Please be patient as the page loads
|
HTRA1_MOUSE
|
||||||
| SwissProt Accessions | Q9R118 | Gene names | Htra1, Htra, Prss11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HTRA1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.990165 (rank : 2) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 121 | |
| SwissProt Accessions | Q92743, Q9UNS5 | Gene names | HTRA1, HTRA, PRSS11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-) (L56). | |||||
|
HTRA1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 140 | |
| SwissProt Accessions | Q9R118 | Gene names | Htra1, Htra, Prss11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-). | |||||
|
HTRA3_HUMAN
|
||||||
| θ value | 2.4321e-138 (rank : 3) | NC score | 0.960360 (rank : 4) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | P83110, Q7Z7A2 | Gene names | HTRA3, PRSP | |||
|
Domain Architecture |
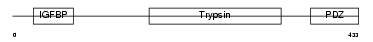
|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease). | |||||
|
HTRA3_MOUSE
|
||||||
| θ value | 1.63136e-134 (rank : 4) | NC score | 0.960934 (rank : 3) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9D236, Q6WLC5, Q6YDR0 | Gene names | Htra3, Prsp, Tasp | |||
|
Domain Architecture |
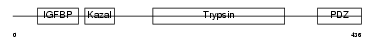
|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease) (Toll-associated serine protease). | |||||
|
HTRA4_HUMAN
|
||||||
| θ value | 1.29261e-131 (rank : 5) | NC score | 0.947567 (rank : 5) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P83105, Q542Z4, Q6PF13 | Gene names | HTRA4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable serine protease HTRA4 precursor (EC 3.4.21.-). | |||||
|
HTRA2_HUMAN
|
||||||
| θ value | 1.03614e-88 (rank : 6) | NC score | 0.871812 (rank : 7) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43464, Q9HBZ4, Q9P0Y3, Q9P0Y4 | Gene names | HTRA2, OMI, PRSS25 | |||
|
Domain Architecture |
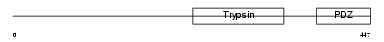
|
|||||
| Description | Serine protease HTRA2, mitochondrial precursor (EC 3.4.21.-) (High temperature requirement protein A2) (HtrA2) (Omi stress-regulated endoprotease) (Serine proteinase OMI). | |||||
|
HTRA2_MOUSE
|
||||||
| θ value | 1.03614e-88 (rank : 7) | NC score | 0.873310 (rank : 6) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JIY5, Q9R108 | Gene names | Htra2, Omi, Prss25 | |||
|
Domain Architecture |
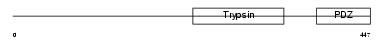
|
|||||
| Description | Serine protease HTRA2, mitochondrial precursor (EC 3.4.21.-) (High temperature requirement protein A2) (HtrA2) (Omi stress-regulated endoprotease) (Serine proteinase OMI). | |||||
|
IBP7_HUMAN
|
||||||
| θ value | 2.43343e-21 (rank : 8) | NC score | 0.533448 (rank : 8) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q16270, Q07822 | Gene names | IGFBP7, MAC25, PSF | |||
|
Domain Architecture |
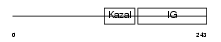
|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein) (Prostacyclin-stimulating factor) (PGI2-stimulating factor) (IGFBP-rP1). | |||||
|
KAZD1_HUMAN
|
||||||
| θ value | 5.07402e-19 (rank : 9) | NC score | 0.469695 (rank : 10) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96I82, Q6ZMB1, Q9BQ73 | Gene names | KAZALD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor. | |||||
|
IBP7_MOUSE
|
||||||
| θ value | 1.13037e-18 (rank : 10) | NC score | 0.531797 (rank : 9) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61581, O88812, Q9EQW0 | Gene names | Igfbp7, Mac25 | |||
|
Domain Architecture |
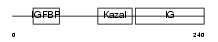
|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein). | |||||
|
KAZD1_MOUSE
|
||||||
| θ value | 1.92812e-18 (rank : 11) | NC score | 0.449320 (rank : 11) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8BJ66 | Gene names | Kazald1, Bono1, Igfbprp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor (IGFBP-related protein 10) (IGFBP-rP10) (Bone and odontoblast expressed protein 1). | |||||
|
ESM1_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 12) | NC score | 0.406793 (rank : 12) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QYY7 | Gene names | Esm1 | |||
|
Domain Architecture |
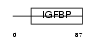
|
|||||
| Description | Endothelial cell-specific molecule 1 precursor (ESM-1 secretory protein) (ESM-1). | |||||
|
IBP3_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 13) | NC score | 0.360498 (rank : 14) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P17936, Q2V509, Q6P1M6 | Gene names | IGFBP3, IBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
ESM1_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 14) | NC score | 0.382938 (rank : 13) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NQ30, Q15330, Q96ES3 | Gene names | ESM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-specific molecule 1 precursor (ESM-1 secretory protein) (ESM-1). | |||||
|
IBP2_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 15) | NC score | 0.339092 (rank : 15) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P18065, Q14619, Q9UCL3 | Gene names | IGFBP2, BP2 | |||
|
Domain Architecture |
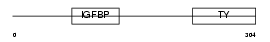
|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
IBP1_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 16) | NC score | 0.339042 (rank : 16) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
|
CRIM1_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 17) | NC score | 0.193842 (rank : 39) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
IBP3_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 18) | NC score | 0.310904 (rank : 20) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P47878 | Gene names | Igfbp3, Igfbp-3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
IBP5_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 19) | NC score | 0.335645 (rank : 18) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P24593 | Gene names | IGFBP5, IBP5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP5_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 20) | NC score | 0.336098 (rank : 17) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q07079 | Gene names | Igfbp5, Igfbp-5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP4_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 21) | NC score | 0.300563 (rank : 22) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P22692 | Gene names | IGFBP4, IBP4 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
IBP4_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 22) | NC score | 0.295479 (rank : 24) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P47879, O35666 | Gene names | Igfbp4, Igfbp-4 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
IBP2_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 23) | NC score | 0.327496 (rank : 19) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P47877, Q61722 | Gene names | Igfbp2, Igfbp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
CYR61_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 24) | NC score | 0.253219 (rank : 29) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O00622, O14934, O43775, Q9BZL7 | Gene names | CYR61, CCN1, GIG1, IGFBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CYR61 precursor (Cysteine-rich, angiogenic inducer, 61) (Insulin-like growth factor-binding protein 10) (Protein GIG1). | |||||
|
CYR61_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 25) | NC score | 0.253766 (rank : 27) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P18406 | Gene names | Cyr61, Ccn1, Igfbp10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CYR61 precursor (Cysteine-rich, angiogenic inducer, 61) (Insulin-like growth factor-binding protein 10) (3CH61). | |||||
|
PDZD1_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 26) | NC score | 0.161367 (rank : 44) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9JIL4, Q8CDP5, Q8R4G2, Q9CQ72 | Gene names | Pdzk1, Cap70, Nherf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 27) | NC score | 0.120997 (rank : 61) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
NOV_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 28) | NC score | 0.246927 (rank : 32) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P48745, Q96BY5 | Gene names | NOV, CCN3, IGFBP9, NOVH | |||
|
Domain Architecture |
|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
IBP1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 29) | NC score | 0.298342 (rank : 23) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P08833, Q8IYP5 | Gene names | IGFBP1, IBP1 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1) (Placental protein 12) (PP12). | |||||
|
PDZD1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 30) | NC score | 0.159160 (rank : 46) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5T2W1, O60450, Q5T5P6, Q9BQ41 | Gene names | PDZK1, CAP70, NHERF3, PDZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
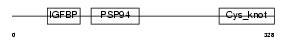
NOV_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 31) | NC score | 0.230570 (rank : 33) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q64299, Q8CA67 | Gene names | Nov, Ccn3 | |||
|
Domain Architecture |
|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
NHERF_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 32) | NC score | 0.137021 (rank : 56) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P70441 | Gene names | Slc9a3r1, Nherf | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
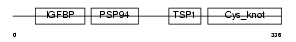
CTGF_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 33) | NC score | 0.252472 (rank : 30) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P29279, Q96QX2 | Gene names | CTGF, CCN2, HCS24 | |||
|
Domain Architecture |
|
|||||
| Description | Connective tissue growth factor precursor (Hypertrophic chondrocyte- specific protein 24). | |||||
|
WISP2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 34) | NC score | 0.258561 (rank : 26) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O76076 | Gene names | WISP2, CCN5, CT58, CTGFL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WNT1-inducible signaling pathway protein 2 precursor (WISP-2) (Connective tissue growth factor-like protein) (CTGF-L) (Connective tissue growth factor-related protein 58). | |||||
|
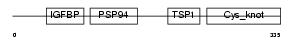
CTGF_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 35) | NC score | 0.211000 (rank : 37) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P29268, Q922U0 | Gene names | Ctgf, Ccn2, Fisp-12, Fisp12, Hcs24 | |||
|
Domain Architecture |
|
|||||
| Description | Connective tissue growth factor precursor (Protein FISP-12) (Hypertrophic chondrocyte-specific protein 24). | |||||
|
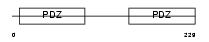
NHERF_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 36) | NC score | 0.140117 (rank : 54) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O14745, O43552, Q86WQ5 | Gene names | SLC9A3R1, NHERF | |||
|
Domain Architecture |
|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
FSTL3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 37) | NC score | 0.168448 (rank : 41) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O95633 | Gene names | FSTL3, FLRG | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
KRA56_HUMAN
|
||||||
| θ value | 0.125558 (rank : 38) | NC score | 0.047842 (rank : 136) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
RHPN1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 39) | NC score | 0.095710 (rank : 79) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TCX5, Q8TAV1 | Gene names | RHPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Rhophilin-1 (GTP-Rho-binding protein 1). | |||||
|
TEFF2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 40) | NC score | 0.115987 (rank : 65) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QYM9, Q3UY20, Q8CDH1, Q9JJE3 | Gene names | Tmeff2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Tomoregulin) (TR). | |||||
|
FSTL5_HUMAN
|
||||||
| θ value | 0.21417 (rank : 41) | NC score | 0.066350 (rank : 112) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N475, Q9NSW7, Q9ULF7 | Gene names | FSTL5, KIAA1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5). | |||||
|
IBP6_MOUSE
|
||||||
| θ value | 0.21417 (rank : 42) | NC score | 0.206947 (rank : 38) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P47880 | Gene names | Igfbp6, Igfbp-6 | |||
|
Domain Architecture |
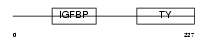
|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
RHPN1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 43) | NC score | 0.094578 (rank : 80) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61085 | Gene names | Rhpn1, Grbp | |||
|
Domain Architecture |

|
|||||
| Description | Rhophilin-1 (GTP-Rho-binding protein 1). | |||||
|
WISP2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 44) | NC score | 0.222282 (rank : 34) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Z0G4, Q8CIC8 | Gene names | Wisp2, Ccn5, Ctgfl | |||
|
Domain Architecture |
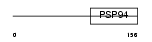
|
|||||
| Description | WNT1-inducible signaling pathway protein 2 precursor (WISP-2) (Connective tissue growth factor-like protein) (CTGF-L). | |||||
|
KRA53_HUMAN
|
||||||
| θ value | 0.279714 (rank : 45) | NC score | 0.040288 (rank : 141) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
TEFF2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 46) | NC score | 0.112763 (rank : 68) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UIK5, Q4ZFW4, Q53H90, Q53RE1, Q8N2R5, Q9NR15, Q9NSS5, Q9P2Y9, Q9UK65 | Gene names | TMEFF2, HPP1, TENB2, TPEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Transmembrane protein containing epidermal growth factor and follistatin domains) (Tomoregulin) (TR) (Hyperplastic polyposis protein 1). | |||||
|
FSTL5_MOUSE
|
||||||
| θ value | 0.365318 (rank : 47) | NC score | 0.064631 (rank : 113) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BFR2, Q80TG3, Q8C4T3 | Gene names | Fstl5, Kiaa1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5) (m- D/Bsp120I 1-2). | |||||
|
LCE1B_HUMAN
|
||||||
| θ value | 0.365318 (rank : 48) | NC score | 0.253674 (rank : 28) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5T7P3 | Gene names | LCE1B, LEP2, SPRL2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1B (Late envelope protein 2) (Small proline-rich-like epidermal differentiation complex protein 2A). | |||||
|
LCE1C_HUMAN
|
||||||
| θ value | 0.365318 (rank : 49) | NC score | 0.282716 (rank : 25) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
WISP3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 50) | NC score | 0.220588 (rank : 35) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O95389, Q6UXH6 | Gene names | WISP3, CCN6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WNT1-inducible signaling pathway protein 3 precursor (WISP-3). | |||||
|
FSTL3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 51) | NC score | 0.160083 (rank : 45) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9EQC7 | Gene names | Fstl3, Flrg | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
FST_HUMAN
|
||||||
| θ value | 0.47712 (rank : 52) | NC score | 0.145881 (rank : 49) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P19883, Q9BTH0 | Gene names | FST | |||
|
Domain Architecture |
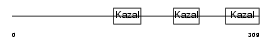
|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
FST_MOUSE
|
||||||
| θ value | 0.47712 (rank : 53) | NC score | 0.145234 (rank : 50) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P47931 | Gene names | Fst | |||
|
Domain Architecture |
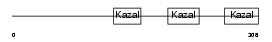
|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
NHRF2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 54) | NC score | 0.118720 (rank : 62) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15599, O00272, O00556 | Gene names | SLC9A3R2, NHERF2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2). | |||||
|
PDZ11_HUMAN
|
||||||
| θ value | 0.47712 (rank : 55) | NC score | 0.144783 (rank : 51) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5EBL8, Q6UWE1, Q9P0Q1 | Gene names | PDZD11, PDZK11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
PDZ11_MOUSE
|
||||||
| θ value | 0.47712 (rank : 56) | NC score | 0.143952 (rank : 52) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9CZG9 | Gene names | Pdzd11, Pdzk11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
PDZD3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 57) | NC score | 0.137537 (rank : 55) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99MJ6, Q8BWE5 | Gene names | Pdzd3, Pdzk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Natrium-phosphate cotransporter IIa C-terminal-associated protein 2) (NaPi-Cap2). | |||||
|
ZO3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 58) | NC score | 0.075090 (rank : 100) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9QXY1 | Gene names | Tjp3, Zo3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
FSTL4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 59) | NC score | 0.083659 (rank : 92) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5STE3, Q5DU03 | Gene names | Fstl4, Kiaa1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4) (m- D/Bsp120I 1-1). | |||||
|
NHRF2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 60) | NC score | 0.113040 (rank : 67) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JHL1, Q3TDR3, Q8BGL9, Q8BW05, Q9DCR6 | Gene names | Slc9a3r2, Nherf2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2) (Octs2). | |||||
|
SMOC1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 61) | NC score | 0.117170 (rank : 64) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BLY1, Q9WVN9 | Gene names | Smoc1, Srg | |||
|
Domain Architecture |
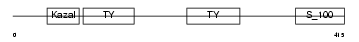
|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1) (SPARC-related gene protein). | |||||
|
GRASP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 62) | NC score | 0.092316 (rank : 85) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7Z6J2, Q6PIF8, Q7Z741 | Gene names | GRASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General receptor for phosphoinositides 1-associated scaffold protein (GRP1-associated scaffold protein). | |||||
|
MEGF6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 63) | NC score | 0.031939 (rank : 142) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
WHRN_MOUSE
|
||||||
| θ value | 0.813845 (rank : 64) | NC score | 0.094398 (rank : 81) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
ADM1B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.012002 (rank : 150) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R534, Q9R156 | Gene names | Adam1b | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 1b precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 1b) (Fertilin alpha 1b subunit). | |||||
|
FSTL4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.068935 (rank : 105) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6MZW2, Q8TBU0, Q9UPU1 | Gene names | FSTL4, KIAA1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4). | |||||
|
IBP6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.211900 (rank : 36) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P24592, Q14492 | Gene names | IGFBP6, IBP6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
LAP4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 68) | NC score | 0.055947 (rank : 127) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
ADA12_MOUSE
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.011072 (rank : 153) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
LCE1E_HUMAN
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.303293 (rank : 21) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
LCE2B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.129317 (rank : 60) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14633 | Gene names | LCE2B, LEP10, SPRL1B, XP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2B (Late envelope protein 10) (Small proline-rich-like epidermal differentiation complex protein 1B) (Skin- specific protein Xp5). | |||||
|
PDZK4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.071449 (rank : 102) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QY39, Q6PHP2 | Gene names | Pdzk4, Pdzrn4l, Xlu | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 4 (PDZ domain-containing RING finger protein 4-like protein). | |||||
|
SMOC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.108623 (rank : 71) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H4F8, Q96F78 | Gene names | SMOC1 | |||
|
Domain Architecture |
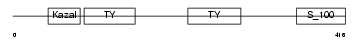
|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1). | |||||
|
USH1C_HUMAN
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.093204 (rank : 84) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y6N9, Q96B29, Q9UM04, Q9UM17, Q9UPC3 | Gene names | USH1C, AIE75 | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein) (Autoimmune enteropathy- related antigen AIE-75) (Antigen NY-CO-38/NY-CO-37) (PDZ-73 protein) (NY-REN-3 antigen). | |||||
|
USH1C_MOUSE
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.086862 (rank : 90) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
WHRN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 76) | NC score | 0.094110 (rank : 82) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
WISP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 77) | NC score | 0.165641 (rank : 43) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O95388, Q9HCS3 | Gene names | WISP1, CCN4 | |||
|
Domain Architecture |
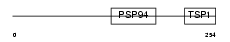
|
|||||
| Description | WNT1-inducible signaling pathway protein 1 precursor (WISP-1) (Wnt-1- induced secreted protein). | |||||
|
WISP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 78) | NC score | 0.176329 (rank : 40) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O54775, Q80ZL1 | Gene names | Wisp1, Ccn4, Elm1 | |||
|
Domain Architecture |

|
|||||
| Description | WNT1-inducible signaling pathway protein 1 precursor (WISP-1) (ELM-1). | |||||
|
CUBN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.011228 (rank : 152) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60494, Q5VTA6, Q96RU9 | Gene names | CUBN, IFCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor) (Intrinsic factor-vitamin B12 receptor) (460 kDa receptor) (Intestinal intrinsic factor receptor). | |||||
|
FSTL1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 80) | NC score | 0.108430 (rank : 72) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q12841 | Gene names | FSTL1, FRP | |||
|
Domain Architecture |
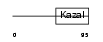
|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1). | |||||
|
FSTL1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 81) | NC score | 0.106749 (rank : 73) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q62356, Q99JI9 | Gene names | Fstl1, Frp, Fstl, Tsc36 | |||
|
Domain Architecture |
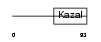
|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1) (TGF- beta-inducible protein TSC-36). | |||||
|
GRASP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 82) | NC score | 0.088039 (rank : 89) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JJA9, Q9JKL0 | Gene names | Grasp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General receptor for phosphoinositides 1-associated scaffold protein (GRP1-associated scaffold protein). | |||||
|
MPDZ_HUMAN
|
||||||
| θ value | 1.81305 (rank : 83) | NC score | 0.068323 (rank : 107) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O75970, O43798, Q5CZ80, Q5JTX3, Q5JTX6, Q5JTX7, Q5JUC3, Q5JUC4, Q5VZ62, Q8N790 | Gene names | MPDZ, MUPP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ- domain protein 1). | |||||
|
ZO1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 84) | NC score | 0.062366 (rank : 119) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
ZO1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | 0.061851 (rank : 121) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P39447 | Gene names | Tjp1, Zo1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
ITB2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | 0.016272 (rank : 146) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P11835, Q64482 | Gene names | Itgb2 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-2 precursor (Cell surface adhesion glycoproteins LFA- 1/CR3/P150,95 subunit beta) (Complement receptor C3 subunit beta) (CD18 antigen). | |||||
|
KR511_HUMAN
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.041971 (rank : 140) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
MPDZ_MOUSE
|
||||||
| θ value | 2.36792 (rank : 88) | NC score | 0.072359 (rank : 101) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8VBX6, O08783, Q6P7U4, Q80ZY8, Q8BKJ1, Q8C0H8, Q8VBV5, Q8VBY0, Q9Z1K3 | Gene names | Mpdz, Mupp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ domain protein 1). | |||||
|
PDZK4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 89) | NC score | 0.068947 (rank : 104) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q76G19, Q8NB75, Q9BUH9, Q9P284 | Gene names | PDZK4, KIAA1444, PDZRN4L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 4 (PDZ domain-containing RING finger protein 4-like protein). | |||||
|
TICN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.093344 (rank : 83) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q08629 | Gene names | SPOCK1, SPOCK, TIC1, TICN1 | |||
|
Domain Architecture |
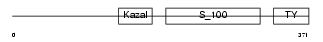
|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
ZO3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.060582 (rank : 122) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95049 | Gene names | TJP3, ZO3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
IPK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.133707 (rank : 58) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P00995 | Gene names | SPINK1, PSTI | |||
|
Domain Architecture |
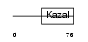
|
|||||
| Description | Pancreatic secretory trypsin inhibitor precursor (Tumor-associated trypsin inhibitor) (TATI) (Serine protease inhibitor Kazal-type 1). | |||||
|
ISK3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.146196 (rank : 48) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P09036, Q5M9M3 | Gene names | Spink3 | |||
|
Domain Architecture |
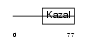
|
|||||
| Description | Serine protease inhibitor Kazal-type 3 precursor (Prostatic secretory glycoprotein) (P12). | |||||
|
LIN7A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.068124 (rank : 110) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O14910, Q6LES3, Q7LDS4 | Gene names | LIN7A, MALS1, VELI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog A (Lin-7A) (hLin-7) (Mammalian lin-seven protein 1) (MALS-1) (Vertebrate lin-7 homolog 1) (Veli-1 protein) (Tax interaction protein 33) (TIP-33). | |||||
|
LIN7A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.068240 (rank : 108) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8JZS0 | Gene names | Lin7a, Mals1, Veli1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog A (Lin-7A) (mLin-7) (Mammalian lin-seven protein 1) (MALS-1) (Vertebrate lin-7 homolog 1) (Veli-1 protein). | |||||
|
MAGI1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.055468 (rank : 129) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
MAGI1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.055808 (rank : 128) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
PDZD3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.131097 (rank : 59) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q86UT5, Q8N6R4, Q8NAW7, Q8NEX7, Q9H5Z3 | Gene names | PDZD3, IKEPP, PDZK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Intestinal and kidney-enriched PDZ protein) (Protein DLNB27). | |||||
|
PSCBP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.076086 (rank : 98) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O60759, Q15630, Q8NE32 | Gene names | PSCDBP, CYTIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology Sec7 and coiled-coil domains-binding protein (Cytohesin-binding protein HE) (CYBR) (Cytohesin binder and regulator) (Cytohesin-interacting protein). | |||||
|
PZRN4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 100) | NC score | 0.075669 (rank : 99) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6ZMN7, Q6N052, Q8IUU1, Q9NTP7 | Gene names | PDZRN4, LNX4, SEMCAP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 4 (Ligand of Numb-protein X 4) (SEMACAP3-like protein). | |||||
|
TENX_HUMAN
|
||||||
| θ value | 3.0926 (rank : 101) | NC score | 0.023726 (rank : 145) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
TICN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 102) | NC score | 0.102837 (rank : 74) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q62288 | Gene names | Spock1, Spock, Ticn1 | |||
|
Domain Architecture |
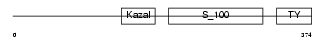
|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
ATX2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.012187 (rank : 149) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
LCE1D_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.252378 (rank : 31) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
LCE2A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.089017 (rank : 87) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5TA79 | Gene names | LCE2A, LEP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2A (Late envelope protein 9). | |||||
|
SMOC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.101106 (rank : 76) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8CD91, Q8VCM1, Q9D9K2, Q9ER95 | Gene names | Smoc2 | |||
|
Domain Architecture |
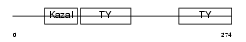
|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2). | |||||
|
TENR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.009602 (rank : 155) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92752, Q15568, Q5R3G0 | Gene names | TNR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin). | |||||
|
TNR14_HUMAN
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.026027 (rank : 143) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92956, Q8WXR1, Q96J31, Q9UM65 | Gene names | TNFRSF14, HVEA, HVEM | |||
|
Domain Architecture |
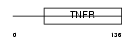
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 14 precursor (Herpesvirus entry mediator A) (Tumor necrosis factor receptor-like 2) (TR2). | |||||
|
ZDHC3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.006817 (rank : 156) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R173, Q6EMK3 | Gene names | Zdhhc3, Godz, Gramp1 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC3 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 3) (DHHC-3) (Golgi-specific DHHC zinc finger protein) (GABA-A receptor-associated membrane protein 1). | |||||
|
ISK5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.084972 (rank : 91) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NQ38, O75770, Q96PP2, Q96PP3 | Gene names | SPINK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 5 precursor (Lympho-epithelial Kazal-type-related inhibitor) (LEKTI) [Contains: Hemofiltrate peptide HF6478; Hemofiltrate peptide HF7665]. | |||||
|
LAP4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.050228 (rank : 133) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
LCE2C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.112606 (rank : 69) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5TA81 | Gene names | LCE2C, LEP11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2C (Late envelope protein 11). | |||||
|
LIN7B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.069843 (rank : 103) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HAP6 | Gene names | LIN7B, MALS2, VELI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog B (Lin-7B) (hLin7B) (Mammalian lin-seven protein 2) (MALS-2) (Vertebrate lin-7 homolog 2) (Veli-2 protein) (hVeli2). | |||||
|
LIN7B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.068541 (rank : 106) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O88951 | Gene names | Lin7b, Mals2, Veli2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog B (Lin-7B) (Mammalian lin-seven protein 2) (MALS-2) (Vertebrate lin-7 homolog 2) (Veli-2 protein). | |||||
|
SMOC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.097749 (rank : 77) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9H3U7, Q5TAT7, Q5TAT8, Q86VV9, Q96SF3, Q9H1L3, Q9H1L4, Q9H3U0, Q9H4F7, Q9HCV2 | Gene names | SMOC2, SMAP2 | |||
|
Domain Architecture |
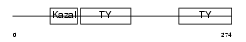
|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2) (Smooth muscle-associated protein 2) (SMAP-2). | |||||
|
SPR2B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.045491 (rank : 137) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70554, Q7TSB2 | Gene names | Sprr2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B. | |||||
|
CREL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.011342 (rank : 151) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91XD7, Q8BGJ8 | Gene names | Creld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
GOPC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.062077 (rank : 120) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HD26, Q59FS4, Q969U8 | Gene names | GOPC, CAL, FIG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi-associated PDZ and coiled-coil motif-containing protein (PDZ protein interacting specifically with TC10) (PIST) (CFTR-associated ligand) (Fused in glioblastoma). | |||||
|
GOPC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.063085 (rank : 117) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8BH60, Q8BSV4, Q920R1, Q9ET11 | Gene names | Gopc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi-associated PDZ and coiled-coil motif-containing protein (PDZ protein interacting specifically with TC10) (PIST). | |||||
|
ISK4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.063506 (rank : 116) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60575 | Gene names | SPINK4 | |||
|
Domain Architecture |
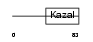
|
|||||
| Description | Serine protease inhibitor Kazal-type 4 precursor (Peptide PEC-60 homolog). | |||||
|
ISK6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.101765 (rank : 75) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BT20 | Gene names | Spink6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
JAG2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.013515 (rank : 148) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
LCE2D_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.136981 (rank : 57) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5TA82 | Gene names | LCE2D, LEP12, SPRL1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2D (Late envelope protein 12) (Small proline-rich-like epidermal differentiation complex protein 1A). | |||||
|
PSMD9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.042199 (rank : 139) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CR00, Q3TG66 | Gene names | Psmd9 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 9 (26S proteasome regulatory subunit p27). | |||||
|
RECK_HUMAN
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.081624 (rank : 93) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O95980, Q8WX37 | Gene names | RECK | |||
|
Domain Architecture |

|
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (hRECK) (Suppressor of tumorigenicity 15) (ST15). | |||||
|
RECK_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.078585 (rank : 97) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z0J1 | Gene names | Reck | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (mRECK). | |||||
|
TICN3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.097009 (rank : 78) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BKV0, Q9ER59 | Gene names | Spock3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
ATX2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.010988 (rank : 154) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
KR122_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.026016 (rank : 144) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59991 | Gene names | KRTAP12-2, KAP12.2, KRTAP12.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-2 (Keratin-associated protein 12.2) (High sulfur keratin-associated protein 12.2). | |||||
|
LCE4A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.118671 (rank : 63) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5TA78 | Gene names | LCE4A, LEP8, SPRL4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 4A (Late envelope protein 8) (Small proline-rich-like epidermal differentiation complex protein 4A). | |||||
|
LCE5A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.142546 (rank : 53) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5TCM9 | Gene names | LCE5A, LEP18, SPRL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 5A (Late envelope protein 18) (Small proline-rich-like epidermal differentiation complex protein 5A). | |||||
|
LIN7C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.064483 (rank : 114) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NUP9 | Gene names | LIN7C, MALS3, VELI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog C (Lin-7C) (Mammalian lin-seven protein 3) (MALS-3) (Vertebrate lin-7 homolog 3) (Veli-3 protein). | |||||
|
LIN7C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.064483 (rank : 115) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O88952 | Gene names | Lin7c, Mals3, Veli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog C (Lin-7C) (mLin7C) (Mammalian lin-seven protein 3) (MALS-3) (Vertebrate lin-7 homolog 3) (Veli-3 protein). | |||||
|
PSCBP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.062564 (rank : 118) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q91VY6, Q3TZH1, Q8R4T4, Q9QZA9 | Gene names | Pscdbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology Sec7 and coiled-coil domains-binding protein (Cytohesin-binding protein HE) (Cbp). | |||||
|
PSMD9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.042417 (rank : 138) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00233, Q9BQ42 | Gene names | PSMD9 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 9 (26S proteasome regulatory subunit p27). | |||||
|
PZRN3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.067591 (rank : 111) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UPQ7, Q8N2N7, Q96CC2, Q9NSQ2 | Gene names | PDZRN3, KIAA1095, LNX3, SEMCAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Ligand of Numb-protein X 3) (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
PZRN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.068194 (rank : 109) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q69ZS0, Q91Z03, Q9QY54, Q9QY55 | Gene names | Pdzrn3, Kiaa1095, Semcap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
SO5A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.016153 (rank : 147) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H2Y9 | Gene names | SLCO5A1, SLC21A15 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 5A1 (Solute carrier family 21 member 15) (Organic anion transporter polypeptide- related protein 4) (OATP-RP4) (OATPRP4). | |||||
|
TICN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.110102 (rank : 70) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q92563, Q6UW87 | Gene names | SPOCK2, KIAA0275, TICN2 | |||
|
Domain Architecture |
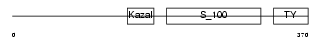
|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
TICN2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.113661 (rank : 66) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9ER58 | Gene names | Spock2, Ticn2 | |||
|
Domain Architecture |
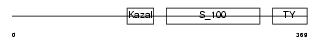
|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
ARHGB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.050010 (rank : 135) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
GRIP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.057512 (rank : 124) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
HG2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.056348 (rank : 125) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P04441, O19452 | Gene names | Cd74, Ii | |||
|
Domain Architecture |
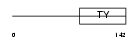
|
|||||
| Description | H-2 class II histocompatibility antigen gamma chain (MHC class II- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (CD74 antigen). | |||||
|
INADL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.053613 (rank : 130) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q63ZW7, O70471, Q5PRG3, Q6P6J1, Q80YR8, Q8BPB9 | Gene names | Inadl, Cipp, Patj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (Pals1-associated tight junction protein) (Protein associated to tight junctions) (Channel-interacting PDZ domain-containing protein). | |||||
|
IPK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.050998 (rank : 132) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P20155 | Gene names | SPINK2 | |||
|
Domain Architecture |
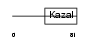
|
|||||
| Description | Serine protease inhibitor Kazal-type 2 precursor (Acrosin-trypsin inhibitor) (HUSI-II). | |||||
|
ISK6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.088432 (rank : 88) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6UWN8, Q8N5P0 | Gene names | SPINK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
LCE1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.167108 (rank : 42) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T7P2 | Gene names | LCE1A, LEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1A (Late envelope protein 1). | |||||
|
LCE1F_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.150459 (rank : 47) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5T754 | Gene names | LCE1F, LEP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1F (Late envelope protein 6). | |||||
|
PDZD7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.080112 (rank : 96) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9H5P4, Q8N321 | Gene names | PDZD7, PDZK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 7. | |||||
|
RPGF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.051050 (rank : 131) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8TEU7, Q8NI21, Q8TEU6, Q96PC1 | Gene names | RAPGEF6, PDZGEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 6 (PDZ domain-containing guanine nucleotide exchange factor 2) (PDZ-GEF2) (RA-GEF-2). | |||||
|
SDCB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.058333 (rank : 123) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O00560, O00173, O43391 | Gene names | SDCBP, MDA9, SYCL | |||
|
Domain Architecture |
|
|||||
| Description | Syntenin-1 (Syndecan-binding protein 1) (Melanoma differentiation- associated protein 9) (MDA-9) (Scaffold protein Pbp1) (Pro-TGF-alpha cytoplasmic domain-interacting protein 18) (TACIP18). | |||||
|
SDCB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.056341 (rank : 126) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O08992, Q544P5 | Gene names | Sdcbp | |||
|
Domain Architecture |
|
|||||
| Description | Syntenin-1 (Syndecan-binding protein 1) (Scaffold protein Pbp1). | |||||
|
SYJ2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.050124 (rank : 134) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P57105, Q96IA4 | Gene names | SYNJ2BP, OMP25 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptojanin-2-binding protein (Mitochondrial outer membrane protein 25). | |||||
|
TICN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.089309 (rank : 86) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9BQ16, O75705, Q6UW53, Q96Q26 | Gene names | SPOCK3, TICN3 | |||
|
Domain Architecture |
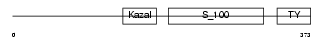
|
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
TX1B3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.081357 (rank : 94) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O14907, Q7LCQ4 | Gene names | TAX1BP3, TIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 3 (Tax interaction protein 1) (TIP-1) (Glutaminase-interacting protein 3). | |||||
|
TX1B3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.080853 (rank : 95) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9DBG9 | Gene names | Tax1bp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 3 (Tax interaction protein 1) (TIP-1). | |||||
|
HTRA1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 140 | |
| SwissProt Accessions | Q9R118 | Gene names | Htra1, Htra, Prss11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-). | |||||
|
HTRA1_HUMAN
|
||||||
| NC score | 0.990165 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 121 | |
| SwissProt Accessions | Q92743, Q9UNS5 | Gene names | HTRA1, HTRA, PRSS11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-) (L56). | |||||
|
HTRA3_MOUSE
|
||||||
| NC score | 0.960934 (rank : 3) | θ value | 1.63136e-134 (rank : 4) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9D236, Q6WLC5, Q6YDR0 | Gene names | Htra3, Prsp, Tasp | |||
|
Domain Architecture |

|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease) (Toll-associated serine protease). | |||||
|
HTRA3_HUMAN
|
||||||
| NC score | 0.960360 (rank : 4) | θ value | 2.4321e-138 (rank : 3) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | P83110, Q7Z7A2 | Gene names | HTRA3, PRSP | |||
|
Domain Architecture |

|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease). | |||||
|
HTRA4_HUMAN
|
||||||
| NC score | 0.947567 (rank : 5) | θ value | 1.29261e-131 (rank : 5) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P83105, Q542Z4, Q6PF13 | Gene names | HTRA4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable serine protease HTRA4 precursor (EC 3.4.21.-). | |||||
|
HTRA2_MOUSE
|
||||||
| NC score | 0.873310 (rank : 6) | θ value | 1.03614e-88 (rank : 7) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JIY5, Q9R108 | Gene names | Htra2, Omi, Prss25 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA2, mitochondrial precursor (EC 3.4.21.-) (High temperature requirement protein A2) (HtrA2) (Omi stress-regulated endoprotease) (Serine proteinase OMI). | |||||
|
HTRA2_HUMAN
|
||||||
| NC score | 0.871812 (rank : 7) | θ value | 1.03614e-88 (rank : 6) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43464, Q9HBZ4, Q9P0Y3, Q9P0Y4 | Gene names | HTRA2, OMI, PRSS25 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA2, mitochondrial precursor (EC 3.4.21.-) (High temperature requirement protein A2) (HtrA2) (Omi stress-regulated endoprotease) (Serine proteinase OMI). | |||||
|
IBP7_HUMAN
|
||||||
| NC score | 0.533448 (rank : 8) | θ value | 2.43343e-21 (rank : 8) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q16270, Q07822 | Gene names | IGFBP7, MAC25, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein) (Prostacyclin-stimulating factor) (PGI2-stimulating factor) (IGFBP-rP1). | |||||
|
IBP7_MOUSE
|
||||||
| NC score | 0.531797 (rank : 9) | θ value | 1.13037e-18 (rank : 10) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q61581, O88812, Q9EQW0 | Gene names | Igfbp7, Mac25 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein). | |||||
|
KAZD1_HUMAN
|
||||||
| NC score | 0.469695 (rank : 10) | θ value | 5.07402e-19 (rank : 9) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96I82, Q6ZMB1, Q9BQ73 | Gene names | KAZALD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor. | |||||
|
KAZD1_MOUSE
|
||||||
| NC score | 0.449320 (rank : 11) | θ value | 1.92812e-18 (rank : 11) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8BJ66 | Gene names | Kazald1, Bono1, Igfbprp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor (IGFBP-related protein 10) (IGFBP-rP10) (Bone and odontoblast expressed protein 1). | |||||
|
ESM1_MOUSE
|
||||||
| NC score | 0.406793 (rank : 12) | θ value | 3.41135e-07 (rank : 12) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QYY7 | Gene names | Esm1 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial cell-specific molecule 1 precursor (ESM-1 secretory protein) (ESM-1). | |||||
|
ESM1_HUMAN
|
||||||
| NC score | 0.382938 (rank : 13) | θ value | 1.69304e-06 (rank : 14) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NQ30, Q15330, Q96ES3 | Gene names | ESM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-specific molecule 1 precursor (ESM-1 secretory protein) (ESM-1). | |||||
|
IBP3_HUMAN
|
||||||
| NC score | 0.360498 (rank : 14) | θ value | 9.92553e-07 (rank : 13) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P17936, Q2V509, Q6P1M6 | Gene names | IGFBP3, IBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
IBP2_HUMAN
|
||||||
| NC score | 0.339092 (rank : 15) | θ value | 1.69304e-06 (rank : 15) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P18065, Q14619, Q9UCL3 | Gene names | IGFBP2, BP2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
IBP1_MOUSE
|
||||||
| NC score | 0.339042 (rank : 16) | θ value | 2.21117e-06 (rank : 16) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
|
IBP5_MOUSE
|
||||||
| NC score | 0.336098 (rank : 17) | θ value | 1.43324e-05 (rank : 20) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q07079 | Gene names | Igfbp5, Igfbp-5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP5_HUMAN
|
||||||
| NC score | 0.335645 (rank : 18) | θ value | 1.43324e-05 (rank : 19) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P24593 | Gene names | IGFBP5, IBP5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP2_MOUSE
|
||||||
| NC score | 0.327496 (rank : 19) | θ value | 5.44631e-05 (rank : 23) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P47877, Q61722 | Gene names | Igfbp2, Igfbp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
IBP3_MOUSE
|
||||||
| NC score | 0.310904 (rank : 20) | θ value | 8.40245e-06 (rank : 18) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P47878 | Gene names | Igfbp3, Igfbp-3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
LCE1E_HUMAN
|
||||||
| NC score | 0.303293 (rank : 21) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
IBP4_HUMAN
|
||||||
| NC score | 0.300563 (rank : 22) | θ value | 1.87187e-05 (rank : 21) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P22692 | Gene names | IGFBP4, IBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
IBP1_HUMAN
|
||||||
| NC score | 0.298342 (rank : 23) | θ value | 0.00134147 (rank : 29) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P08833, Q8IYP5 | Gene names | IGFBP1, IBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1) (Placental protein 12) (PP12). | |||||
|
IBP4_MOUSE
|
||||||
| NC score | 0.295479 (rank : 24) | θ value | 3.19293e-05 (rank : 22) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P47879, O35666 | Gene names | Igfbp4, Igfbp-4 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
LCE1C_HUMAN
|
||||||
| NC score | 0.282716 (rank : 25) | θ value | 0.365318 (rank : 49) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
WISP2_HUMAN
|
||||||
| NC score | 0.258561 (rank : 26) | θ value | 0.0113563 (rank : 34) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O76076 | Gene names | WISP2, CCN5, CT58, CTGFL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WNT1-inducible signaling pathway protein 2 precursor (WISP-2) (Connective tissue growth factor-like protein) (CTGF-L) (Connective tissue growth factor-related protein 58). | |||||
|
CYR61_MOUSE
|
||||||
| NC score | 0.253766 (rank : 27) | θ value | 9.29e-05 (rank : 25) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P18406 | Gene names | Cyr61, Ccn1, Igfbp10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CYR61 precursor (Cysteine-rich, angiogenic inducer, 61) (Insulin-like growth factor-binding protein 10) (3CH61). | |||||
|
LCE1B_HUMAN
|
||||||
| NC score | 0.253674 (rank : 28) | θ value | 0.365318 (rank : 48) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5T7P3 | Gene names | LCE1B, LEP2, SPRL2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1B (Late envelope protein 2) (Small proline-rich-like epidermal differentiation complex protein 2A). | |||||
|
CYR61_HUMAN
|
||||||
| NC score | 0.253219 (rank : 29) | θ value | 9.29e-05 (rank : 24) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O00622, O14934, O43775, Q9BZL7 | Gene names | CYR61, CCN1, GIG1, IGFBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CYR61 precursor (Cysteine-rich, angiogenic inducer, 61) (Insulin-like growth factor-binding protein 10) (Protein GIG1). | |||||
|
CTGF_HUMAN
|
||||||
| NC score | 0.252472 (rank : 30) | θ value | 0.0113563 (rank : 33) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P29279, Q96QX2 | Gene names | CTGF, CCN2, HCS24 | |||
|
Domain Architecture |

|
|||||
| Description | Connective tissue growth factor precursor (Hypertrophic chondrocyte- specific protein 24). | |||||
|
LCE1D_HUMAN
|
||||||
| NC score | 0.252378 (rank : 31) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
NOV_HUMAN
|
||||||
| NC score | 0.246927 (rank : 32) | θ value | 0.000461057 (rank : 28) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P48745, Q96BY5 | Gene names | NOV, CCN3, IGFBP9, NOVH | |||
|
Domain Architecture |

|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
NOV_MOUSE
|
||||||
| NC score | 0.230570 (rank : 33) | θ value | 0.00228821 (rank : 31) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q64299, Q8CA67 | Gene names | Nov, Ccn3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
WISP2_MOUSE
|
||||||
| NC score | 0.222282 (rank : 34) | θ value | 0.21417 (rank : 44) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Z0G4, Q8CIC8 | Gene names | Wisp2, Ccn5, Ctgfl | |||
|
Domain Architecture |

|
|||||
| Description | WNT1-inducible signaling pathway protein 2 precursor (WISP-2) (Connective tissue growth factor-like protein) (CTGF-L). | |||||
|
WISP3_HUMAN
|
||||||
| NC score | 0.220588 (rank : 35) | θ value | 0.365318 (rank : 50) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O95389, Q6UXH6 | Gene names | WISP3, CCN6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WNT1-inducible signaling pathway protein 3 precursor (WISP-3). | |||||
|
IBP6_HUMAN
|
||||||
| NC score | 0.211900 (rank : 36) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P24592, Q14492 | Gene names | IGFBP6, IBP6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
CTGF_MOUSE
|
||||||
| NC score | 0.211000 (rank : 37) | θ value | 0.0193708 (rank : 35) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P29268, Q922U0 | Gene names | Ctgf, Ccn2, Fisp-12, Fisp12, Hcs24 | |||
|
Domain Architecture |

|
|||||
| Description | Connective tissue growth factor precursor (Protein FISP-12) (Hypertrophic chondrocyte-specific protein 24). | |||||
|
IBP6_MOUSE
|
||||||
| NC score | 0.206947 (rank : 38) | θ value | 0.21417 (rank : 42) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P47880 | Gene names | Igfbp6, Igfbp-6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
CRIM1_HUMAN
|
||||||
| NC score | 0.193842 (rank : 39) | θ value | 8.40245e-06 (rank : 17) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
WISP1_MOUSE
|
||||||
| NC score | 0.176329 (rank : 40) | θ value | 1.38821 (rank : 78) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O54775, Q80ZL1 | Gene names | Wisp1, Ccn4, Elm1 | |||
|
Domain Architecture |

|
|||||
| Description | WNT1-inducible signaling pathway protein 1 precursor (WISP-1) (ELM-1). | |||||
|
FSTL3_HUMAN
|
||||||
| NC score | 0.168448 (rank : 41) | θ value | 0.0431538 (rank : 37) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O95633 | Gene names | FSTL3, FLRG | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
LCE1A_HUMAN
|
||||||
| NC score | 0.167108 (rank : 42) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5T7P2 | Gene names | LCE1A, LEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1A (Late envelope protein 1). | |||||
|
WISP1_HUMAN
|
||||||
| NC score | 0.165641 (rank : 43) | θ value | 1.38821 (rank : 77) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O95388, Q9HCS3 | Gene names | WISP1, CCN4 | |||
|
Domain Architecture |

|
|||||
| Description | WNT1-inducible signaling pathway protein 1 precursor (WISP-1) (Wnt-1- induced secreted protein). | |||||
|
PDZD1_MOUSE
|
||||||
| NC score | 0.161367 (rank : 44) | θ value | 0.000270298 (rank : 26) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9JIL4, Q8CDP5, Q8R4G2, Q9CQ72 | Gene names | Pdzk1, Cap70, Nherf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
FSTL3_MOUSE
|
||||||
| NC score | 0.160083 (rank : 45) | θ value | 0.47712 (rank : 51) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9EQC7 | Gene names | Fstl3, Flrg | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
PDZD1_HUMAN
|
||||||
| NC score | 0.159160 (rank : 46) | θ value | 0.00134147 (rank : 30) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5T2W1, O60450, Q5T5P6, Q9BQ41 | Gene names | PDZK1, CAP70, NHERF3, PDZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
LCE1F_HUMAN
|
||||||
| NC score | 0.150459 (rank : 47) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5T754 | Gene names | LCE1F, LEP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1F (Late envelope protein 6). | |||||
|
ISK3_MOUSE
|
||||||
| NC score | 0.146196 (rank : 48) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P09036, Q5M9M3 | Gene names | Spink3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 3 precursor (Prostatic secretory glycoprotein) (P12). | |||||
|
FST_HUMAN
|
||||||
| NC score | 0.145881 (rank : 49) | θ value | 0.47712 (rank : 52) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P19883, Q9BTH0 | Gene names | FST | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
FST_MOUSE
|
||||||
| NC score | 0.145234 (rank : 50) | θ value | 0.47712 (rank : 53) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P47931 | Gene names | Fst | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
PDZ11_HUMAN
|
||||||
| NC score | 0.144783 (rank : 51) | θ value | 0.47712 (rank : 55) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5EBL8, Q6UWE1, Q9P0Q1 | Gene names | PDZD11, PDZK11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
PDZ11_MOUSE
|
||||||
| NC score | 0.143952 (rank : 52) | θ value | 0.47712 (rank : 56) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9CZG9 | Gene names | Pdzd11, Pdzk11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
LCE5A_HUMAN
|
||||||
| NC score | 0.142546 (rank : 53) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5TCM9 | Gene names | LCE5A, LEP18, SPRL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 5A (Late envelope protein 18) (Small proline-rich-like epidermal differentiation complex protein 5A). | |||||
|
NHERF_HUMAN
|
||||||
| NC score | 0.140117 (rank : 54) | θ value | 0.0193708 (rank : 36) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O14745, O43552, Q86WQ5 | Gene names | SLC9A3R1, NHERF | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
PDZD3_MOUSE
|
||||||
| NC score | 0.137537 (rank : 55) | θ value | 0.47712 (rank : 57) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99MJ6, Q8BWE5 | Gene names | Pdzd3, Pdzk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Natrium-phosphate cotransporter IIa C-terminal-associated protein 2) (NaPi-Cap2). | |||||
|
NHERF_MOUSE
|
||||||
| NC score | 0.137021 (rank : 56) | θ value | 0.00869519 (rank : 32) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P70441 | Gene names | Slc9a3r1, Nherf | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
LCE2D_HUMAN
|
||||||
| NC score | 0.136981 (rank : 57) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5TA82 | Gene names | LCE2D, LEP12, SPRL1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2D (Late envelope protein 12) (Small proline-rich-like epidermal differentiation complex protein 1A). | |||||
|
IPK1_HUMAN
|
||||||
| NC score | 0.133707 (rank : 58) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P00995 | Gene names | SPINK1, PSTI | |||
|
Domain Architecture |

|
|||||
| Description | Pancreatic secretory trypsin inhibitor precursor (Tumor-associated trypsin inhibitor) (TATI) (Serine protease inhibitor Kazal-type 1). | |||||
|
PDZD3_HUMAN
|
||||||
| NC score | 0.131097 (rank : 59) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q86UT5, Q8N6R4, Q8NAW7, Q8NEX7, Q9H5Z3 | Gene names | PDZD3, IKEPP, PDZK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Intestinal and kidney-enriched PDZ protein) (Protein DLNB27). | |||||
|
LCE2B_HUMAN
|
||||||
| NC score | 0.129317 (rank : 60) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14633 | Gene names | LCE2B, LEP10, SPRL1B, XP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2B (Late envelope protein 10) (Small proline-rich-like epidermal differentiation complex protein 1B) (Skin- specific protein Xp5). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.120997 (rank : 61) | θ value | 0.000461057 (rank : 27) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
NHRF2_HUMAN
|
||||||
| NC score | 0.118720 (rank : 62) | θ value | 0.47712 (rank : 54) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15599, O00272, O00556 | Gene names | SLC9A3R2, NHERF2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2). | |||||
|
LCE4A_HUMAN
|
||||||
| NC score | 0.118671 (rank : 63) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5TA78 | Gene names | LCE4A, LEP8, SPRL4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 4A (Late envelope protein 8) (Small proline-rich-like epidermal differentiation complex protein 4A). | |||||
|
SMOC1_MOUSE
|
||||||
| NC score | 0.117170 (rank : 64) | θ value | 0.62314 (rank : 61) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BLY1, Q9WVN9 | Gene names | Smoc1, Srg | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1) (SPARC-related gene protein). | |||||
|
TEFF2_MOUSE
|
||||||
| NC score | 0.115987 (rank : 65) | θ value | 0.163984 (rank : 40) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QYM9, Q3UY20, Q8CDH1, Q9JJE3 | Gene names | Tmeff2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Tomoregulin) (TR). | |||||
|
TICN2_MOUSE
|
||||||
| NC score | 0.113661 (rank : 66) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9ER58 | Gene names | Spock2, Ticn2 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
NHRF2_MOUSE
|
||||||
| NC score | 0.113040 (rank : 67) | θ value | 0.62314 (rank : 60) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JHL1, Q3TDR3, Q8BGL9, Q8BW05, Q9DCR6 | Gene names | Slc9a3r2, Nherf2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2) (Octs2). | |||||
|
TEFF2_HUMAN
|
||||||
| NC score | 0.112763 (rank : 68) | θ value | 0.279714 (rank : 46) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UIK5, Q4ZFW4, Q53H90, Q53RE1, Q8N2R5, Q9NR15, Q9NSS5, Q9P2Y9, Q9UK65 | Gene names | TMEFF2, HPP1, TENB2, TPEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Transmembrane protein containing epidermal growth factor and follistatin domains) (Tomoregulin) (TR) (Hyperplastic polyposis protein 1). | |||||
|
LCE2C_HUMAN
|
||||||
| NC score | 0.112606 (rank : 69) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5TA81 | Gene names | LCE2C, LEP11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2C (Late envelope protein 11). | |||||
|
TICN2_HUMAN
|
||||||
| NC score | 0.110102 (rank : 70) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q92563, Q6UW87 | Gene names | SPOCK2, KIAA0275, TICN2 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
SMOC1_HUMAN
|
||||||
| NC score | 0.108623 (rank : 71) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H4F8, Q96F78 | Gene names | SMOC1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1). | |||||
|
FSTL1_HUMAN
|
||||||
| NC score | 0.108430 (rank : 72) | θ value | 1.81305 (rank : 80) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q12841 | Gene names | FSTL1, FRP | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1). | |||||
|
FSTL1_MOUSE
|
||||||
| NC score | 0.106749 (rank : 73) | θ value | 1.81305 (rank : 81) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q62356, Q99JI9 | Gene names | Fstl1, Frp, Fstl, Tsc36 | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1) (TGF- beta-inducible protein TSC-36). | |||||
|
TICN1_MOUSE
|
||||||
| NC score | 0.102837 (rank : 74) | θ value | 3.0926 (rank : 102) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q62288 | Gene names | Spock1, Spock, Ticn1 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
ISK6_MOUSE
|
||||||
| NC score | 0.101765 (rank : 75) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BT20 | Gene names | Spink6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
SMOC2_MOUSE
|
||||||
| NC score | 0.101106 (rank : 76) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8CD91, Q8VCM1, Q9D9K2, Q9ER95 | Gene names | Smoc2 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2). | |||||
|
SMOC2_HUMAN
|
||||||
| NC score | 0.097749 (rank : 77) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9H3U7, Q5TAT7, Q5TAT8, Q86VV9, Q96SF3, Q9H1L3, Q9H1L4, Q9H3U0, Q9H4F7, Q9HCV2 | Gene names | SMOC2, SMAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2) (Smooth muscle-associated protein 2) (SMAP-2). | |||||
|
TICN3_MOUSE
|
||||||
| NC score | 0.097009 (rank : 78) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BKV0, Q9ER59 | Gene names | Spock3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
RHPN1_HUMAN
|
||||||
| NC score | 0.095710 (rank : 79) | θ value | 0.163984 (rank : 39) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TCX5, Q8TAV1 | Gene names | RHPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Rhophilin-1 (GTP-Rho-binding protein 1). | |||||
|
RHPN1_MOUSE
|
||||||
| NC score | 0.094578 (rank : 80) | θ value | 0.21417 (rank : 43) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q61085 | Gene names | Rhpn1, Grbp | |||
|
Domain Architecture |

|
|||||
| Description | Rhophilin-1 (GTP-Rho-binding protein 1). | |||||
|
WHRN_MOUSE
|
||||||
| NC score | 0.094398 (rank : 81) | θ value | 0.813845 (rank : 64) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
WHRN_HUMAN
|
||||||
| NC score | 0.094110 (rank : 82) | θ value | 1.38821 (rank : 76) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
TICN1_HUMAN
|
||||||
| NC score | 0.093344 (rank : 83) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q08629 | Gene names | SPOCK1, SPOCK, TIC1, TICN1 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
USH1C_HUMAN
|
||||||
| NC score | 0.093204 (rank : 84) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y6N9, Q96B29, Q9UM04, Q9UM17, Q9UPC3 | Gene names | USH1C, AIE75 | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein) (Autoimmune enteropathy- related antigen AIE-75) (Antigen NY-CO-38/NY-CO-37) (PDZ-73 protein) (NY-REN-3 antigen). | |||||
|
GRASP_HUMAN
|
||||||
| NC score | 0.092316 (rank : 85) | θ value | 0.813845 (rank : 62) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7Z6J2, Q6PIF8, Q7Z741 | Gene names | GRASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General receptor for phosphoinositides 1-associated scaffold protein (GRP1-associated scaffold protein). | |||||
|
TICN3_HUMAN
|
||||||
| NC score | 0.089309 (rank : 86) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9BQ16, O75705, Q6UW53, Q96Q26 | Gene names | SPOCK3, TICN3 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
LCE2A_HUMAN
|
||||||
| NC score | 0.089017 (rank : 87) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5TA79 | Gene names | LCE2A, LEP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2A (Late envelope protein 9). | |||||
|
ISK6_HUMAN
|
||||||
| NC score | 0.088432 (rank : 88) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6UWN8, Q8N5P0 | Gene names | SPINK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
GRASP_MOUSE
|
||||||
| NC score | 0.088039 (rank : 89) | θ value | 1.81305 (rank : 82) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JJA9, Q9JKL0 | Gene names | Grasp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General receptor for phosphoinositides 1-associated scaffold protein (GRP1-associated scaffold protein). | |||||
|
USH1C_MOUSE
|
||||||
| NC score | 0.086862 (rank : 90) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
ISK5_HUMAN
|
||||||
| NC score | 0.084972 (rank : 91) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NQ38, O75770, Q96PP2, Q96PP3 | Gene names | SPINK5 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 5 precursor (Lympho-epithelial Kazal-type-related inhibitor) (LEKTI) [Contains: Hemofiltrate peptide HF6478; Hemofiltrate peptide HF7665]. | |||||
|
FSTL4_MOUSE
|
||||||
| NC score | 0.083659 (rank : 92) | θ value | 0.62314 (rank : 59) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5STE3, Q5DU03 | Gene names | Fstl4, Kiaa1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4) (m- D/Bsp120I 1-1). | |||||
|
RECK_HUMAN
|
||||||
| NC score | 0.081624 (rank : 93) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O95980, Q8WX37 | Gene names | RECK | |||
|
Domain Architecture |

|
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (hRECK) (Suppressor of tumorigenicity 15) (ST15). | |||||
|
TX1B3_HUMAN
|
||||||
| NC score | 0.081357 (rank : 94) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O14907, Q7LCQ4 | Gene names | TAX1BP3, TIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 3 (Tax interaction protein 1) (TIP-1) (Glutaminase-interacting protein 3). | |||||
|
TX1B3_MOUSE
|
||||||
| NC score | 0.080853 (rank : 95) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9DBG9 | Gene names | Tax1bp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 3 (Tax interaction protein 1) (TIP-1). | |||||
|
PDZD7_HUMAN
|
||||||
| NC score | 0.080112 (rank : 96) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9H5P4, Q8N321 | Gene names | PDZD7, PDZK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 7. | |||||
|
RECK_MOUSE
|
||||||
| NC score | 0.078585 (rank : 97) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z0J1 | Gene names | Reck | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (mRECK). | |||||
|
PSCBP_HUMAN
|
||||||
| NC score | 0.076086 (rank : 98) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O60759, Q15630, Q8NE32 | Gene names | PSCDBP, CYTIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology Sec7 and coiled-coil domains-binding protein (Cytohesin-binding protein HE) (CYBR) (Cytohesin binder and regulator) (Cytohesin-interacting protein). | |||||
|
PZRN4_HUMAN
|
||||||
| NC score | 0.075669 (rank : 99) | θ value | 3.0926 (rank : 100) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6ZMN7, Q6N052, Q8IUU1, Q9NTP7 | Gene names | PDZRN4, LNX4, SEMCAP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 4 (Ligand of Numb-protein X 4) (SEMACAP3-like protein). | |||||
|
ZO3_MOUSE
|
||||||
| NC score | 0.075090 (rank : 100) | θ value | 0.47712 (rank : 58) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9QXY1 | Gene names | Tjp3, Zo3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
MPDZ_MOUSE
|
||||||
| NC score | 0.072359 (rank : 101) | θ value | 2.36792 (rank : 88) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8VBX6, O08783, Q6P7U4, Q80ZY8, Q8BKJ1, Q8C0H8, Q8VBV5, Q8VBY0, Q9Z1K3 | Gene names | Mpdz, Mupp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ domain protein 1). | |||||
|
PDZK4_MOUSE
|
||||||
| NC score | 0.071449 (rank : 102) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QY39, Q6PHP2 | Gene names | Pdzk4, Pdzrn4l, Xlu | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 4 (PDZ domain-containing RING finger protein 4-like protein). | |||||
|
LIN7B_HUMAN
|
||||||
| NC score | 0.069843 (rank : 103) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HAP6 | Gene names | LIN7B, MALS2, VELI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog B (Lin-7B) (hLin7B) (Mammalian lin-seven protein 2) (MALS-2) (Vertebrate lin-7 homolog 2) (Veli-2 protein) (hVeli2). | |||||
|
PDZK4_HUMAN
|
||||||
| NC score | 0.068947 (rank : 104) | θ value | 2.36792 (rank : 89) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q76G19, Q8NB75, Q9BUH9, Q9P284 | Gene names | PDZK4, KIAA1444, PDZRN4L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 4 (PDZ domain-containing RING finger protein 4-like protein). | |||||
|
FSTL4_HUMAN
|
||||||
| NC score | 0.068935 (rank : 105) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6MZW2, Q8TBU0, Q9UPU1 | Gene names | FSTL4, KIAA1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4). | |||||
|
LIN7B_MOUSE
|
||||||
| NC score | 0.068541 (rank : 106) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O88951 | Gene names | Lin7b, Mals2, Veli2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog B (Lin-7B) (Mammalian lin-seven protein 2) (MALS-2) (Vertebrate lin-7 homolog 2) (Veli-2 protein). | |||||
|
MPDZ_HUMAN
|
||||||
| NC score | 0.068323 (rank : 107) | θ value | 1.81305 (rank : 83) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O75970, O43798, Q5CZ80, Q5JTX3, Q5JTX6, Q5JTX7, Q5JUC3, Q5JUC4, Q5VZ62, Q8N790 | Gene names | MPDZ, MUPP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ- domain protein 1). | |||||
|
LIN7A_MOUSE
|
||||||
| NC score | 0.068240 (rank : 108) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8JZS0 | Gene names | Lin7a, Mals1, Veli1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog A (Lin-7A) (mLin-7) (Mammalian lin-seven protein 1) (MALS-1) (Vertebrate lin-7 homolog 1) (Veli-1 protein). | |||||
|
PZRN3_MOUSE
|
||||||
| NC score | 0.068194 (rank : 109) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q69ZS0, Q91Z03, Q9QY54, Q9QY55 | Gene names | Pdzrn3, Kiaa1095, Semcap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
LIN7A_HUMAN
|
||||||
| NC score | 0.068124 (rank : 110) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O14910, Q6LES3, Q7LDS4 | Gene names | LIN7A, MALS1, VELI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog A (Lin-7A) (hLin-7) (Mammalian lin-seven protein 1) (MALS-1) (Vertebrate lin-7 homolog 1) (Veli-1 protein) (Tax interaction protein 33) (TIP-33). | |||||
|
PZRN3_HUMAN
|
||||||
| NC score | 0.067591 (rank : 111) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UPQ7, Q8N2N7, Q96CC2, Q9NSQ2 | Gene names | PDZRN3, KIAA1095, LNX3, SEMCAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Ligand of Numb-protein X 3) (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
FSTL5_HUMAN
|
||||||
| NC score | 0.066350 (rank : 112) | θ value | 0.21417 (rank : 41) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N475, Q9NSW7, Q9ULF7 | Gene names | FSTL5, KIAA1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5). | |||||
|
FSTL5_MOUSE
|
||||||
| NC score | 0.064631 (rank : 113) | θ value | 0.365318 (rank : 47) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BFR2, Q80TG3, Q8C4T3 | Gene names | Fstl5, Kiaa1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5) (m- D/Bsp120I 1-2). | |||||
|
LIN7C_HUMAN
|
||||||
| NC score | 0.064483 (rank : 114) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NUP9 | Gene names | LIN7C, MALS3, VELI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog C (Lin-7C) (Mammalian lin-seven protein 3) (MALS-3) (Vertebrate lin-7 homolog 3) (Veli-3 protein). | |||||
|
LIN7C_MOUSE
|
||||||
| NC score | 0.064483 (rank : 115) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O88952 | Gene names | Lin7c, Mals3, Veli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog C (Lin-7C) (mLin7C) (Mammalian lin-seven protein 3) (MALS-3) (Vertebrate lin-7 homolog 3) (Veli-3 protein). | |||||
|
ISK4_HUMAN
|
||||||
| NC score | 0.063506 (rank : 116) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60575 | Gene names | SPINK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 4 precursor (Peptide PEC-60 homolog). | |||||
|
GOPC_MOUSE
|
||||||
| NC score | 0.063085 (rank : 117) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8BH60, Q8BSV4, Q920R1, Q9ET11 | Gene names | Gopc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi-associated PDZ and coiled-coil motif-containing protein (PDZ protein interacting specifically with TC10) (PIST). | |||||
|
PSCBP_MOUSE
|
||||||
| NC score | 0.062564 (rank : 118) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q91VY6, Q3TZH1, Q8R4T4, Q9QZA9 | Gene names | Pscdbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology Sec7 and coiled-coil domains-binding protein (Cytohesin-binding protein HE) (Cbp). | |||||
|
ZO1_HUMAN
|
||||||
| NC score | 0.062366 (rank : 119) | θ value | 1.81305 (rank : 84) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
GOPC_HUMAN
|
||||||
| NC score | 0.062077 (rank : 120) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HD26, Q59FS4, Q969U8 | Gene names | GOPC, CAL, FIG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi-associated PDZ and coiled-coil motif-containing protein (PDZ protein interacting specifically with TC10) (PIST) (CFTR-associated ligand) (Fused in glioblastoma). | |||||
|
ZO1_MOUSE
|
||||||
| NC score | 0.061851 (rank : 121) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P39447 | Gene names | Tjp1, Zo1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
ZO3_HUMAN
|
||||||
| NC score | 0.060582 (rank : 122) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95049 | Gene names | TJP3, ZO3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
SDCB1_HUMAN
|
||||||
| NC score | 0.058333 (rank : 123) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O00560, O00173, O43391 | Gene names | SDCBP, MDA9, SYCL | |||
|
Domain Architecture |

|
|||||
| Description | Syntenin-1 (Syndecan-binding protein 1) (Melanoma differentiation- associated protein 9) (MDA-9) (Scaffold protein Pbp1) (Pro-TGF-alpha cytoplasmic domain-interacting protein 18) (TACIP18). | |||||
|
GRIP2_HUMAN
|
||||||
| NC score | 0.057512 (rank : 124) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
HG2A_MOUSE
|
||||||
| NC score | 0.056348 (rank : 125) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P04441, O19452 | Gene names | Cd74, Ii | |||
|
Domain Architecture |

|
|||||
| Description | H-2 class II histocompatibility antigen gamma chain (MHC class II- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (CD74 antigen). | |||||
|
SDCB1_MOUSE
|
||||||
| NC score | 0.056341 (rank : 126) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O08992, Q544P5 | Gene names | Sdcbp | |||
|
Domain Architecture |

|
|||||
| Description | Syntenin-1 (Syndecan-binding protein 1) (Scaffold protein Pbp1). | |||||
|
LAP4_HUMAN
|
||||||
| NC score | 0.055947 (rank : 127) | θ value | 1.06291 (rank : 68) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
MAGI1_MOUSE
|
||||||
| NC score | 0.055808 (rank : 128) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
MAGI1_HUMAN
|
||||||
| NC score | 0.055468 (rank : 129) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
INADL_MOUSE
|
||||||
| NC score | 0.053613 (rank : 130) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q63ZW7, O70471, Q5PRG3, Q6P6J1, Q80YR8, Q8BPB9 | Gene names | Inadl, Cipp, Patj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (Pals1-associated tight junction protein) (Protein associated to tight junctions) (Channel-interacting PDZ domain-containing protein). | |||||
|
RPGF6_HUMAN
|
||||||
| NC score | 0.051050 (rank : 131) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8TEU7, Q8NI21, Q8TEU6, Q96PC1 | Gene names | RAPGEF6, PDZGEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 6 (PDZ domain-containing guanine nucleotide exchange factor 2) (PDZ-GEF2) (RA-GEF-2). | |||||
|
IPK2_HUMAN
|
||||||
| NC score | 0.050998 (rank : 132) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P20155 | Gene names | SPINK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease inhibitor Kazal-type 2 precursor (Acrosin-trypsin inhibitor) (HUSI-II). | |||||
|
LAP4_MOUSE
|
||||||
| NC score | 0.050228 (rank : 133) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
SYJ2B_HUMAN
|
||||||
| NC score | 0.050124 (rank : 134) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P57105, Q96IA4 | Gene names | SYNJ2BP, OMP25 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-2-binding protein (Mitochondrial outer membrane protein 25). | |||||
|
ARHGB_HUMAN
|
||||||
| NC score | 0.050010 (rank : 135) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
KRA56_HUMAN
|
||||||
| NC score | 0.047842 (rank : 136) | θ value | 0.125558 (rank : 38) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
SPR2B_MOUSE
|
||||||
| NC score | 0.045491 (rank : 137) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70554, Q7TSB2 | Gene names | Sprr2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small proline-rich protein 2B. | |||||
|
PSMD9_HUMAN
|
||||||
| NC score | 0.042417 (rank : 138) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00233, Q9BQ42 | Gene names | PSMD9 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 9 (26S proteasome regulatory subunit p27). | |||||
|
PSMD9_MOUSE
|
||||||
| NC score | 0.042199 (rank : 139) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CR00, Q3TG66 | Gene names | Psmd9 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 9 (26S proteasome regulatory subunit p27). | |||||
|
KR511_HUMAN
|
||||||
| NC score | 0.041971 (rank : 140) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
KRA53_HUMAN
|
||||||
| NC score | 0.040288 (rank : 141) | θ value | 0.279714 (rank : 45) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
MEGF6_HUMAN
|
||||||
| NC score | 0.031939 (rank : 142) | θ value | 0.813845 (rank : 63) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
TNR14_HUMAN
|
||||||
| NC score | 0.026027 (rank : 143) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92956, Q8WXR1, Q96J31, Q9UM65 | Gene names | TNFRSF14, HVEA, HVEM | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 14 precursor (Herpesvirus entry mediator A) (Tumor necrosis factor receptor-like 2) (TR2). | |||||
|
KR122_HUMAN
|
||||||
| NC score | 0.026016 (rank : 144) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59991 | Gene names | KRTAP12-2, KAP12.2, KRTAP12.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 12-2 (Keratin-associated protein 12.2) (High sulfur keratin-associated protein 12.2). | |||||
|
TENX_HUMAN
|
||||||
| NC score | 0.023726 (rank : 145) | θ value | 3.0926 (rank : 101) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
ITB2_MOUSE
|
||||||
| NC score | 0.016272 (rank : 146) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P11835, Q64482 | Gene names | Itgb2 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-2 precursor (Cell surface adhesion glycoproteins LFA- 1/CR3/P150,95 subunit beta) (Complement receptor C3 subunit beta) (CD18 antigen). | |||||
|
SO5A1_HUMAN
|
||||||
| NC score | 0.016153 (rank : 147) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H2Y9 | Gene names | SLCO5A1, SLC21A15 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier organic anion transporter family member 5A1 (Solute carrier family 21 member 15) (Organic anion transporter polypeptide- related protein 4) (OATP-RP4) (OATPRP4). | |||||
|
JAG2_MOUSE
|
||||||
| NC score | 0.013515 (rank : 148) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
ATX2_MOUSE
|
||||||
| NC score | 0.012187 (rank : 149) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
ADM1B_MOUSE
|
||||||
| NC score | 0.012002 (rank : 150) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R534, Q9R156 | Gene names | Adam1b | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 1b precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 1b) (Fertilin alpha 1b subunit). | |||||
|
CREL1_MOUSE
|
||||||
| NC score | 0.011342 (rank : 151) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91XD7, Q8BGJ8 | Gene names | Creld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
CUBN_HUMAN
|
||||||
| NC score | 0.011228 (rank : 152) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60494, Q5VTA6, Q96RU9 | Gene names | CUBN, IFCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor) (Intrinsic factor-vitamin B12 receptor) (460 kDa receptor) (Intestinal intrinsic factor receptor). | |||||
|
ADA12_MOUSE
|
||||||
| NC score | 0.011072 (rank : 153) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
ATX2_HUMAN
|
||||||
| NC score | 0.010988 (rank : 154) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
TENR_HUMAN
|
||||||
| NC score | 0.009602 (rank : 155) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92752, Q15568, Q5R3G0 | Gene names | TNR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin). | |||||
|
ZDHC3_MOUSE
|
||||||
| NC score | 0.006817 (rank : 156) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 140 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R173, Q6EMK3 | Gene names | Zdhhc3, Godz, Gramp1 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC3 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 3) (DHHC-3) (Golgi-specific DHHC zinc finger protein) (GABA-A receptor-associated membrane protein 1). | |||||