θ value | 0.47712 (rank : 78) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UF33 | Gene names | EPHA6, EHK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ephrin type-A receptor 6 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EHK-2) (EPH homology kinase 2). | |||||
|
TR10C_HUMAN
|
||||||
| NC score | 0.032334 (rank : 150) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14798, O14755, Q6UXM5 | Gene names | TNFRSF10C, DCR1, LIT, TRAILR3, TRID | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10C precursor (Decoy receptor 1) (DcR1) (Decoy TRAIL receptor without death domain) (TNF- related apoptosis-inducing ligand receptor 3) (TRAIL receptor 3) (TRAIL-R3) (Trail receptor without an intracellular domain) (Lymphocyte inhibitor of TRAIL) (Antagonist decoy receptor for TRAIL/Apo-2L) (CD263 antigen). | |||||
|
TNR5_MOUSE
|
||||||
| NC score | 0.031945 (rank : 151) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P27512, Q99NE0, Q99NE1, Q99NE2, Q99NE3 | Gene names | Cd40, Tnfrsf5 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 5 precursor (CD40L receptor) (B-cell surface antigen CD40) (BP50) (CDw40). | |||||
|
TENR_HUMAN
|
||||||
| NC score | 0.031018 (rank : 152) | θ value | 2.36792 (rank : 107) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q92752, Q15568, Q5R3G0 | Gene names | TNR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin). | |||||
|
EPHA2_MOUSE
|
||||||
| NC score | 0.030967 (rank : 153) | θ value | 0.125558 (rank : 66) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q03145, Q60633, Q62212 | Gene names | Epha2, Eck, Sek2 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 2 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor ECK) (Epithelial cell kinase) (MPK-5) (SEK-2). | |||||
|
WBP1_HUMAN
|
||||||
| NC score | 0.030914 (rank : 154) | θ value | 0.365318 (rank : 76) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96G27, O95637 | Gene names | WBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 1 (WBP-1). | |||||
|
EPHA4_MOUSE
|
||||||
| NC score | 0.030791 (rank : 155) | θ value | 0.365318 (rank : 74) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 982 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q03137 | Gene names | Epha4, Sek, Sek1 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 4 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor SEK) (MPK-3). | |||||
|
TR19L_HUMAN
|
||||||
| NC score | 0.030657 (rank : 156) | θ value | 3.0926 (rank : 116) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q969Z4, Q86V34, Q96JU1, Q9BUX7 | Gene names | TNFRSF19L, RELT | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 19L precursor (Receptor expressed in lymphoid tissues). | |||||
|
PKHG4_HUMAN
|
||||||
| NC score | 0.030390 (rank : 157) | θ value | 0.0330416 (rank : 55) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q58EX7, Q4G0J8, Q4H485, Q56A69, Q9H7K4, Q9UFW0 | Gene names | PLEKHG4, PRTPHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Puratrophin-1 (Pleckstrin homology domain-containing family G member 4) (Purkinje cell atrophy-associated protein 1). | |||||
|
TENR_MOUSE
|
||||||
| NC score | 0.030295 (rank : 158) | θ value | 4.03905 (rank : 125) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8BYI9, O88717 | Gene names | Tnr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin) (Neural recognition molecule J1-160/180). | |||||
|
EPHA1_HUMAN
|
||||||
| NC score | 0.029742 (rank : 159) | θ value | 2.36792 (rank : 102) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P21709, Q15405 | Gene names | EPHA1, EPH, EPHT, EPHT1 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 1 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EPH). | |||||
|
EPHA1_MOUSE
|
||||||
| NC score | 0.029313 (rank : 160) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q60750, Q8CED9, Q9ESJ2 | Gene names | Epha1, Esk | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 1 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor ESK). | |||||
|
EPHA4_HUMAN
|
||||||
| NC score | 0.029116 (rank : 161) | θ value | 1.81305 (rank : 97) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P54764 | Gene names | EPHA4, HEK8, SEK | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 4 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor SEK) (Receptor protein-tyrosine kinase HEK8). | |||||
|
CJ047_MOUSE
|
||||||
| NC score | 0.027839 (rank : 162) | θ value | 0.62314 (rank : 82) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C5R2, Q8R017, Q8R2Z9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47 homolog. | |||||
|
EPHB1_HUMAN
|
||||||
| NC score | 0.027271 (rank : 163) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 988 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P54762, O43569, O95142, O95143 | Gene names | EPHB1, EPHT2, NET | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 1 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EPH-2) (NET) (HEK6) (ELK). | |||||
|
NLTP_HUMAN
|
||||||
| NC score | 0.027102 (rank : 164) | θ value | 0.813845 (rank : 84) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22307, Q15432, Q16622, Q99430 | Gene names | SCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Nonspecific lipid-transfer protein (EC 2.3.1.176) (Propanoyl-CoA C- acyltransferase) (NSL-TP) (Sterol carrier protein 2) (SCP-2) (Sterol carrier protein X) (SCP-X) (SCP-chi) (SCPX). | |||||
|
EPHA2_HUMAN
|
||||||
| NC score | 0.027020 (rank : 165) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P29317 | Gene names | EPHA2, ECK | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 2 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor ECK) (Epithelial cell kinase). | |||||
|
NLTP_MOUSE
|
||||||
| NC score | 0.026957 (rank : 166) | θ value | 0.813845 (rank : 85) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32020, Q9DBM7 | Gene names | Scp2, Scp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Nonspecific lipid-transfer protein (EC 2.3.1.176) (Propanoyl-CoA C- acyltransferase) (NSL-TP) (Sterol carrier protein 2) (SCP-2) (Sterol carrier protein X) (SCP-X) (SCP-chi) (SCPX). | |||||
|
MKS3_MOUSE
|
||||||
| NC score | 0.026275 (rank : 167) | θ value | 4.03905 (rank : 124) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BR76, Q78U07 | Gene names | Tmem67, Mks3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Meckelin (Meckel syndrome type 3 protein homolog) (Transmembrane protein 67). | |||||
|
KR102_HUMAN
|
||||||
| NC score | 0.025992 (rank : 168) | θ value | 4.03905 (rank : 123) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
TR10B_HUMAN
|
||||||
| NC score | 0.025891 (rank : 169) | θ value | 3.0926 (rank : 115) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O14763, O14720, O15508, O15517, O15531, Q9BVE0 | Gene names | TNFRSF10B, DR5, KILLER, TRAILR2, TRICK2, ZTNFR9 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10B precursor (Death receptor 5) (TNF-related apoptosis-inducing ligand receptor 2) (TRAIL receptor 2) (TRAIL-R2) (CD262 antigen). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.022168 (rank : 170) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
AASS_HUMAN
|
||||||
| NC score | 0.020346 (rank : 171) | θ value | 1.06291 (rank : 87) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UDR5, O95462 | Gene names | AASS | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-aminoadipic semialdehyde synthase, mitochondrial precursor (LKR/SDH) [Includes: Lysine ketoglutarate reductase (EC 1.5.1.8) (LOR) (LKR); Saccharopine dehydrogenase (EC 1.5.1.9) (SDH)]. | |||||
|
RSPO1_HUMAN
|
||||||
| NC score | 0.017820 (rank : 172) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q2MKA7, Q5T0F2, Q8N7L5 | Gene names | RSPO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-1 precursor (Roof plate-specific spondin-1) (hRspo1). | |||||
|
FUT1_MOUSE
|
||||||
| NC score | 0.016497 (rank : 173) | θ value | 1.06291 (rank : 90) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09160 | Gene names | Fut1 | |||
|
Domain Architecture |

|
|||||
| Description | Galactoside 2-alpha-L-fucosyltransferase 1 (EC 2.4.1.69) (GDP-L- fucose:beta-D-galactoside 2-alpha-L-fucosyltransferase 1) (Alpha(1,2)FT 1) (Fucosyltransferase 1). | |||||
|
CELR1_HUMAN
|
||||||
| NC score | 0.015192 (rank : 174) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NYQ6, O95722, Q5TH47, Q9BWQ5, Q9Y506, Q9Y526 | Gene names | CELSR1, CDHF9, FMI2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor (Flamingo homolog 2) (hFmi2). | |||||
|
NDUB9_HUMAN
|
||||||
| NC score | 0.014169 (rank : 175) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y6M9, Q9UQE8 | Gene names | NDUFB9, UQOR22 | |||
|
Domain Architecture |
|
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9 (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase B22 subunit) (Complex I-B22) (CI-B22). | |||||
|
TNR27_MOUSE
|
||||||
| NC score | 0.013917 (rank : 176) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BX35, Q8BM50 | Gene names | Eda2r, Tnfrsf27, Xedar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 27 (X-linked ectodysplasin-A2 receptor) (EDA-A2 receptor). | |||||
|
KR261_HUMAN
|
||||||
| NC score | 0.013561 (rank : 177) | θ value | 3.0926 (rank : 110) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PEX3 | Gene names | KRTAP26-1, KAP26.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 26-1. | |||||
|
APBB2_HUMAN
|
||||||
| NC score | 0.012860 (rank : 178) | θ value | 4.03905 (rank : 119) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
APBB2_MOUSE
|
||||||
| NC score | 0.011557 (rank : 179) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBR4, Q6DFX8 | Gene names | Apbb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2. | |||||
|
DIAP2_HUMAN
|
||||||
| NC score | 0.011052 (rank : 180) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60879, O60878, Q8WX06, Q8WX48, Q9UJL2 | Gene names | DIAPH2, DIA | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2). | |||||
|
PUS7_HUMAN
|
||||||
| NC score | 0.010666 (rank : 181) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96PZ0, Q75MG4, Q9NX19 | Gene names | PUS7, KIAA1897 | |||
|
Domain Architecture |

|
|||||
| Description | Pseudouridylate synthase 7 homolog (EC 5.4.99.-). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.009688 (rank : 182) | θ value | 0.21417 (rank : 71) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
LRP12_HUMAN
|
||||||
| NC score | 0.009461 (rank : 183) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y561 | Gene names | LRP12, ST7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor (Suppressor of tumorigenicity protein 7). | |||||
|
SNX6_HUMAN
|
||||||
| NC score | 0.009045 (rank : 184) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UNH7, Q9Y449 | Gene names | SNX6 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-6 (TRAF4-associated factor 2). | |||||
|
SNX6_MOUSE
|
||||||
| NC score | 0.009036 (rank : 185) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P8X1 | Gene names | Snx6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-6. | |||||
|
RABX5_HUMAN
|
||||||
| NC score | 0.008273 (rank : 186) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJ41 | Gene names | RABGEF1, RABEX5 | |||
|
Domain Architecture |

|
|||||
| Description | Rab5 GDP/GTP exchange factor (Rabex-5) (Rabaptin-5-associated exchange factor for Rab5). | |||||
|
CSTN2_HUMAN
|
||||||
| NC score | 0.007137 (rank : 187) | θ value | 2.36792 (rank : 101) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H4D0, Q9BSS0 | Gene names | CLSTN2, CS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calsyntenin-2 precursor. | |||||
|
CASR_HUMAN
|
||||||
| NC score | 0.007091 (rank : 188) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P41180, Q13912, Q16108, Q16109, Q16110, Q16379, Q4PJ19 | Gene names | CASR, GPRC2A, PCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Extracellular calcium-sensing receptor precursor (CaSR) (Parathyroid Cell calcium-sensing receptor). | |||||
|
ARI4A_HUMAN
|
||||||
| NC score | 0.006795 (rank : 189) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
CRIS1_MOUSE
|
||||||
| NC score | 0.005049 (rank : 190) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q03401 | Gene names | Crisp1, Aeg-1, Aeg1 | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine-rich secretory protein 1 precursor (CRISP-1) (Acidic epididymal glycoprotein 1) (Sperm-coating glycoprotein 1) (SCP 1). | |||||
|
TEP1_HUMAN
|
||||||
| NC score | 0.004658 (rank : 191) | θ value | 1.81305 (rank : 100) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99973 | Gene names | TEP1, TLP1, TP1 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase protein component 1 (Telomerase-associated protein 1) (Telomerase protein 1) (p240) (p80 telomerase homolog). | |||||
|
GAK8_HUMAN
|
||||||
| NC score | 0.003838 (rank : 192) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HDB9 | Gene names | ERVK5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q12.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(II) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
AN32A_HUMAN
|
||||||
| NC score | 0.002572 (rank : 193) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P39687 | Gene names | ANP32A, C15orf1, LANP, MAPM, PHAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member A (Potent heat-stable protein phosphatase 2A inhibitor I1PP2A) (Acidic nuclear phosphoprotein pp32) (Leucine-rich acidic nuclear protein) (Lanp) (Putative HLA-DR-associated protein I) (PHAPI) (Mapmodulin). | |||||
|
MMP17_MOUSE
|
||||||
| NC score | 0.001133 (rank : 194) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0S3 | Gene names | Mmp17, Mt4mmp | |||
|
Domain Architecture |

|
|||||
| Description | Matrix metalloproteinase-17 precursor (EC 3.4.24.-) (MMP-17) (Membrane-type matrix metalloproteinase 4) (MT-MMP 4) (Membrane-type-4 matrix metalloproteinase) (MT4-MMP). | |||||
|
PCDG4_HUMAN
|
||||||
| NC score | -0.000202 (rank : 195) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y5G9, Q9Y5D3 | Gene names | PCDHGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A4 precursor (PCDH-gamma-A4). | |||||
|
PCDG5_HUMAN
|
||||||
| NC score | -0.000218 (rank : 196) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y5G8, Q9Y5D2 | Gene names | PCDHGA5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A5 precursor (PCDH-gamma-A5). | |||||
|
ANR26_HUMAN
|
||||||
| NC score | -0.004241 (rank : 197) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||