Please be patient as the page loads
|
TICN3_MOUSE
|
||||||
| SwissProt Accessions | Q8BKV0, Q9ER59 | Gene names | Spock3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TICN3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.996990 (rank : 2) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9BQ16, O75705, Q6UW53, Q96Q26 | Gene names | SPOCK3, TICN3 | |||
|
Domain Architecture |
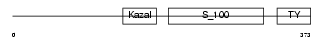
|
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
TICN3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8BKV0, Q9ER59 | Gene names | Spock3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
TICN1_MOUSE
|
||||||
| θ value | 1.63136e-134 (rank : 3) | NC score | 0.980192 (rank : 3) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q62288 | Gene names | Spock1, Spock, Ticn1 | |||
|
Domain Architecture |
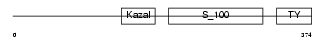
|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
TICN1_HUMAN
|
||||||
| θ value | 8.37844e-131 (rank : 4) | NC score | 0.978388 (rank : 4) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q08629 | Gene names | SPOCK1, SPOCK, TIC1, TICN1 | |||
|
Domain Architecture |
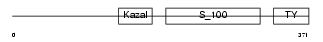
|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
TICN2_HUMAN
|
||||||
| θ value | 6.90186e-109 (rank : 5) | NC score | 0.974619 (rank : 6) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q92563, Q6UW87 | Gene names | SPOCK2, KIAA0275, TICN2 | |||
|
Domain Architecture |
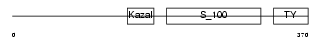
|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
TICN2_MOUSE
|
||||||
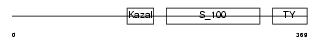
| θ value | 1.30163e-107 (rank : 6) | NC score | 0.975033 (rank : 5) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9ER58 | Gene names | Spock2, Ticn2 | |||
|
Domain Architecture |
|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
SPRL1_MOUSE
|
||||||
| θ value | 8.67504e-11 (rank : 7) | NC score | 0.416573 (rank : 19) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
NID1_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 8) | NC score | 0.236181 (rank : 39) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P14543, Q14942, Q59FL2, Q5TAF2, Q5TAF3, Q86XD7 | Gene names | NID1, NID | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
NID1_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 9) | NC score | 0.232867 (rank : 40) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P10493, Q8BQI3, Q8C3U8, Q8C9P6 | Gene names | Nid1, Ent | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
SPRC_HUMAN
|
||||||
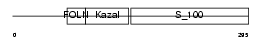
| θ value | 3.29651e-10 (rank : 10) | NC score | 0.493575 (rank : 9) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P09486 | Gene names | SPARC, ON | |||
|
Domain Architecture |
|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
SPRL1_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 11) | NC score | 0.458201 (rank : 16) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
SPRC_MOUSE
|
||||||
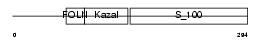
| θ value | 7.34386e-10 (rank : 12) | NC score | 0.480702 (rank : 11) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P07214 | Gene names | Sparc | |||
|
Domain Architecture |
|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
HG2A_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 13) | NC score | 0.555450 (rank : 8) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P04233, Q14597, Q29832, Q5U0J8, Q8WLP6 | Gene names | CD74, DHLAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HLA class II histocompatibility antigen gamma chain (HLA-DR antigens- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (p33) (CD74 antigen). | |||||
|
THYG_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 14) | NC score | 0.277570 (rank : 37) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08710, O88590, Q9QWY7 | Gene names | Tg, Tgn | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
NID2_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 15) | NC score | 0.222502 (rank : 45) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
NID2_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 16) | NC score | 0.223351 (rank : 44) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O88322 | Gene names | Nid2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Entactin-2). | |||||
|
HG2A_MOUSE
|
||||||
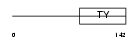
| θ value | 4.0297e-08 (rank : 17) | NC score | 0.571321 (rank : 7) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P04441, O19452 | Gene names | Cd74, Ii | |||
|
Domain Architecture |
|
|||||
| Description | H-2 class II histocompatibility antigen gamma chain (MHC class II- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (CD74 antigen). | |||||
|
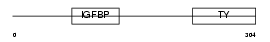
IBP3_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 18) | NC score | 0.445982 (rank : 17) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P17936, Q2V509, Q6P1M6 | Gene names | IGFBP3, IBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
IBP3_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 19) | NC score | 0.439499 (rank : 18) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P47878 | Gene names | Igfbp3, Igfbp-3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
THYG_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 20) | NC score | 0.270708 (rank : 38) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P01266, O15274, O43899, Q15593, Q15948, Q9NYR1, Q9NYR2, Q9UMZ0, Q9UNY3 | Gene names | TG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 21) | NC score | 0.184340 (rank : 47) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
IBP6_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 22) | NC score | 0.467714 (rank : 14) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P24592, Q14492 | Gene names | IGFBP6, IBP6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
IBP5_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 23) | NC score | 0.408239 (rank : 21) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P24593 | Gene names | IGFBP5, IBP5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP5_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 24) | NC score | 0.404087 (rank : 22) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q07079 | Gene names | Igfbp5, Igfbp-5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
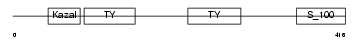
SMOC1_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 25) | NC score | 0.468514 (rank : 13) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9H4F8, Q96F78 | Gene names | SMOC1 | |||
|
Domain Architecture |
|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1). | |||||
|
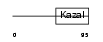
FSTL1_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 26) | NC score | 0.383245 (rank : 24) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q12841 | Gene names | FSTL1, FRP | |||
|
Domain Architecture |
|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1). | |||||
|
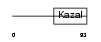
FSTL1_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 27) | NC score | 0.384403 (rank : 23) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62356, Q99JI9 | Gene names | Fstl1, Frp, Fstl, Tsc36 | |||
|
Domain Architecture |
|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1) (TGF- beta-inducible protein TSC-36). | |||||
|
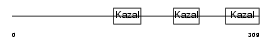
FST_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 28) | NC score | 0.346642 (rank : 30) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P19883, Q9BTH0 | Gene names | FST | |||
|
Domain Architecture |
|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
SMOC2_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 29) | NC score | 0.481074 (rank : 10) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 52 | |
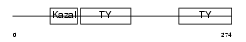
| SwissProt Accessions | Q9H3U7, Q5TAT7, Q5TAT8, Q86VV9, Q96SF3, Q9H1L3, Q9H1L4, Q9H3U0, Q9H4F7, Q9HCV2 | Gene names | SMOC2, SMAP2 | |||
|
Domain Architecture |
|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2) (Smooth muscle-associated protein 2) (SMAP-2). | |||||
|
SMOC2_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 30) | NC score | 0.476554 (rank : 12) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 55 | |
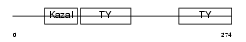
| SwissProt Accessions | Q8CD91, Q8VCM1, Q9D9K2, Q9ER95 | Gene names | Smoc2 | |||
|
Domain Architecture |
|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2). | |||||
|
FST_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 31) | NC score | 0.340751 (rank : 33) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 42 | |
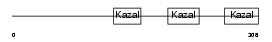
| SwissProt Accessions | P47931 | Gene names | Fst | |||
|
Domain Architecture |
|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
FSTL4_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 32) | NC score | 0.169738 (rank : 48) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6MZW2, Q8TBU0, Q9UPU1 | Gene names | FSTL4, KIAA1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4). | |||||
|
SMOC1_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 33) | NC score | 0.465245 (rank : 15) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 55 | |
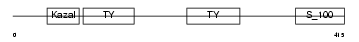
| SwissProt Accessions | Q8BLY1, Q9WVN9 | Gene names | Smoc1, Srg | |||
|
Domain Architecture |
|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1) (SPARC-related gene protein). | |||||
|
FSTL4_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 34) | NC score | 0.156981 (rank : 54) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5STE3, Q5DU03 | Gene names | Fstl4, Kiaa1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4) (m- D/Bsp120I 1-1). | |||||
|
IBP1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 35) | NC score | 0.375528 (rank : 25) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P08833, Q8IYP5 | Gene names | IGFBP1, IBP1 | |||
|
Domain Architecture |
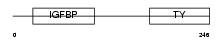
|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1) (Placental protein 12) (PP12). | |||||
|
IBP4_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 36) | NC score | 0.370555 (rank : 27) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P22692 | Gene names | IGFBP4, IBP4 | |||
|
Domain Architecture |
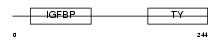
|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
IBP4_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 37) | NC score | 0.373993 (rank : 26) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P47879, O35666 | Gene names | Igfbp4, Igfbp-4 | |||
|
Domain Architecture |
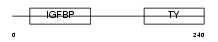
|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
FSTL5_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 38) | NC score | 0.133955 (rank : 56) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N475, Q9NSW7, Q9ULF7 | Gene names | FSTL5, KIAA1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5). | |||||
|
IBP6_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 39) | NC score | 0.408897 (rank : 20) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P47880 | Gene names | Igfbp6, Igfbp-6 | |||
|
Domain Architecture |
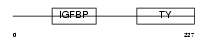
|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
ISK5_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 40) | NC score | 0.231905 (rank : 42) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NQ38, O75770, Q96PP2, Q96PP3 | Gene names | SPINK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 5 precursor (Lympho-epithelial Kazal-type-related inhibitor) (LEKTI) [Contains: Hemofiltrate peptide HF6478; Hemofiltrate peptide HF7665]. | |||||
|
TEFF2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 41) | NC score | 0.231519 (rank : 43) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UIK5, Q4ZFW4, Q53H90, Q53RE1, Q8N2R5, Q9NR15, Q9NSS5, Q9P2Y9, Q9UK65 | Gene names | TMEFF2, HPP1, TENB2, TPEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Transmembrane protein containing epidermal growth factor and follistatin domains) (Tomoregulin) (TR) (Hyperplastic polyposis protein 1). | |||||
|
TEFF2_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 42) | NC score | 0.232212 (rank : 41) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QYM9, Q3UY20, Q8CDH1, Q9JJE3 | Gene names | Tmeff2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Tomoregulin) (TR). | |||||
|
IBP2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 43) | NC score | 0.348146 (rank : 29) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P18065, Q14619, Q9UCL3 | Gene names | IGFBP2, BP2 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
IBP2_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 44) | NC score | 0.360173 (rank : 28) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P47877, Q61722 | Gene names | Igfbp2, Igfbp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
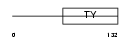
TACD1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 45) | NC score | 0.345007 (rank : 32) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P16422, P18180, Q96C47 | Gene names | TACSTD1, GA733-2, M1S2, TROP1 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor-associated calcium signal transducer 1 precursor (Major gastrointestinal tumor-associated protein GA733-2) (Epithelial cell surface antigen) (Epithelial glycoprotein) (EGP) (Adenocarcinoma- associated antigen) (KSA) (KS 1/4 antigen) (Cell surface glycoprotein Trop-1) (CD326 antigen). | |||||
|
FSTL3_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 46) | NC score | 0.281668 (rank : 35) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9EQC7 | Gene names | Fstl3, Flrg | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
FSTL5_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 47) | NC score | 0.126710 (rank : 57) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BFR2, Q80TG3, Q8C4T3 | Gene names | Fstl5, Kiaa1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5) (m- D/Bsp120I 1-2). | |||||
|
FSTL3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 48) | NC score | 0.291248 (rank : 34) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O95633 | Gene names | FSTL3, FLRG | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
IBP1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 49) | NC score | 0.346390 (rank : 31) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
|
KAZD1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 50) | NC score | 0.159989 (rank : 50) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96I82, Q6ZMB1, Q9BQ73 | Gene names | KAZALD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor. | |||||
|
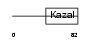
ISK7_HUMAN
|
||||||
| θ value | 0.21417 (rank : 51) | NC score | 0.160285 (rank : 49) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P58062 | Gene names | SPINK7, ECG2 | |||
|
Domain Architecture |
|
|||||
| Description | Serine protease inhibitor Kazal-type 7 precursor (Esophagus cancer- related gene 2 protein) (ECRG-2). | |||||
|
ISK7_MOUSE
|
||||||
| θ value | 0.365318 (rank : 52) | NC score | 0.207484 (rank : 46) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6IE32 | Gene names | Spink7, Ecg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 7 precursor (Esophagus cancer- related gene 2 protein) (ECRG-2). | |||||
|
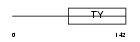
TACD2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 53) | NC score | 0.280269 (rank : 36) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P09758, Q15658, Q96QD2 | Gene names | TACSTD2, GA733-1, M1S1, TROP2 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor-associated calcium signal transducer 2 precursor (Pancreatic carcinoma marker protein GA733-1) (Cell surface glycoprotein Trop-2). | |||||
|
ISK6_HUMAN
|
||||||
| θ value | 0.62314 (rank : 54) | NC score | 0.159571 (rank : 52) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6UWN8, Q8N5P0 | Gene names | SPINK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
TRI17_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.011433 (rank : 83) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y577 | Gene names | TRIM17, RBCC, RNF16, TERF | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 17 (Testis RING finger protein) (RING finger protein 16). | |||||
|
STAB2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.045417 (rank : 71) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
SO4A1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.019334 (rank : 75) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96BD0, Q9H4T7, Q9H4T8, Q9H8P2, Q9NWX8, Q9UI35, Q9UIG7 | Gene names | SLCO4A1, OATP1, OATPE, SLC21A12 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 4A1 (Solute carrier family 21 member 12) (Sodium-independent organic anion transporter E) (Organic anion-transporting polypeptide E) (OATP-E) (Colon organic anion transporter) (Organic anion transporter polypeptide-related protein 1) (OATP-RP1) (OATPRP1) (POAT). | |||||
|
ZAN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.019332 (rank : 76) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
APBB2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.017110 (rank : 78) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
ISK3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.124421 (rank : 60) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P09036, Q5M9M3 | Gene names | Spink3 | |||
|
Domain Architecture |
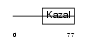
|
|||||
| Description | Serine protease inhibitor Kazal-type 3 precursor (Prostatic secretory glycoprotein) (P12). | |||||
|
TRI17_MOUSE
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.007558 (rank : 87) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 620 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TPM3, Q99PP8 | Gene names | Trim17 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 17. | |||||
|
HTRA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.084283 (rank : 64) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q92743, Q9UNS5 | Gene names | HTRA1, HTRA, PRSS11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-) (L56). | |||||
|
HTRA3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.078748 (rank : 65) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P83110, Q7Z7A2 | Gene names | HTRA3, PRSP | |||
|
Domain Architecture |
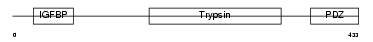
|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease). | |||||
|
HTRA3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.075057 (rank : 67) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D236, Q6WLC5, Q6YDR0 | Gene names | Htra3, Prsp, Tasp | |||
|
Domain Architecture |
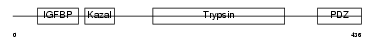
|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease) (Toll-associated serine protease). | |||||
|
P85B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.011425 (rank : 84) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
SO4A1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.016619 (rank : 79) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K078, Q8BZT4 | Gene names | Slco4a1, Oatpe, Slc21a12 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 4A1 (Solute carrier family 21 member 12) (Sodium-independent organic anion transporter E) (Organic anion-transporting polypeptide E) (OATP-E). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.013752 (rank : 81) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TF2AY_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.014169 (rank : 80) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UNN4 | Gene names | GTF2A1LF, ALF | |||
|
Domain Architecture |

|
|||||
| Description | TFIIA-alpha and beta-like factor (General transcription factor II A, 1-like factor). | |||||
|
VPS72_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.028556 (rank : 72) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15906, Q53GJ2, Q5U0R4 | Gene names | VPS72, TCFL1, YL1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
VPS72_MOUSE
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.028220 (rank : 73) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
BRCA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.017136 (rank : 77) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51587, O00183, O15008, Q13879 | Gene names | BRCA2, FANCD1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein (Fanconi anemia group D1 protein). | |||||
|
CAC1C_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.003603 (rank : 88) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
IPK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.143397 (rank : 55) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P00995 | Gene names | SPINK1, PSTI | |||
|
Domain Architecture |
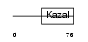
|
|||||
| Description | Pancreatic secretory trypsin inhibitor precursor (Tumor-associated trypsin inhibitor) (TATI) (Serine protease inhibitor Kazal-type 1). | |||||
|
ISK4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.159595 (rank : 51) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60575 | Gene names | SPINK4 | |||
|
Domain Architecture |
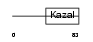
|
|||||
| Description | Serine protease inhibitor Kazal-type 4 precursor (Peptide PEC-60 homolog). | |||||
|
ISK6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.125186 (rank : 58) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BT20 | Gene names | Spink6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
HTRA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.097009 (rank : 62) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9R118 | Gene names | Htra1, Htra, Prss11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-). | |||||
|
IPK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.104236 (rank : 61) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P20155 | Gene names | SPINK2 | |||
|
Domain Architecture |
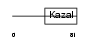
|
|||||
| Description | Serine protease inhibitor Kazal-type 2 precursor (Acrosin-trypsin inhibitor) (HUSI-II). | |||||
|
LAMB2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.022677 (rank : 74) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61292, Q62182 | Gene names | Lamb2, Lams | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (S-LAM). | |||||
|
TCAL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.012785 (rank : 82) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q969E4 | Gene names | TCEAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
KAZD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.125081 (rank : 59) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BJ66 | Gene names | Kazald1, Bono1, Igfbprp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor (IGFBP-related protein 10) (IGFBP-rP10) (Bone and odontoblast expressed protein 1). | |||||
|
P85B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.008874 (rank : 85) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08908 | Gene names | Pik3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
TNR11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.008385 (rank : 86) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y6Q6 | Gene names | TNFRSF11A, RANK | |||
|
Domain Architecture |
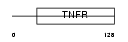
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11A precursor (Receptor activator of NF-KB) (Osteoclast differentiation factor receptor) (ODFR) (CD265 antigen). | |||||
|
UBP48_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.003213 (rank : 89) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
IBP7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.076874 (rank : 66) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q16270, Q07822 | Gene names | IGFBP7, MAC25, PSF | |||
|
Domain Architecture |
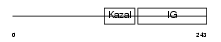
|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein) (Prostacyclin-stimulating factor) (PGI2-stimulating factor) (IGFBP-rP1). | |||||
|
IBP7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.074421 (rank : 68) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61581, O88812, Q9EQW0 | Gene names | Igfbp7, Mac25 | |||
|
Domain Architecture |
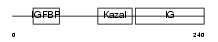
|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein). | |||||
|
IPK2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.085101 (rank : 63) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BMY7 | Gene names | Spink2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 2 precursor. | |||||
|
ISK4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.158245 (rank : 53) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O35679 | Gene names | Spink4, Mpgc60 | |||
|
Domain Architecture |
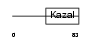
|
|||||
| Description | Serine protease inhibitor Kazal-type 4 precursor (Peptide PEC-60 homolog) (MPGC60 protein). | |||||
|
RECK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.066675 (rank : 69) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O95980, Q8WX37 | Gene names | RECK | |||
|
Domain Architecture |

|
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (hRECK) (Suppressor of tumorigenicity 15) (ST15). | |||||
|
RECK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.060898 (rank : 70) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z0J1 | Gene names | Reck | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (mRECK). | |||||
|
TICN3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q8BKV0, Q9ER59 | Gene names | Spock3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
TICN3_HUMAN
|
||||||
| NC score | 0.996990 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9BQ16, O75705, Q6UW53, Q96Q26 | Gene names | SPOCK3, TICN3 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
TICN1_MOUSE
|
||||||
| NC score | 0.980192 (rank : 3) | θ value | 1.63136e-134 (rank : 3) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q62288 | Gene names | Spock1, Spock, Ticn1 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
TICN1_HUMAN
|
||||||
| NC score | 0.978388 (rank : 4) | θ value | 8.37844e-131 (rank : 4) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q08629 | Gene names | SPOCK1, SPOCK, TIC1, TICN1 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
TICN2_MOUSE
|
||||||
| NC score | 0.975033 (rank : 5) | θ value | 1.30163e-107 (rank : 6) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9ER58 | Gene names | Spock2, Ticn2 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
TICN2_HUMAN
|
||||||
| NC score | 0.974619 (rank : 6) | θ value | 6.90186e-109 (rank : 5) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q92563, Q6UW87 | Gene names | SPOCK2, KIAA0275, TICN2 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
HG2A_MOUSE
|
||||||
| NC score | 0.571321 (rank : 7) | θ value | 4.0297e-08 (rank : 17) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P04441, O19452 | Gene names | Cd74, Ii | |||
|
Domain Architecture |

|
|||||
| Description | H-2 class II histocompatibility antigen gamma chain (MHC class II- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (CD74 antigen). | |||||
|
HG2A_HUMAN
|
||||||
| NC score | 0.555450 (rank : 8) | θ value | 1.38499e-08 (rank : 13) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P04233, Q14597, Q29832, Q5U0J8, Q8WLP6 | Gene names | CD74, DHLAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HLA class II histocompatibility antigen gamma chain (HLA-DR antigens- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (p33) (CD74 antigen). | |||||
|
SPRC_HUMAN
|
||||||
| NC score | 0.493575 (rank : 9) | θ value | 3.29651e-10 (rank : 10) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P09486 | Gene names | SPARC, ON | |||
|
Domain Architecture |

|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
SMOC2_HUMAN
|
||||||
| NC score | 0.481074 (rank : 10) | θ value | 5.44631e-05 (rank : 29) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H3U7, Q5TAT7, Q5TAT8, Q86VV9, Q96SF3, Q9H1L3, Q9H1L4, Q9H3U0, Q9H4F7, Q9HCV2 | Gene names | SMOC2, SMAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2) (Smooth muscle-associated protein 2) (SMAP-2). | |||||
|
SPRC_MOUSE
|
||||||
| NC score | 0.480702 (rank : 11) | θ value | 7.34386e-10 (rank : 12) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P07214 | Gene names | Sparc | |||
|
Domain Architecture |

|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
SMOC2_MOUSE
|
||||||
| NC score | 0.476554 (rank : 12) | θ value | 5.44631e-05 (rank : 30) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8CD91, Q8VCM1, Q9D9K2, Q9ER95 | Gene names | Smoc2 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2). | |||||
|
SMOC1_HUMAN
|
||||||
| NC score | 0.468514 (rank : 13) | θ value | 1.87187e-05 (rank : 25) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9H4F8, Q96F78 | Gene names | SMOC1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1). | |||||
|
IBP6_HUMAN
|
||||||
| NC score | 0.467714 (rank : 14) | θ value | 2.88788e-06 (rank : 22) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P24592, Q14492 | Gene names | IGFBP6, IBP6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
SMOC1_MOUSE
|
||||||
| NC score | 0.465245 (rank : 15) | θ value | 0.000461057 (rank : 33) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8BLY1, Q9WVN9 | Gene names | Smoc1, Srg | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1) (SPARC-related gene protein). | |||||
|
SPRL1_HUMAN
|
||||||
| NC score | 0.458201 (rank : 16) | θ value | 4.30538e-10 (rank : 11) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
IBP3_HUMAN
|
||||||
| NC score | 0.445982 (rank : 17) | θ value | 1.99992e-07 (rank : 18) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P17936, Q2V509, Q6P1M6 | Gene names | IGFBP3, IBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
IBP3_MOUSE
|
||||||
| NC score | 0.439499 (rank : 18) | θ value | 7.59969e-07 (rank : 19) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P47878 | Gene names | Igfbp3, Igfbp-3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
SPRL1_MOUSE
|
||||||
| NC score | 0.416573 (rank : 19) | θ value | 8.67504e-11 (rank : 7) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
IBP6_MOUSE
|
||||||
| NC score | 0.408897 (rank : 20) | θ value | 0.00509761 (rank : 39) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P47880 | Gene names | Igfbp6, Igfbp-6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
IBP5_HUMAN
|
||||||
| NC score | 0.408239 (rank : 21) | θ value | 1.43324e-05 (rank : 23) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P24593 | Gene names | IGFBP5, IBP5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP5_MOUSE
|
||||||
| NC score | 0.404087 (rank : 22) | θ value | 1.87187e-05 (rank : 24) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q07079 | Gene names | Igfbp5, Igfbp-5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
FSTL1_MOUSE
|
||||||
| NC score | 0.384403 (rank : 23) | θ value | 5.44631e-05 (rank : 27) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62356, Q99JI9 | Gene names | Fstl1, Frp, Fstl, Tsc36 | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1) (TGF- beta-inducible protein TSC-36). | |||||
|
FSTL1_HUMAN
|
||||||
| NC score | 0.383245 (rank : 24) | θ value | 4.1701e-05 (rank : 26) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q12841 | Gene names | FSTL1, FRP | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1). | |||||
|
IBP1_HUMAN
|
||||||
| NC score | 0.375528 (rank : 25) | θ value | 0.00134147 (rank : 35) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P08833, Q8IYP5 | Gene names | IGFBP1, IBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1) (Placental protein 12) (PP12). | |||||
|
IBP4_MOUSE
|
||||||
| NC score | 0.373993 (rank : 26) | θ value | 0.00175202 (rank : 37) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P47879, O35666 | Gene names | Igfbp4, Igfbp-4 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
IBP4_HUMAN
|
||||||
| NC score | 0.370555 (rank : 27) | θ value | 0.00175202 (rank : 36) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P22692 | Gene names | IGFBP4, IBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
IBP2_MOUSE
|
||||||
| NC score | 0.360173 (rank : 28) | θ value | 0.0113563 (rank : 44) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P47877, Q61722 | Gene names | Igfbp2, Igfbp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
IBP2_HUMAN
|
||||||
| NC score | 0.348146 (rank : 29) | θ value | 0.0113563 (rank : 43) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P18065, Q14619, Q9UCL3 | Gene names | IGFBP2, BP2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
FST_HUMAN
|
||||||
| NC score | 0.346642 (rank : 30) | θ value | 5.44631e-05 (rank : 28) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P19883, Q9BTH0 | Gene names | FST | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
IBP1_MOUSE
|
||||||
| NC score | 0.346390 (rank : 31) | θ value | 0.0431538 (rank : 49) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
|
TACD1_HUMAN
|
||||||
| NC score | 0.345007 (rank : 32) | θ value | 0.0113563 (rank : 45) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P16422, P18180, Q96C47 | Gene names | TACSTD1, GA733-2, M1S2, TROP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor-associated calcium signal transducer 1 precursor (Major gastrointestinal tumor-associated protein GA733-2) (Epithelial cell surface antigen) (Epithelial glycoprotein) (EGP) (Adenocarcinoma- associated antigen) (KSA) (KS 1/4 antigen) (Cell surface glycoprotein Trop-1) (CD326 antigen). | |||||
|
FST_MOUSE
|
||||||
| NC score | 0.340751 (rank : 33) | θ value | 7.1131e-05 (rank : 31) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P47931 | Gene names | Fst | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
FSTL3_HUMAN
|
||||||
| NC score | 0.291248 (rank : 34) | θ value | 0.0431538 (rank : 48) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O95633 | Gene names | FSTL3, FLRG | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
FSTL3_MOUSE
|
||||||
| NC score | 0.281668 (rank : 35) | θ value | 0.0330416 (rank : 46) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9EQC7 | Gene names | Fstl3, Flrg | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
TACD2_HUMAN
|
||||||
| NC score | 0.280269 (rank : 36) | θ value | 0.47712 (rank : 53) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P09758, Q15658, Q96QD2 | Gene names | TACSTD2, GA733-1, M1S1, TROP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor-associated calcium signal transducer 2 precursor (Pancreatic carcinoma marker protein GA733-1) (Cell surface glycoprotein Trop-2). | |||||
|
THYG_MOUSE
|
||||||
| NC score | 0.277570 (rank : 37) | θ value | 1.38499e-08 (rank : 14) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08710, O88590, Q9QWY7 | Gene names | Tg, Tgn | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
THYG_HUMAN
|
||||||
| NC score | 0.270708 (rank : 38) | θ value | 9.92553e-07 (rank : 20) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P01266, O15274, O43899, Q15593, Q15948, Q9NYR1, Q9NYR2, Q9UMZ0, Q9UNY3 | Gene names | TG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
NID1_HUMAN
|
||||||
| NC score | 0.236181 (rank : 39) | θ value | 2.52405e-10 (rank : 8) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P14543, Q14942, Q59FL2, Q5TAF2, Q5TAF3, Q86XD7 | Gene names | NID1, NID | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
NID1_MOUSE
|
||||||
| NC score | 0.232867 (rank : 40) | θ value | 2.52405e-10 (rank : 9) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P10493, Q8BQI3, Q8C3U8, Q8C9P6 | Gene names | Nid1, Ent | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
TEFF2_MOUSE
|
||||||
| NC score | 0.232212 (rank : 41) | θ value | 0.00869519 (rank : 42) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QYM9, Q3UY20, Q8CDH1, Q9JJE3 | Gene names | Tmeff2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Tomoregulin) (TR). | |||||
|
ISK5_HUMAN
|
||||||
| NC score | 0.231905 (rank : 42) | θ value | 0.00665767 (rank : 40) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NQ38, O75770, Q96PP2, Q96PP3 | Gene names | SPINK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 5 precursor (Lympho-epithelial Kazal-type-related inhibitor) (LEKTI) [Contains: Hemofiltrate peptide HF6478; Hemofiltrate peptide HF7665]. | |||||
|
TEFF2_HUMAN
|
||||||
| NC score | 0.231519 (rank : 43) | θ value | 0.00869519 (rank : 41) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UIK5, Q4ZFW4, Q53H90, Q53RE1, Q8N2R5, Q9NR15, Q9NSS5, Q9P2Y9, Q9UK65 | Gene names | TMEFF2, HPP1, TENB2, TPEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Transmembrane protein containing epidermal growth factor and follistatin domains) (Tomoregulin) (TR) (Hyperplastic polyposis protein 1). | |||||
|
NID2_MOUSE
|
||||||
| NC score | 0.223351 (rank : 44) | θ value | 3.08544e-08 (rank : 16) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O88322 | Gene names | Nid2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Entactin-2). | |||||
|
NID2_HUMAN
|
||||||
| NC score | 0.222502 (rank : 45) | θ value | 3.08544e-08 (rank : 15) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
ISK7_MOUSE
|
||||||
| NC score | 0.207484 (rank : 46) | θ value | 0.365318 (rank : 52) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6IE32 | Gene names | Spink7, Ecg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 7 precursor (Esophagus cancer- related gene 2 protein) (ECRG-2). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.184340 (rank : 47) | θ value | 2.21117e-06 (rank : 21) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
FSTL4_HUMAN
|
||||||
| NC score | 0.169738 (rank : 48) | θ value | 0.000158464 (rank : 32) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6MZW2, Q8TBU0, Q9UPU1 | Gene names | FSTL4, KIAA1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4). | |||||
|
ISK7_HUMAN
|
||||||
| NC score | 0.160285 (rank : 49) | θ value | 0.21417 (rank : 51) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P58062 | Gene names | SPINK7, ECG2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 7 precursor (Esophagus cancer- related gene 2 protein) (ECRG-2). | |||||
|
KAZD1_HUMAN
|
||||||
| NC score | 0.159989 (rank : 50) | θ value | 0.125558 (rank : 50) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96I82, Q6ZMB1, Q9BQ73 | Gene names | KAZALD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor. | |||||
|
ISK4_HUMAN
|
||||||
| NC score | 0.159595 (rank : 51) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60575 | Gene names | SPINK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 4 precursor (Peptide PEC-60 homolog). | |||||
|
ISK6_HUMAN
|
||||||
| NC score | 0.159571 (rank : 52) | θ value | 0.62314 (rank : 54) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6UWN8, Q8N5P0 | Gene names | SPINK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
ISK4_MOUSE
|
||||||
| NC score | 0.158245 (rank : 53) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O35679 | Gene names | Spink4, Mpgc60 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 4 precursor (Peptide PEC-60 homolog) (MPGC60 protein). | |||||
|
FSTL4_MOUSE
|
||||||
| NC score | 0.156981 (rank : 54) | θ value | 0.00102713 (rank : 34) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5STE3, Q5DU03 | Gene names | Fstl4, Kiaa1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4) (m- D/Bsp120I 1-1). | |||||
|
IPK1_HUMAN
|
||||||
| NC score | 0.143397 (rank : 55) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P00995 | Gene names | SPINK1, PSTI | |||
|
Domain Architecture |

|
|||||
| Description | Pancreatic secretory trypsin inhibitor precursor (Tumor-associated trypsin inhibitor) (TATI) (Serine protease inhibitor Kazal-type 1). | |||||
|
FSTL5_HUMAN
|
||||||
| NC score | 0.133955 (rank : 56) | θ value | 0.00228821 (rank : 38) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N475, Q9NSW7, Q9ULF7 | Gene names | FSTL5, KIAA1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5). | |||||
|
FSTL5_MOUSE
|
||||||
| NC score | 0.126710 (rank : 57) | θ value | 0.0330416 (rank : 47) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BFR2, Q80TG3, Q8C4T3 | Gene names | Fstl5, Kiaa1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5) (m- D/Bsp120I 1-2). | |||||
|
ISK6_MOUSE
|
||||||
| NC score | 0.125186 (rank : 58) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BT20 | Gene names | Spink6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
KAZD1_MOUSE
|
||||||
| NC score | 0.125081 (rank : 59) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BJ66 | Gene names | Kazald1, Bono1, Igfbprp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor (IGFBP-related protein 10) (IGFBP-rP10) (Bone and odontoblast expressed protein 1). | |||||
|
ISK3_MOUSE
|
||||||
| NC score | 0.124421 (rank : 60) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P09036, Q5M9M3 | Gene names | Spink3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 3 precursor (Prostatic secretory glycoprotein) (P12). | |||||
|
IPK2_HUMAN
|
||||||
| NC score | 0.104236 (rank : 61) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P20155 | Gene names | SPINK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 2 precursor (Acrosin-trypsin inhibitor) (HUSI-II). | |||||
|
HTRA1_MOUSE
|
||||||
| NC score | 0.097009 (rank : 62) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9R118 | Gene names | Htra1, Htra, Prss11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-). | |||||
|
IPK2_MOUSE
|
||||||
| NC score | 0.085101 (rank : 63) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BMY7 | Gene names | Spink2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 2 precursor. | |||||
|
HTRA1_HUMAN
|
||||||
| NC score | 0.084283 (rank : 64) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q92743, Q9UNS5 | Gene names | HTRA1, HTRA, PRSS11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-) (L56). | |||||
|
HTRA3_HUMAN
|
||||||
| NC score | 0.078748 (rank : 65) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P83110, Q7Z7A2 | Gene names | HTRA3, PRSP | |||
|
Domain Architecture |

|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease). | |||||
|
IBP7_HUMAN
|
||||||
| NC score | 0.076874 (rank : 66) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q16270, Q07822 | Gene names | IGFBP7, MAC25, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein) (Prostacyclin-stimulating factor) (PGI2-stimulating factor) (IGFBP-rP1). | |||||
|
HTRA3_MOUSE
|
||||||
| NC score | 0.075057 (rank : 67) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D236, Q6WLC5, Q6YDR0 | Gene names | Htra3, Prsp, Tasp | |||
|
Domain Architecture |

|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease) (Toll-associated serine protease). | |||||
|
IBP7_MOUSE
|
||||||
| NC score | 0.074421 (rank : 68) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61581, O88812, Q9EQW0 | Gene names | Igfbp7, Mac25 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein). | |||||
|
RECK_HUMAN
|
||||||
| NC score | 0.066675 (rank : 69) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O95980, Q8WX37 | Gene names | RECK | |||
|
Domain Architecture |

|
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (hRECK) (Suppressor of tumorigenicity 15) (ST15). | |||||
|
RECK_MOUSE
|
||||||
| NC score | 0.060898 (rank : 70) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z0J1 | Gene names | Reck | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (mRECK). | |||||
|
STAB2_MOUSE
|
||||||
| NC score | 0.045417 (rank : 71) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
VPS72_HUMAN
|
||||||
| NC score | 0.028556 (rank : 72) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15906, Q53GJ2, Q5U0R4 | Gene names | VPS72, TCFL1, YL1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
VPS72_MOUSE
|
||||||
| NC score | 0.028220 (rank : 73) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
LAMB2_MOUSE
|
||||||
| NC score | 0.022677 (rank : 74) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61292, Q62182 | Gene names | Lamb2, Lams | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (S-LAM). | |||||
|
SO4A1_HUMAN
|
||||||
| NC score | 0.019334 (rank : 75) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96BD0, Q9H4T7, Q9H4T8, Q9H8P2, Q9NWX8, Q9UI35, Q9UIG7 | Gene names | SLCO4A1, OATP1, OATPE, SLC21A12 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 4A1 (Solute carrier family 21 member 12) (Sodium-independent organic anion transporter E) (Organic anion-transporting polypeptide E) (OATP-E) (Colon organic anion transporter) (Organic anion transporter polypeptide-related protein 1) (OATP-RP1) (OATPRP1) (POAT). | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.019332 (rank : 76) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
BRCA2_HUMAN
|
||||||
| NC score | 0.017136 (rank : 77) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51587, O00183, O15008, Q13879 | Gene names | BRCA2, FANCD1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein (Fanconi anemia group D1 protein). | |||||
|
APBB2_HUMAN
|
||||||
| NC score | 0.017110 (rank : 78) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
SO4A1_MOUSE
|
||||||
| NC score | 0.016619 (rank : 79) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K078, Q8BZT4 | Gene names | Slco4a1, Oatpe, Slc21a12 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 4A1 (Solute carrier family 21 member 12) (Sodium-independent organic anion transporter E) (Organic anion-transporting polypeptide E) (OATP-E). | |||||
|
TF2AY_HUMAN
|
||||||
| NC score | 0.014169 (rank : 80) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UNN4 | Gene names | GTF2A1LF, ALF | |||
|
Domain Architecture |

|
|||||
| Description | TFIIA-alpha and beta-like factor (General transcription factor II A, 1-like factor). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.013752 (rank : 81) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TCAL3_HUMAN
|
||||||
| NC score | 0.012785 (rank : 82) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q969E4 | Gene names | TCEAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
TRI17_HUMAN
|
||||||
| NC score | 0.011433 (rank : 83) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y577 | Gene names | TRIM17, RBCC, RNF16, TERF | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 17 (Testis RING finger protein) (RING finger protein 16). | |||||
|
P85B_HUMAN
|
||||||
| NC score | 0.011425 (rank : 84) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00459, Q9UPH9 | Gene names | PIK3R2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
P85B_MOUSE
|
||||||
| NC score | 0.008874 (rank : 85) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08908 | Gene names | Pik3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
TNR11_HUMAN
|
||||||
| NC score | 0.008385 (rank : 86) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y6Q6 | Gene names | TNFRSF11A, RANK | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11A precursor (Receptor activator of NF-KB) (Osteoclast differentiation factor receptor) (ODFR) (CD265 antigen). | |||||
|
TRI17_MOUSE
|
||||||
| NC score | 0.007558 (rank : 87) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 620 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TPM3, Q99PP8 | Gene names | Trim17 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 17. | |||||
|
CAC1C_MOUSE
|
||||||
| NC score | 0.003603 (rank : 88) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
UBP48_MOUSE
|
||||||
| NC score | 0.003213 (rank : 89) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 83 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||