Please be patient as the page loads
|
IBP3_MOUSE
|
||||||
| SwissProt Accessions | P47878 | Gene names | Igfbp3, Igfbp-3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
IBP3_MOUSE
|
||||||
| θ value | 7.27209e-159 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P47878 | Gene names | Igfbp3, Igfbp-3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
IBP3_HUMAN
|
||||||
| θ value | 7.59556e-124 (rank : 2) | NC score | 0.989791 (rank : 2) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P17936, Q2V509, Q6P1M6 | Gene names | IGFBP3, IBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
IBP5_MOUSE
|
||||||
| θ value | 1.0458e-56 (rank : 3) | NC score | 0.961473 (rank : 3) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q07079 | Gene names | Igfbp5, Igfbp-5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP5_HUMAN
|
||||||
| θ value | 1.36585e-56 (rank : 4) | NC score | 0.960989 (rank : 4) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P24593 | Gene names | IGFBP5, IBP5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP2_HUMAN
|
||||||
| θ value | 2.86786e-30 (rank : 5) | NC score | 0.903467 (rank : 9) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P18065, Q14619, Q9UCL3 | Gene names | IGFBP2, BP2 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
IBP2_MOUSE
|
||||||
| θ value | 6.38894e-30 (rank : 6) | NC score | 0.905614 (rank : 8) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P47877, Q61722 | Gene names | Igfbp2, Igfbp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
IBP6_HUMAN
|
||||||
| θ value | 1.85889e-29 (rank : 7) | NC score | 0.911026 (rank : 7) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P24592, Q14492 | Gene names | IGFBP6, IBP6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
IBP1_MOUSE
|
||||||
| θ value | 4.14116e-29 (rank : 8) | NC score | 0.885102 (rank : 12) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
|
IBP4_MOUSE
|
||||||
| θ value | 5.40856e-29 (rank : 9) | NC score | 0.912039 (rank : 5) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P47879, O35666 | Gene names | Igfbp4, Igfbp-4 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
IBP4_HUMAN
|
||||||
| θ value | 7.80994e-28 (rank : 10) | NC score | 0.911063 (rank : 6) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P22692 | Gene names | IGFBP4, IBP4 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
IBP6_MOUSE
|
||||||
| θ value | 4.73814e-25 (rank : 11) | NC score | 0.897120 (rank : 10) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P47880 | Gene names | Igfbp6, Igfbp-6 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
IBP1_HUMAN
|
||||||
| θ value | 1.80048e-24 (rank : 12) | NC score | 0.886101 (rank : 11) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P08833, Q8IYP5 | Gene names | IGFBP1, IBP1 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1) (Placental protein 12) (PP12). | |||||
|
TICN1_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 13) | NC score | 0.467123 (rank : 14) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q08629 | Gene names | SPOCK1, SPOCK, TIC1, TICN1 | |||
|
Domain Architecture |
|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
TICN1_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 14) | NC score | 0.467373 (rank : 13) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62288 | Gene names | Spock1, Spock, Ticn1 | |||
|
Domain Architecture |
|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
TICN3_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 15) | NC score | 0.439499 (rank : 18) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BKV0, Q9ER59 | Gene names | Spock3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
TICN2_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 16) | NC score | 0.460791 (rank : 15) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q92563, Q6UW87 | Gene names | SPOCK2, KIAA0275, TICN2 | |||
|
Domain Architecture |
|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
TICN2_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 17) | NC score | 0.460719 (rank : 16) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9ER58 | Gene names | Spock2, Ticn2 | |||
|
Domain Architecture |
|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
TICN3_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 18) | NC score | 0.441913 (rank : 17) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BQ16, O75705, Q6UW53, Q96Q26 | Gene names | SPOCK3, TICN3 | |||
|
Domain Architecture |
|
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
HTRA1_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 19) | NC score | 0.310904 (rank : 25) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9R118 | Gene names | Htra1, Htra, Prss11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-). | |||||
|
NID2_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 20) | NC score | 0.186873 (rank : 39) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O88322 | Gene names | Nid2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Entactin-2). | |||||
|
HTRA1_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 21) | NC score | 0.306999 (rank : 27) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q92743, Q9UNS5 | Gene names | HTRA1, HTRA, PRSS11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-) (L56). | |||||
|
NID2_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 22) | NC score | 0.184747 (rank : 40) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
NID1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 23) | NC score | 0.183623 (rank : 42) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P14543, Q14942, Q59FL2, Q5TAF2, Q5TAF3, Q86XD7 | Gene names | NID1, NID | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
CRIM1_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 24) | NC score | 0.149503 (rank : 46) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
NID1_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 25) | NC score | 0.172781 (rank : 44) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P10493, Q8BQI3, Q8C3U8, Q8C9P6 | Gene names | Nid1, Ent | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
SMOC2_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 26) | NC score | 0.366622 (rank : 20) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CD91, Q8VCM1, Q9D9K2, Q9ER95 | Gene names | Smoc2 | |||
|
Domain Architecture |
|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2). | |||||
|
ESM1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 27) | NC score | 0.290716 (rank : 28) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NQ30, Q15330, Q96ES3 | Gene names | ESM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-specific molecule 1 precursor (ESM-1 secretory protein) (ESM-1). | |||||
|
SMOC2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 28) | NC score | 0.364015 (rank : 21) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H3U7, Q5TAT7, Q5TAT8, Q86VV9, Q96SF3, Q9H1L3, Q9H1L4, Q9H3U0, Q9H4F7, Q9HCV2 | Gene names | SMOC2, SMAP2 | |||
|
Domain Architecture |
|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2) (Smooth muscle-associated protein 2) (SMAP-2). | |||||
|
TACD1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 29) | NC score | 0.307292 (rank : 26) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P16422, P18180, Q96C47 | Gene names | TACSTD1, GA733-2, M1S2, TROP1 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor-associated calcium signal transducer 1 precursor (Major gastrointestinal tumor-associated protein GA733-2) (Epithelial cell surface antigen) (Epithelial glycoprotein) (EGP) (Adenocarcinoma- associated antigen) (KSA) (KS 1/4 antigen) (Cell surface glycoprotein Trop-1) (CD326 antigen). | |||||
|
THYG_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 30) | NC score | 0.208775 (rank : 32) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08710, O88590, Q9QWY7 | Gene names | Tg, Tgn | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
THYG_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 31) | NC score | 0.217543 (rank : 31) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P01266, O15274, O43899, Q15593, Q15948, Q9NYR1, Q9NYR2, Q9UMZ0, Q9UNY3 | Gene names | TG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
SMOC1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 32) | NC score | 0.343549 (rank : 23) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BLY1, Q9WVN9 | Gene names | Smoc1, Srg | |||
|
Domain Architecture |
|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1) (SPARC-related gene protein). | |||||
|
SMOC1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 33) | NC score | 0.345883 (rank : 22) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H4F8, Q96F78 | Gene names | SMOC1 | |||
|
Domain Architecture |
|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1). | |||||
|
HG2A_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 34) | NC score | 0.388622 (rank : 19) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P04441, O19452 | Gene names | Cd74, Ii | |||
|
Domain Architecture |
|
|||||
| Description | H-2 class II histocompatibility antigen gamma chain (MHC class II- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (CD74 antigen). | |||||
|
HTRA3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 35) | NC score | 0.192190 (rank : 37) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P83110, Q7Z7A2 | Gene names | HTRA3, PRSP | |||
|
Domain Architecture |
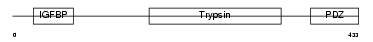
|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease). | |||||
|
HTRA3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 36) | NC score | 0.190200 (rank : 38) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9D236, Q6WLC5, Q6YDR0 | Gene names | Htra3, Prsp, Tasp | |||
|
Domain Architecture |
|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease) (Toll-associated serine protease). | |||||
|
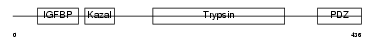
KAZD1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 37) | NC score | 0.201607 (rank : 35) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BJ66 | Gene names | Kazald1, Bono1, Igfbprp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor (IGFBP-related protein 10) (IGFBP-rP10) (Bone and odontoblast expressed protein 1). | |||||
|
TACD2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 38) | NC score | 0.273681 (rank : 29) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P09758, Q15658, Q96QD2 | Gene names | TACSTD2, GA733-1, M1S1, TROP2 | |||
|
Domain Architecture |
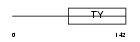
|
|||||
| Description | Tumor-associated calcium signal transducer 2 precursor (Pancreatic carcinoma marker protein GA733-1) (Cell surface glycoprotein Trop-2). | |||||
|
CTGF_HUMAN
|
||||||
| θ value | 0.21417 (rank : 39) | NC score | 0.140832 (rank : 49) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P29279, Q96QX2 | Gene names | CTGF, CCN2, HCS24 | |||
|
Domain Architecture |
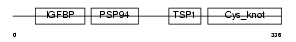
|
|||||
| Description | Connective tissue growth factor precursor (Hypertrophic chondrocyte- specific protein 24). | |||||
|
ESM1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 40) | NC score | 0.253077 (rank : 30) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QYY7 | Gene names | Esm1 | |||
|
Domain Architecture |
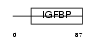
|
|||||
| Description | Endothelial cell-specific molecule 1 precursor (ESM-1 secretory protein) (ESM-1). | |||||
|
NOV_HUMAN
|
||||||
| θ value | 0.365318 (rank : 41) | NC score | 0.143166 (rank : 48) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P48745, Q96BY5 | Gene names | NOV, CCN3, IGFBP9, NOVH | |||
|
Domain Architecture |
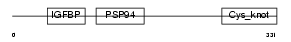
|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
WISP2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 42) | NC score | 0.184405 (rank : 41) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O76076 | Gene names | WISP2, CCN5, CT58, CTGFL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WNT1-inducible signaling pathway protein 2 precursor (WISP-2) (Connective tissue growth factor-like protein) (CTGF-L) (Connective tissue growth factor-related protein 58). | |||||
|
WISP2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 43) | NC score | 0.153945 (rank : 45) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Z0G4, Q8CIC8 | Gene names | Wisp2, Ccn5, Ctgfl | |||
|
Domain Architecture |
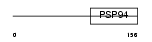
|
|||||
| Description | WNT1-inducible signaling pathway protein 2 precursor (WISP-2) (Connective tissue growth factor-like protein) (CTGF-L). | |||||
|
CN120_MOUSE
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.033291 (rank : 74) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DB96, Q8CIJ2 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C14orf120 homolog. | |||||
|
CYR61_HUMAN
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | 0.127598 (rank : 51) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00622, O14934, O43775, Q9BZL7 | Gene names | CYR61, CCN1, GIG1, IGFBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CYR61 precursor (Cysteine-rich, angiogenic inducer, 61) (Insulin-like growth factor-binding protein 10) (Protein GIG1). | |||||
|
CYR61_MOUSE
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.127605 (rank : 50) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P18406 | Gene names | Cyr61, Ccn1, Igfbp10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CYR61 precursor (Cysteine-rich, angiogenic inducer, 61) (Insulin-like growth factor-binding protein 10) (3CH61). | |||||
|
HG2A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 47) | NC score | 0.341289 (rank : 24) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P04233, Q14597, Q29832, Q5U0J8, Q8WLP6 | Gene names | CD74, DHLAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HLA class II histocompatibility antigen gamma chain (HLA-DR antigens- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (p33) (CD74 antigen). | |||||
|
KAZD1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 48) | NC score | 0.197640 (rank : 36) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96I82, Q6ZMB1, Q9BQ73 | Gene names | KAZALD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor. | |||||
|
IBP7_MOUSE
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.203269 (rank : 34) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61581, O88812, Q9EQW0 | Gene names | Igfbp7, Mac25 | |||
|
Domain Architecture |
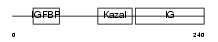
|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein). | |||||
|
LAMC3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.018392 (rank : 77) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6N6 | Gene names | LAMC3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.020090 (rank : 76) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
TNR14_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.020946 (rank : 75) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92956, Q8WXR1, Q96J31, Q9UM65 | Gene names | TNFRSF14, HVEA, HVEM | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 14 precursor (Herpesvirus entry mediator A) (Tumor necrosis factor receptor-like 2) (TR2). | |||||
|
IBP7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.207172 (rank : 33) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q16270, Q07822 | Gene names | IGFBP7, MAC25, PSF | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein) (Prostacyclin-stimulating factor) (PGI2-stimulating factor) (IGFBP-rP1). | |||||
|
LAMA5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.017130 (rank : 79) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61001, Q9JHQ6 | Gene names | Lama5 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
WISP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.082154 (rank : 56) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O54775, Q80ZL1 | Gene names | Wisp1, Ccn4, Elm1 | |||
|
Domain Architecture |

|
|||||
| Description | WNT1-inducible signaling pathway protein 1 precursor (WISP-1) (ELM-1). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.000685 (rank : 81) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RNF12_MOUSE
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.007149 (rank : 80) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WTV7 | Gene names | Rnf12, Rlim | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM). | |||||
|
WISP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.117132 (rank : 53) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O95389, Q6UXH6 | Gene names | WISP3, CCN6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WNT1-inducible signaling pathway protein 3 precursor (WISP-3). | |||||
|
ZHANG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.018224 (rank : 78) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NS37 | Gene names | ZF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor-binding transcription factor Zhangfei (HCF-binding transcription factor Zhangfei). | |||||
|
HTRA4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.148020 (rank : 47) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P83105, Q542Z4, Q6PF13 | Gene names | HTRA4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable serine protease HTRA4 precursor (EC 3.4.21.-). | |||||
|
CTGF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.099633 (rank : 54) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P29268, Q922U0 | Gene names | Ctgf, Ccn2, Fisp-12, Fisp12, Hcs24 | |||
|
Domain Architecture |
|
|||||
| Description | Connective tissue growth factor precursor (Protein FISP-12) (Hypertrophic chondrocyte-specific protein 24). | |||||
|
FSTL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.058961 (rank : 67) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q12841 | Gene names | FSTL1, FRP | |||
|
Domain Architecture |
|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1). | |||||
|
FSTL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.058953 (rank : 68) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62356, Q99JI9 | Gene names | Fstl1, Frp, Fstl, Tsc36 | |||
|
Domain Architecture |
|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1) (TGF- beta-inducible protein TSC-36). | |||||
|
FSTL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.059491 (rank : 64) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95633 | Gene names | FSTL3, FLRG | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
FSTL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.057122 (rank : 70) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9EQC7 | Gene names | Fstl3, Flrg | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
FST_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.059340 (rank : 65) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P19883, Q9BTH0 | Gene names | FST | |||
|
Domain Architecture |
|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
FST_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.057390 (rank : 69) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P47931 | Gene names | Fst | |||
|
Domain Architecture |
|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
HTRA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.078539 (rank : 58) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43464, Q9HBZ4, Q9P0Y3, Q9P0Y4 | Gene names | HTRA2, OMI, PRSS25 | |||
|
Domain Architecture |
|
|||||
| Description | Serine protease HTRA2, mitochondrial precursor (EC 3.4.21.-) (High temperature requirement protein A2) (HtrA2) (Omi stress-regulated endoprotease) (Serine proteinase OMI). | |||||
|
HTRA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.078777 (rank : 57) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JIY5, Q9R108 | Gene names | Htra2, Omi, Prss25 | |||
|
Domain Architecture |
|
|||||
| Description | Serine protease HTRA2, mitochondrial precursor (EC 3.4.21.-) (High temperature requirement protein A2) (HtrA2) (Omi stress-regulated endoprotease) (Serine proteinase OMI). | |||||
|
LCE1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.059558 (rank : 63) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5T7P3 | Gene names | LCE1B, LEP2, SPRL2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1B (Late envelope protein 2) (Small proline-rich-like epidermal differentiation complex protein 2A). | |||||
|
LCE1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.059184 (rank : 66) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
LCE1D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.055604 (rank : 72) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
LCE1E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.055793 (rank : 71) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
LCE2D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.054017 (rank : 73) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5TA82 | Gene names | LCE2D, LEP12, SPRL1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2D (Late envelope protein 12) (Small proline-rich-like epidermal differentiation complex protein 1A). | |||||
|
LCE4A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.179087 (rank : 43) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5TA78 | Gene names | LCE4A, LEP8, SPRL4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 4A (Late envelope protein 8) (Small proline-rich-like epidermal differentiation complex protein 4A). | |||||
|
NOV_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.118959 (rank : 52) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q64299, Q8CA67 | Gene names | Nov, Ccn3 | |||
|
Domain Architecture |
|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
SPRC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.077084 (rank : 59) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P09486 | Gene names | SPARC, ON | |||
|
Domain Architecture |
|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
SPRC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.074007 (rank : 60) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P07214 | Gene names | Sparc | |||
|
Domain Architecture |
|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
SPRL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.069100 (rank : 61) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
SPRL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.061929 (rank : 62) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
WISP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.082465 (rank : 55) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95388, Q9HCS3 | Gene names | WISP1, CCN4 | |||
|
Domain Architecture |
|
|||||
| Description | WNT1-inducible signaling pathway protein 1 precursor (WISP-1) (Wnt-1- induced secreted protein). | |||||
|
IBP3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 7.27209e-159 (rank : 1) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P47878 | Gene names | Igfbp3, Igfbp-3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
IBP3_HUMAN
|
||||||
| NC score | 0.989791 (rank : 2) | θ value | 7.59556e-124 (rank : 2) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P17936, Q2V509, Q6P1M6 | Gene names | IGFBP3, IBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
IBP5_MOUSE
|
||||||
| NC score | 0.961473 (rank : 3) | θ value | 1.0458e-56 (rank : 3) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q07079 | Gene names | Igfbp5, Igfbp-5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP5_HUMAN
|
||||||
| NC score | 0.960989 (rank : 4) | θ value | 1.36585e-56 (rank : 4) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P24593 | Gene names | IGFBP5, IBP5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP4_MOUSE
|
||||||
| NC score | 0.912039 (rank : 5) | θ value | 5.40856e-29 (rank : 9) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P47879, O35666 | Gene names | Igfbp4, Igfbp-4 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
IBP4_HUMAN
|
||||||
| NC score | 0.911063 (rank : 6) | θ value | 7.80994e-28 (rank : 10) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P22692 | Gene names | IGFBP4, IBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
IBP6_HUMAN
|
||||||
| NC score | 0.911026 (rank : 7) | θ value | 1.85889e-29 (rank : 7) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P24592, Q14492 | Gene names | IGFBP6, IBP6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
IBP2_MOUSE
|
||||||
| NC score | 0.905614 (rank : 8) | θ value | 6.38894e-30 (rank : 6) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P47877, Q61722 | Gene names | Igfbp2, Igfbp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
IBP2_HUMAN
|
||||||
| NC score | 0.903467 (rank : 9) | θ value | 2.86786e-30 (rank : 5) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P18065, Q14619, Q9UCL3 | Gene names | IGFBP2, BP2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
IBP6_MOUSE
|
||||||
| NC score | 0.897120 (rank : 10) | θ value | 4.73814e-25 (rank : 11) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P47880 | Gene names | Igfbp6, Igfbp-6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
IBP1_HUMAN
|
||||||
| NC score | 0.886101 (rank : 11) | θ value | 1.80048e-24 (rank : 12) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P08833, Q8IYP5 | Gene names | IGFBP1, IBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1) (Placental protein 12) (PP12). | |||||
|
IBP1_MOUSE
|
||||||
| NC score | 0.885102 (rank : 12) | θ value | 4.14116e-29 (rank : 8) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
|
TICN1_MOUSE
|
||||||
| NC score | 0.467373 (rank : 13) | θ value | 5.81887e-07 (rank : 14) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62288 | Gene names | Spock1, Spock, Ticn1 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
TICN1_HUMAN
|
||||||
| NC score | 0.467123 (rank : 14) | θ value | 5.81887e-07 (rank : 13) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q08629 | Gene names | SPOCK1, SPOCK, TIC1, TICN1 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
TICN2_HUMAN
|
||||||
| NC score | 0.460791 (rank : 15) | θ value | 3.77169e-06 (rank : 16) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q92563, Q6UW87 | Gene names | SPOCK2, KIAA0275, TICN2 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
TICN2_MOUSE
|
||||||
| NC score | 0.460719 (rank : 16) | θ value | 3.77169e-06 (rank : 17) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9ER58 | Gene names | Spock2, Ticn2 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
TICN3_HUMAN
|
||||||
| NC score | 0.441913 (rank : 17) | θ value | 4.92598e-06 (rank : 18) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BQ16, O75705, Q6UW53, Q96Q26 | Gene names | SPOCK3, TICN3 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
TICN3_MOUSE
|
||||||
| NC score | 0.439499 (rank : 18) | θ value | 7.59969e-07 (rank : 15) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BKV0, Q9ER59 | Gene names | Spock3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
HG2A_MOUSE
|
||||||
| NC score | 0.388622 (rank : 19) | θ value | 0.0431538 (rank : 34) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P04441, O19452 | Gene names | Cd74, Ii | |||
|
Domain Architecture |

|
|||||
| Description | H-2 class II histocompatibility antigen gamma chain (MHC class II- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (CD74 antigen). | |||||
|
SMOC2_MOUSE
|
||||||
| NC score | 0.366622 (rank : 20) | θ value | 0.00134147 (rank : 26) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CD91, Q8VCM1, Q9D9K2, Q9ER95 | Gene names | Smoc2 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2). | |||||
|
SMOC2_HUMAN
|
||||||
| NC score | 0.364015 (rank : 21) | θ value | 0.00869519 (rank : 28) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H3U7, Q5TAT7, Q5TAT8, Q86VV9, Q96SF3, Q9H1L3, Q9H1L4, Q9H3U0, Q9H4F7, Q9HCV2 | Gene names | SMOC2, SMAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2) (Smooth muscle-associated protein 2) (SMAP-2). | |||||
|
SMOC1_HUMAN
|
||||||
| NC score | 0.345883 (rank : 22) | θ value | 0.0330416 (rank : 33) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H4F8, Q96F78 | Gene names | SMOC1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1). | |||||
|
SMOC1_MOUSE
|
||||||
| NC score | 0.343549 (rank : 23) | θ value | 0.0252991 (rank : 32) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BLY1, Q9WVN9 | Gene names | Smoc1, Srg | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1) (SPARC-related gene protein). | |||||
|
HG2A_HUMAN
|
||||||
| NC score | 0.341289 (rank : 24) | θ value | 0.813845 (rank : 47) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P04233, Q14597, Q29832, Q5U0J8, Q8WLP6 | Gene names | CD74, DHLAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HLA class II histocompatibility antigen gamma chain (HLA-DR antigens- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (p33) (CD74 antigen). | |||||
|
HTRA1_MOUSE
|
||||||
| NC score | 0.310904 (rank : 25) | θ value | 8.40245e-06 (rank : 19) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9R118 | Gene names | Htra1, Htra, Prss11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-). | |||||
|
TACD1_HUMAN
|
||||||
| NC score | 0.307292 (rank : 26) | θ value | 0.0148317 (rank : 29) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P16422, P18180, Q96C47 | Gene names | TACSTD1, GA733-2, M1S2, TROP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor-associated calcium signal transducer 1 precursor (Major gastrointestinal tumor-associated protein GA733-2) (Epithelial cell surface antigen) (Epithelial glycoprotein) (EGP) (Adenocarcinoma- associated antigen) (KSA) (KS 1/4 antigen) (Cell surface glycoprotein Trop-1) (CD326 antigen). | |||||
|
HTRA1_HUMAN
|
||||||
| NC score | 0.306999 (rank : 27) | θ value | 7.1131e-05 (rank : 21) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q92743, Q9UNS5 | Gene names | HTRA1, HTRA, PRSS11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-) (L56). | |||||
|
ESM1_HUMAN
|
||||||
| NC score | 0.290716 (rank : 28) | θ value | 0.00228821 (rank : 27) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NQ30, Q15330, Q96ES3 | Gene names | ESM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-specific molecule 1 precursor (ESM-1 secretory protein) (ESM-1). | |||||
|
TACD2_HUMAN
|
||||||
| NC score | 0.273681 (rank : 29) | θ value | 0.163984 (rank : 38) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P09758, Q15658, Q96QD2 | Gene names | TACSTD2, GA733-1, M1S1, TROP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor-associated calcium signal transducer 2 precursor (Pancreatic carcinoma marker protein GA733-1) (Cell surface glycoprotein Trop-2). | |||||
|
ESM1_MOUSE
|
||||||
| NC score | 0.253077 (rank : 30) | θ value | 0.279714 (rank : 40) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QYY7 | Gene names | Esm1 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial cell-specific molecule 1 precursor (ESM-1 secretory protein) (ESM-1). | |||||
|
THYG_HUMAN
|
||||||
| NC score | 0.217543 (rank : 31) | θ value | 0.0193708 (rank : 31) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P01266, O15274, O43899, Q15593, Q15948, Q9NYR1, Q9NYR2, Q9UMZ0, Q9UNY3 | Gene names | TG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
THYG_MOUSE
|
||||||
| NC score | 0.208775 (rank : 32) | θ value | 0.0148317 (rank : 30) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08710, O88590, Q9QWY7 | Gene names | Tg, Tgn | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
IBP7_HUMAN
|
||||||
| NC score | 0.207172 (rank : 33) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q16270, Q07822 | Gene names | IGFBP7, MAC25, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein) (Prostacyclin-stimulating factor) (PGI2-stimulating factor) (IGFBP-rP1). | |||||
|
IBP7_MOUSE
|
||||||
| NC score | 0.203269 (rank : 34) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61581, O88812, Q9EQW0 | Gene names | Igfbp7, Mac25 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein). | |||||
|
KAZD1_MOUSE
|
||||||
| NC score | 0.201607 (rank : 35) | θ value | 0.163984 (rank : 37) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BJ66 | Gene names | Kazald1, Bono1, Igfbprp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor (IGFBP-related protein 10) (IGFBP-rP10) (Bone and odontoblast expressed protein 1). | |||||
|
KAZD1_HUMAN
|
||||||
| NC score | 0.197640 (rank : 36) | θ value | 0.813845 (rank : 48) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96I82, Q6ZMB1, Q9BQ73 | Gene names | KAZALD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor. | |||||
|
HTRA3_HUMAN
|
||||||
| NC score | 0.192190 (rank : 37) | θ value | 0.163984 (rank : 35) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P83110, Q7Z7A2 | Gene names | HTRA3, PRSP | |||
|
Domain Architecture |

|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease). | |||||
|
HTRA3_MOUSE
|
||||||
| NC score | 0.190200 (rank : 38) | θ value | 0.163984 (rank : 36) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9D236, Q6WLC5, Q6YDR0 | Gene names | Htra3, Prsp, Tasp | |||
|
Domain Architecture |

|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease) (Toll-associated serine protease). | |||||
|
NID2_MOUSE
|
||||||
| NC score | 0.186873 (rank : 39) | θ value | 1.87187e-05 (rank : 20) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O88322 | Gene names | Nid2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Entactin-2). | |||||
|
NID2_HUMAN
|
||||||
| NC score | 0.184747 (rank : 40) | θ value | 7.1131e-05 (rank : 22) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
WISP2_HUMAN
|
||||||
| NC score | 0.184405 (rank : 41) | θ value | 0.365318 (rank : 42) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O76076 | Gene names | WISP2, CCN5, CT58, CTGFL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WNT1-inducible signaling pathway protein 2 precursor (WISP-2) (Connective tissue growth factor-like protein) (CTGF-L) (Connective tissue growth factor-related protein 58). | |||||
|
NID1_HUMAN
|
||||||
| NC score | 0.183623 (rank : 42) | θ value | 0.000121331 (rank : 23) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P14543, Q14942, Q59FL2, Q5TAF2, Q5TAF3, Q86XD7 | Gene names | NID1, NID | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
LCE4A_HUMAN
|
||||||
| NC score | 0.179087 (rank : 43) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5TA78 | Gene names | LCE4A, LEP8, SPRL4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 4A (Late envelope protein 8) (Small proline-rich-like epidermal differentiation complex protein 4A). | |||||
|
NID1_MOUSE
|
||||||
| NC score | 0.172781 (rank : 44) | θ value | 0.000270298 (rank : 25) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P10493, Q8BQI3, Q8C3U8, Q8C9P6 | Gene names | Nid1, Ent | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
WISP2_MOUSE
|
||||||
| NC score | 0.153945 (rank : 45) | θ value | 0.47712 (rank : 43) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Z0G4, Q8CIC8 | Gene names | Wisp2, Ccn5, Ctgfl | |||
|
Domain Architecture |

|
|||||
| Description | WNT1-inducible signaling pathway protein 2 precursor (WISP-2) (Connective tissue growth factor-like protein) (CTGF-L). | |||||
|
CRIM1_HUMAN
|
||||||
| NC score | 0.149503 (rank : 46) | θ value | 0.000270298 (rank : 24) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
HTRA4_HUMAN
|
||||||
| NC score | 0.148020 (rank : 47) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P83105, Q542Z4, Q6PF13 | Gene names | HTRA4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable serine protease HTRA4 precursor (EC 3.4.21.-). | |||||
|
NOV_HUMAN
|
||||||
| NC score | 0.143166 (rank : 48) | θ value | 0.365318 (rank : 41) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P48745, Q96BY5 | Gene names | NOV, CCN3, IGFBP9, NOVH | |||
|
Domain Architecture |

|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
CTGF_HUMAN
|
||||||
| NC score | 0.140832 (rank : 49) | θ value | 0.21417 (rank : 39) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P29279, Q96QX2 | Gene names | CTGF, CCN2, HCS24 | |||
|
Domain Architecture |

|
|||||
| Description | Connective tissue growth factor precursor (Hypertrophic chondrocyte- specific protein 24). | |||||
|
CYR61_MOUSE
|
||||||
| NC score | 0.127605 (rank : 50) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P18406 | Gene names | Cyr61, Ccn1, Igfbp10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CYR61 precursor (Cysteine-rich, angiogenic inducer, 61) (Insulin-like growth factor-binding protein 10) (3CH61). | |||||
|
CYR61_HUMAN
|
||||||
| NC score | 0.127598 (rank : 51) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00622, O14934, O43775, Q9BZL7 | Gene names | CYR61, CCN1, GIG1, IGFBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CYR61 precursor (Cysteine-rich, angiogenic inducer, 61) (Insulin-like growth factor-binding protein 10) (Protein GIG1). | |||||
|
NOV_MOUSE
|
||||||
| NC score | 0.118959 (rank : 52) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q64299, Q8CA67 | Gene names | Nov, Ccn3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
WISP3_HUMAN
|
||||||
| NC score | 0.117132 (rank : 53) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O95389, Q6UXH6 | Gene names | WISP3, CCN6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WNT1-inducible signaling pathway protein 3 precursor (WISP-3). | |||||
|
CTGF_MOUSE
|
||||||
| NC score | 0.099633 (rank : 54) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P29268, Q922U0 | Gene names | Ctgf, Ccn2, Fisp-12, Fisp12, Hcs24 | |||
|
Domain Architecture |

|
|||||
| Description | Connective tissue growth factor precursor (Protein FISP-12) (Hypertrophic chondrocyte-specific protein 24). | |||||
|
WISP1_HUMAN
|
||||||
| NC score | 0.082465 (rank : 55) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95388, Q9HCS3 | Gene names | WISP1, CCN4 | |||
|
Domain Architecture |

|
|||||
| Description | WNT1-inducible signaling pathway protein 1 precursor (WISP-1) (Wnt-1- induced secreted protein). | |||||
|
WISP1_MOUSE
|
||||||
| NC score | 0.082154 (rank : 56) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O54775, Q80ZL1 | Gene names | Wisp1, Ccn4, Elm1 | |||
|
Domain Architecture |

|
|||||
| Description | WNT1-inducible signaling pathway protein 1 precursor (WISP-1) (ELM-1). | |||||
|
HTRA2_MOUSE
|
||||||
| NC score | 0.078777 (rank : 57) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JIY5, Q9R108 | Gene names | Htra2, Omi, Prss25 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA2, mitochondrial precursor (EC 3.4.21.-) (High temperature requirement protein A2) (HtrA2) (Omi stress-regulated endoprotease) (Serine proteinase OMI). | |||||
|
HTRA2_HUMAN
|
||||||
| NC score | 0.078539 (rank : 58) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43464, Q9HBZ4, Q9P0Y3, Q9P0Y4 | Gene names | HTRA2, OMI, PRSS25 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA2, mitochondrial precursor (EC 3.4.21.-) (High temperature requirement protein A2) (HtrA2) (Omi stress-regulated endoprotease) (Serine proteinase OMI). | |||||
|
SPRC_HUMAN
|
||||||
| NC score | 0.077084 (rank : 59) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P09486 | Gene names | SPARC, ON | |||
|
Domain Architecture |

|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
SPRC_MOUSE
|
||||||
| NC score | 0.074007 (rank : 60) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P07214 | Gene names | Sparc | |||
|
Domain Architecture |

|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
SPRL1_HUMAN
|
||||||
| NC score | 0.069100 (rank : 61) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
SPRL1_MOUSE
|
||||||
| NC score | 0.061929 (rank : 62) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
LCE1B_HUMAN
|
||||||
| NC score | 0.059558 (rank : 63) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5T7P3 | Gene names | LCE1B, LEP2, SPRL2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1B (Late envelope protein 2) (Small proline-rich-like epidermal differentiation complex protein 2A). | |||||
|
FSTL3_HUMAN
|
||||||
| NC score | 0.059491 (rank : 64) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95633 | Gene names | FSTL3, FLRG | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
FST_HUMAN
|
||||||
| NC score | 0.059340 (rank : 65) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P19883, Q9BTH0 | Gene names | FST | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
LCE1C_HUMAN
|
||||||
| NC score | 0.059184 (rank : 66) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
FSTL1_HUMAN
|
||||||
| NC score | 0.058961 (rank : 67) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q12841 | Gene names | FSTL1, FRP | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1). | |||||
|
FSTL1_MOUSE
|
||||||
| NC score | 0.058953 (rank : 68) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62356, Q99JI9 | Gene names | Fstl1, Frp, Fstl, Tsc36 | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1) (TGF- beta-inducible protein TSC-36). | |||||
|
FST_MOUSE
|
||||||
| NC score | 0.057390 (rank : 69) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P47931 | Gene names | Fst | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
FSTL3_MOUSE
|
||||||
| NC score | 0.057122 (rank : 70) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9EQC7 | Gene names | Fstl3, Flrg | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
LCE1E_HUMAN
|
||||||
| NC score | 0.055793 (rank : 71) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
LCE1D_HUMAN
|
||||||
| NC score | 0.055604 (rank : 72) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
LCE2D_HUMAN
|
||||||
| NC score | 0.054017 (rank : 73) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5TA82 | Gene names | LCE2D, LEP12, SPRL1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2D (Late envelope protein 12) (Small proline-rich-like epidermal differentiation complex protein 1A). | |||||
|
CN120_MOUSE
|
||||||
| NC score | 0.033291 (rank : 74) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DB96, Q8CIJ2 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C14orf120 homolog. | |||||
|
TNR14_HUMAN
|
||||||
| NC score | 0.020946 (rank : 75) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92956, Q8WXR1, Q96J31, Q9UM65 | Gene names | TNFRSF14, HVEA, HVEM | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 14 precursor (Herpesvirus entry mediator A) (Tumor necrosis factor receptor-like 2) (TR2). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.020090 (rank : 76) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
LAMC3_HUMAN
|
||||||
| NC score | 0.018392 (rank : 77) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y6N6 | Gene names | LAMC3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
ZHANG_HUMAN
|
||||||
| NC score | 0.018224 (rank : 78) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NS37 | Gene names | ZF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor-binding transcription factor Zhangfei (HCF-binding transcription factor Zhangfei). | |||||
|
LAMA5_MOUSE
|
||||||
| NC score | 0.017130 (rank : 79) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61001, Q9JHQ6 | Gene names | Lama5 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
RNF12_MOUSE
|
||||||
| NC score | 0.007149 (rank : 80) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WTV7 | Gene names | Rnf12, Rlim | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.000685 (rank : 81) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||