Please be patient as the page loads
|
IBP1_MOUSE
|
||||||
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
IBP1_MOUSE
|
||||||
| θ value | 4.70271e-166 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
|
IBP1_HUMAN
|
||||||
| θ value | 4.1872e-106 (rank : 2) | NC score | 0.982076 (rank : 2) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P08833, Q8IYP5 | Gene names | IGFBP1, IBP1 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1) (Placental protein 12) (PP12). | |||||
|
IBP4_HUMAN
|
||||||
| θ value | 8.89434e-40 (rank : 3) | NC score | 0.918668 (rank : 3) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P22692 | Gene names | IGFBP4, IBP4 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
IBP4_MOUSE
|
||||||
| θ value | 1.98146e-39 (rank : 4) | NC score | 0.917393 (rank : 4) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P47879, O35666 | Gene names | Igfbp4, Igfbp-4 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
IBP2_MOUSE
|
||||||
| θ value | 2.42216e-37 (rank : 5) | NC score | 0.914044 (rank : 5) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P47877, Q61722 | Gene names | Igfbp2, Igfbp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
IBP2_HUMAN
|
||||||
| θ value | 3.4976e-36 (rank : 6) | NC score | 0.913710 (rank : 6) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P18065, Q14619, Q9UCL3 | Gene names | IGFBP2, BP2 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
IBP5_HUMAN
|
||||||
| θ value | 6.59618e-35 (rank : 7) | NC score | 0.904623 (rank : 7) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P24593 | Gene names | IGFBP5, IBP5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP5_MOUSE
|
||||||
| θ value | 1.46948e-34 (rank : 8) | NC score | 0.903815 (rank : 8) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q07079 | Gene names | Igfbp5, Igfbp-5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP3_HUMAN
|
||||||
| θ value | 5.77852e-31 (rank : 9) | NC score | 0.896221 (rank : 9) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P17936, Q2V509, Q6P1M6 | Gene names | IGFBP3, IBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
IBP3_MOUSE
|
||||||
| θ value | 4.14116e-29 (rank : 10) | NC score | 0.885102 (rank : 10) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P47878 | Gene names | Igfbp3, Igfbp-3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
IBP6_HUMAN
|
||||||
| θ value | 1.47631e-18 (rank : 11) | NC score | 0.839644 (rank : 11) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P24592, Q14492 | Gene names | IGFBP6, IBP6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
IBP6_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 12) | NC score | 0.826219 (rank : 12) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P47880 | Gene names | Igfbp6, Igfbp-6 | |||
|
Domain Architecture |
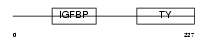
|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
HTRA1_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 13) | NC score | 0.349144 (rank : 23) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92743, Q9UNS5 | Gene names | HTRA1, HTRA, PRSS11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-) (L56). | |||||
|
HTRA1_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 14) | NC score | 0.339042 (rank : 26) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9R118 | Gene names | Htra1, Htra, Prss11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-). | |||||
|
SMOC2_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 15) | NC score | 0.379833 (rank : 16) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8CD91, Q8VCM1, Q9D9K2, Q9ER95 | Gene names | Smoc2 | |||
|
Domain Architecture |
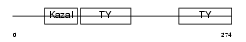
|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2). | |||||
|
SMOC2_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 16) | NC score | 0.374843 (rank : 19) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9H3U7, Q5TAT7, Q5TAT8, Q86VV9, Q96SF3, Q9H1L3, Q9H1L4, Q9H3U0, Q9H4F7, Q9HCV2 | Gene names | SMOC2, SMAP2 | |||
|
Domain Architecture |
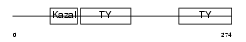
|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2) (Smooth muscle-associated protein 2) (SMAP-2). | |||||
|
IBP7_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 17) | NC score | 0.256872 (rank : 27) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61581, O88812, Q9EQW0 | Gene names | Igfbp7, Mac25 | |||
|
Domain Architecture |
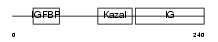
|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein). | |||||
|
THYG_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 18) | NC score | 0.213767 (rank : 34) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O08710, O88590, Q9QWY7 | Gene names | Tg, Tgn | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
TICN2_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 19) | NC score | 0.379598 (rank : 17) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q92563, Q6UW87 | Gene names | SPOCK2, KIAA0275, TICN2 | |||
|
Domain Architecture |
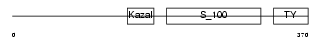
|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
THYG_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 20) | NC score | 0.227822 (rank : 31) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P01266, O15274, O43899, Q15593, Q15948, Q9NYR1, Q9NYR2, Q9UMZ0, Q9UNY3 | Gene names | TG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
TICN1_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 21) | NC score | 0.382433 (rank : 13) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q62288 | Gene names | Spock1, Spock, Ticn1 | |||
|
Domain Architecture |
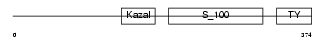
|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
TICN1_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 22) | NC score | 0.380532 (rank : 14) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q08629 | Gene names | SPOCK1, SPOCK, TIC1, TICN1 | |||
|
Domain Architecture |
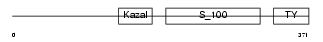
|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
HG2A_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 23) | NC score | 0.380054 (rank : 15) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P04441, O19452 | Gene names | Cd74, Ii | |||
|
Domain Architecture |
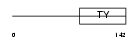
|
|||||
| Description | H-2 class II histocompatibility antigen gamma chain (MHC class II- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (CD74 antigen). | |||||
|
IBP7_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 24) | NC score | 0.255629 (rank : 28) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q16270, Q07822 | Gene names | IGFBP7, MAC25, PSF | |||
|
Domain Architecture |
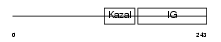
|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein) (Prostacyclin-stimulating factor) (PGI2-stimulating factor) (IGFBP-rP1). | |||||
|
HG2A_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 25) | NC score | 0.346452 (rank : 24) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P04233, Q14597, Q29832, Q5U0J8, Q8WLP6 | Gene names | CD74, DHLAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HLA class II histocompatibility antigen gamma chain (HLA-DR antigens- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (p33) (CD74 antigen). | |||||
|
SMOC1_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 26) | NC score | 0.349846 (rank : 22) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9H4F8, Q96F78 | Gene names | SMOC1 | |||
|
Domain Architecture |
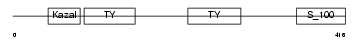
|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1). | |||||
|
SMOC1_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 27) | NC score | 0.350417 (rank : 21) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BLY1, Q9WVN9 | Gene names | Smoc1, Srg | |||
|
Domain Architecture |
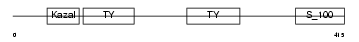
|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1) (SPARC-related gene protein). | |||||
|
TICN2_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 28) | NC score | 0.376294 (rank : 18) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9ER58 | Gene names | Spock2, Ticn2 | |||
|
Domain Architecture |
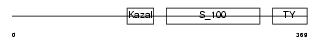
|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
NOTC3_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 29) | NC score | 0.027519 (rank : 83) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
TICN3_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 30) | NC score | 0.353041 (rank : 20) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9BQ16, O75705, Q6UW53, Q96Q26 | Gene names | SPOCK3, TICN3 | |||
|
Domain Architecture |
|
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
NID2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 31) | NC score | 0.149760 (rank : 44) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
WISP2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 32) | NC score | 0.202412 (rank : 37) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O76076 | Gene names | WISP2, CCN5, CT58, CTGFL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WNT1-inducible signaling pathway protein 2 precursor (WISP-2) (Connective tissue growth factor-like protein) (CTGF-L) (Connective tissue growth factor-related protein 58). | |||||
|
MCSP_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 33) | NC score | 0.077956 (rank : 65) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P15265, O70613, Q6P8N3 | Gene names | Smcp, Mcs, Mcsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
GSCL_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 34) | NC score | 0.023342 (rank : 84) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15499 | Gene names | GSCL | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein goosecoid-like (GSC-2). | |||||
|
TICN3_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 35) | NC score | 0.346390 (rank : 25) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BKV0, Q9ER59 | Gene names | Spock3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
CRIM1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 36) | NC score | 0.141521 (rank : 47) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
KAZD1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 37) | NC score | 0.224861 (rank : 32) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BJ66 | Gene names | Kazald1, Bono1, Igfbprp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor (IGFBP-related protein 10) (IGFBP-rP10) (Bone and odontoblast expressed protein 1). | |||||
|
LCE2D_HUMAN
|
||||||
| θ value | 0.125558 (rank : 38) | NC score | 0.112299 (rank : 54) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5TA82 | Gene names | LCE2D, LEP12, SPRL1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2D (Late envelope protein 12) (Small proline-rich-like epidermal differentiation complex protein 1A). | |||||
|
WISP2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 39) | NC score | 0.164022 (rank : 42) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z0G4, Q8CIC8 | Gene names | Wisp2, Ccn5, Ctgfl | |||
|
Domain Architecture |
|
|||||
| Description | WNT1-inducible signaling pathway protein 2 precursor (WISP-2) (Connective tissue growth factor-like protein) (CTGF-L). | |||||
|
ZN639_HUMAN
|
||||||
| θ value | 0.125558 (rank : 40) | NC score | 0.004930 (rank : 114) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UID6 | Gene names | ZNF639, ZASC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 639 (Zinc finger protein ZASC1) (Zinc finger protein ANC_2H01). | |||||
|
KAZD1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 41) | NC score | 0.220726 (rank : 33) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96I82, Q6ZMB1, Q9BQ73 | Gene names | KAZALD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor. | |||||
|
RRMJ3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 42) | NC score | 0.043954 (rank : 78) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBE9, Q3ULI1, Q921I7 | Gene names | Ftsj3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative rRNA methyltransferase 3 (EC 2.1.1.-) (rRNA (uridine-2'-O-)- methyltransferase 3). | |||||
|
HTRA3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 43) | NC score | 0.202340 (rank : 38) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9D236, Q6WLC5, Q6YDR0 | Gene names | Htra3, Prsp, Tasp | |||
|
Domain Architecture |
|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease) (Toll-associated serine protease). | |||||
|
HTRA3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 44) | NC score | 0.201438 (rank : 39) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P83110, Q7Z7A2 | Gene names | HTRA3, PRSP | |||
|
Domain Architecture |
|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease). | |||||
|
ESM1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.244929 (rank : 29) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NQ30, Q15330, Q96ES3 | Gene names | ESM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-specific molecule 1 precursor (ESM-1 secretory protein) (ESM-1). | |||||
|
HTRA4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.176540 (rank : 41) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P83105, Q542Z4, Q6PF13 | Gene names | HTRA4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable serine protease HTRA4 precursor (EC 3.4.21.-). | |||||
|
NID2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.146473 (rank : 45) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O88322 | Gene names | Nid2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Entactin-2). | |||||
|
NOV_HUMAN
|
||||||
| θ value | 0.62314 (rank : 48) | NC score | 0.142155 (rank : 46) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P48745, Q96BY5 | Gene names | NOV, CCN3, IGFBP9, NOVH | |||
|
Domain Architecture |
|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
TRIM7_HUMAN
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.008100 (rank : 110) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9C029, Q969F5, Q96F67, Q96J89, Q96J90 | Gene names | TRIM7, GNIP, RNF90 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 7 (Glycogenin-interacting protein) (RING finger protein 90). | |||||
|
LCE2C_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.090901 (rank : 62) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5TA81 | Gene names | LCE2C, LEP11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2C (Late envelope protein 11). | |||||
|
NOV_MOUSE
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.134578 (rank : 48) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q64299, Q8CA67 | Gene names | Nov, Ccn3 | |||
|
Domain Architecture |
|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
PHX2B_HUMAN
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.014406 (rank : 97) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99453, Q6PJD9 | Gene names | PHOX2B, PMX2B | |||
|
Domain Architecture |
|
|||||
| Description | Paired mesoderm homeobox protein 2B (Paired-like homeobox 2B) (PHOX2B homeodomain protein) (Neuroblastoma Phox) (NBPhox). | |||||
|
PHX2B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.014406 (rank : 98) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35690 | Gene names | Phox2b, Pmx2b | |||
|
Domain Architecture |
|
|||||
| Description | Paired mesoderm homeobox protein 2B (Paired-like homeobox 2B) (PHOX2B homeodomain protein) (Neuroblastoma Phox) (NBPhox). | |||||
|
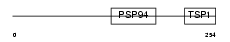
WISP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.093165 (rank : 61) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95388, Q9HCS3 | Gene names | WISP1, CCN4 | |||
|
Domain Architecture |
|
|||||
| Description | WNT1-inducible signaling pathway protein 1 precursor (WISP-1) (Wnt-1- induced secreted protein). | |||||
|
KR103_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.032025 (rank : 81) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P60369, Q70LJ4 | Gene names | KRTAP10-3, KAP10.3, KAP18-3, KRTAP10.3, KRTAP18-3, KRTAP18.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-3 (Keratin-associated protein 10.3) (High sulfur keratin-associated protein 10.3) (Keratin-associated protein 18-3) (Keratin-associated protein 18.3). | |||||
|
MGR4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.014928 (rank : 95) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14833, Q5SZ84 | Gene names | GRM4, GPRC1D, MGLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 4 precursor (mGluR4). | |||||
|
AMOT_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.012805 (rank : 101) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
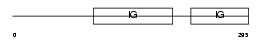
IGHG3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.010228 (rank : 106) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01860 | Gene names | IGHG3 | |||
|
Domain Architecture |
|
|||||
| Description | Ig gamma-3 chain C region (Heavy chain disease protein) (HDC). | |||||
|
NID1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.134528 (rank : 49) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P14543, Q14942, Q59FL2, Q5TAF2, Q5TAF3, Q86XD7 | Gene names | NID1, NID | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
BRE1B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.005611 (rank : 112) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.002235 (rank : 116) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
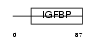
ESM1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.240439 (rank : 30) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QYY7 | Gene names | Esm1 | |||
|
Domain Architecture |
|
|||||
| Description | Endothelial cell-specific molecule 1 precursor (ESM-1 secretory protein) (ESM-1). | |||||
|
MEGF6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.029762 (rank : 82) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
TXD11_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.019690 (rank : 89) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K2W3, Q8BMR8, Q8VCK9 | Gene names | Txndc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 11. | |||||
|
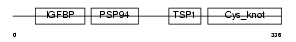
CTGF_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.154986 (rank : 43) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P29279, Q96QX2 | Gene names | CTGF, CCN2, HCS24 | |||
|
Domain Architecture |
|
|||||
| Description | Connective tissue growth factor precursor (Hypertrophic chondrocyte- specific protein 24). | |||||
|
GRN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.034723 (rank : 80) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P28798 | Gene names | Grn | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) (PC cell-derived growth factor) (PCDGF) [Contains: Acrogranin; Granulin-1; Granulin-2; Granulin-3; Granulin-4; Granulin-5; Granulin-6; Granulin-7]. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.004175 (rank : 115) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
RRMJ3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.037251 (rank : 79) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IY81, Q8N3A3, Q8WXX1, Q9BWM4, Q9NXT6 | Gene names | FTSJ3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative rRNA methyltransferase 3 (EC 2.1.1.-) (rRNA (uridine-2'-O-)- methyltransferase 3). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.013679 (rank : 100) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
DLL3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.020656 (rank : 87) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NYJ7 | Gene names | DLL3 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3). | |||||
|
TNR14_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.020939 (rank : 86) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92956, Q8WXR1, Q96J31, Q9UM65 | Gene names | TNFRSF14, HVEA, HVEM | |||
|
Domain Architecture |
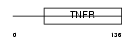
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 14 precursor (Herpesvirus entry mediator A) (Tumor necrosis factor receptor-like 2) (TR2). | |||||
|
WISP3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.121985 (rank : 53) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95389, Q6UXH6 | Gene names | WISP3, CCN6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WNT1-inducible signaling pathway protein 3 precursor (WISP-3). | |||||
|
ABI2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.005568 (rank : 113) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYB9, Q13147, Q13249, Q13801, Q9BV70 | Gene names | ABI2, ARGBPIA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 2 (Abelson interactor 2) (Abi-2) (Abl-binding protein 3) (AblBP3) (Arg-binding protein 1) (ArgBP1). | |||||
|
CABL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.012117 (rank : 104) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TDN4, Q8N3Y8, Q8NA22, Q9BTG1 | Gene names | CABLES1, CABLES | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
|
CELR3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.005738 (rank : 111) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||
|
CXXC6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.018240 (rank : 92) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
|
DMPK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | -0.002901 (rank : 119) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q09013, Q16205 | Gene names | DMPK, MDPK | |||
|
Domain Architecture |

|
|||||
| Description | Myotonin-protein kinase (EC 2.7.11.1) (Myotonic dystrophy protein kinase) (MDPK) (DM-kinase) (DMK) (DMPK) (MT-PK). | |||||
|
EDA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.009684 (rank : 107) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O54693, O35705, Q9QWJ8, Q9QZ01, Q9QZ02 | Gene names | Eda, Ed1, Ta | |||
|
Domain Architecture |

|
|||||
| Description | Ectodysplasin-A (EDA protein homolog) (Tabby protein) [Contains: Ectodysplasin-A, membrane form; Ectodysplasin-A, secreted form]. | |||||
|
NOTC3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.019941 (rank : 88) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
RNF31_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.011963 (rank : 105) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96EP0, Q86VI2, Q8TEI0, Q96GB4, Q96NF1, Q9H5F1, Q9NWD2 | Gene names | RNF31, ZIBRA | |||
|
Domain Architecture |
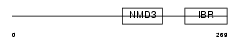
|
|||||
| Description | RING finger protein 31 (Zinc in-between-RING-finger ubiquitin- associated domain protein). | |||||
|
XPC_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.014426 (rank : 96) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51612, P54732, Q3TKI2, Q920M1, Q9DBW7 | Gene names | Xpc | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-C cells homolog (Xeroderma pigmentosum group C-complementing protein homolog) (p125). | |||||
|
ZBTB5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.001252 (rank : 117) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15062 | Gene names | ZBTB5, KIAA0354 | |||
|
Domain Architecture |
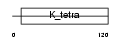
|
|||||
| Description | Zinc finger and BTB domain-containing protein 5. | |||||
|
ZBTB5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.001210 (rank : 118) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 801 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TQG0, Q3TVR7, Q6GV09, Q8BIB5 | Gene names | Zbtb5, Kiaa0354 | |||
|
Domain Architecture |
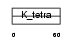
|
|||||
| Description | Zinc finger and BTB domain-containing protein 5 (Transcription factor ZNF-POZ). | |||||
|
ZNHI4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.017330 (rank : 93) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99PT3, Q9CY38 | Gene names | Znhit4, Hmga1l4, Papa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger HIT domain-containing protein 4 (PAP-1-associated protein 1) (PAPA-1) (High mobility group AT-hook 1-like 4). | |||||
|
I10R2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.015485 (rank : 94) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61190 | Gene names | Il10rb, Crfb4 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-10 receptor beta chain precursor (IL-10R-B) (IL-10R2) (Cytokine receptor family 2 member 4) (Cytokine receptor class-II member 4) (CRF2-4) (CDw210b antigen). | |||||
|
LTBP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.018896 (rank : 91) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
MGR6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.012353 (rank : 103) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15303 | Gene names | GRM6, GPRC1F, MGLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 6 precursor (mGluR6). | |||||
|
MGR6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.012425 (rank : 102) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5NCH9 | Gene names | Grm6, Gprc1f, Mglur6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metabotropic glutamate receptor 6 precursor (mGluR6). | |||||
|
WISP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.089415 (rank : 63) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O54775, Q80ZL1 | Gene names | Wisp1, Ccn4, Elm1 | |||
|
Domain Architecture |

|
|||||
| Description | WNT1-inducible signaling pathway protein 1 precursor (WISP-1) (ELM-1). | |||||
|
ADA11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.008413 (rank : 108) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75078, Q14808, Q14809, Q14810 | Gene names | ADAM11, MDC | |||
|
Domain Architecture |
|
|||||
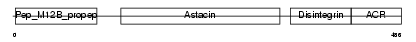
| Description | ADAM 11 precursor (A disintegrin and metalloproteinase domain 11) (Metalloproteinase-like, disintegrin-like, and cysteine-rich protein) (MDC). | |||||
|
ADA11_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.008332 (rank : 109) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R1V4 | Gene names | Adam11, Mdc | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 11 precursor (A disintegrin and metalloproteinase domain 11) (Metalloproteinase-like, disintegrin-like, and cysteine-rich protein) (MDC). | |||||
|
CTGF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.109392 (rank : 56) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P29268, Q922U0 | Gene names | Ctgf, Ccn2, Fisp-12, Fisp12, Hcs24 | |||
|
Domain Architecture |
|
|||||
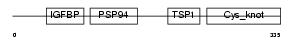
| Description | Connective tissue growth factor precursor (Protein FISP-12) (Hypertrophic chondrocyte-specific protein 24). | |||||
|
I10R2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.014215 (rank : 99) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08334, Q9BUU4 | Gene names | IL10RB, CRFB4 | |||
|
Domain Architecture |
|
|||||
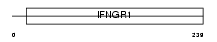
| Description | Interleukin-10 receptor beta chain precursor (IL-10R-B) (IL-10R2) (Cytokine receptor family 2 member 4) (Cytokine receptor class-II member 4) (CRF2-4) (CDw210b antigen). | |||||
|
LCE1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.109804 (rank : 55) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5T7P3 | Gene names | LCE1B, LEP2, SPRL2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1B (Late envelope protein 2) (Small proline-rich-like epidermal differentiation complex protein 2A). | |||||
|
LCE4A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.207713 (rank : 35) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5TA78 | Gene names | LCE4A, LEP8, SPRL4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 4A (Late envelope protein 8) (Small proline-rich-like epidermal differentiation complex protein 4A). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.019354 (rank : 90) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.021512 (rank : 85) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.052029 (rank : 75) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
CYR61_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.129767 (rank : 51) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O00622, O14934, O43775, Q9BZL7 | Gene names | CYR61, CCN1, GIG1, IGFBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CYR61 precursor (Cysteine-rich, angiogenic inducer, 61) (Insulin-like growth factor-binding protein 10) (Protein GIG1). | |||||
|
CYR61_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.129947 (rank : 50) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P18406 | Gene names | Cyr61, Ccn1, Igfbp10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CYR61 precursor (Cysteine-rich, angiogenic inducer, 61) (Insulin-like growth factor-binding protein 10) (3CH61). | |||||
|
FSTL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.051893 (rank : 77) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q12841 | Gene names | FSTL1, FRP | |||
|
Domain Architecture |
|
|||||
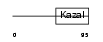
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1). | |||||
|
FSTL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.051977 (rank : 76) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62356, Q99JI9 | Gene names | Fstl1, Frp, Fstl, Tsc36 | |||
|
Domain Architecture |
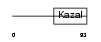
|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1) (TGF- beta-inducible protein TSC-36). | |||||
|
FSTL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.070975 (rank : 66) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95633 | Gene names | FSTL3, FLRG | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
FSTL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.066505 (rank : 67) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9EQC7 | Gene names | Fstl3, Flrg | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
FST_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.058513 (rank : 70) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P19883, Q9BTH0 | Gene names | FST | |||
|
Domain Architecture |
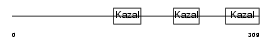
|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
FST_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.056725 (rank : 71) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P47931 | Gene names | Fst | |||
|
Domain Architecture |
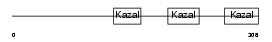
|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
HTRA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.094363 (rank : 60) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43464, Q9HBZ4, Q9P0Y3, Q9P0Y4 | Gene names | HTRA2, OMI, PRSS25 | |||
|
Domain Architecture |
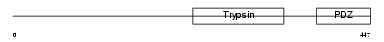
|
|||||
| Description | Serine protease HTRA2, mitochondrial precursor (EC 3.4.21.-) (High temperature requirement protein A2) (HtrA2) (Omi stress-regulated endoprotease) (Serine proteinase OMI). | |||||
|
HTRA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.094675 (rank : 58) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JIY5, Q9R108 | Gene names | Htra2, Omi, Prss25 | |||
|
Domain Architecture |
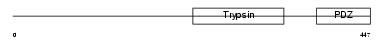
|
|||||
| Description | Serine protease HTRA2, mitochondrial precursor (EC 3.4.21.-) (High temperature requirement protein A2) (HtrA2) (Omi stress-regulated endoprotease) (Serine proteinase OMI). | |||||
|
LCE1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.094624 (rank : 59) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
LCE1D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.095572 (rank : 57) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
LCE1E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.087865 (rank : 64) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
NID1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.122672 (rank : 52) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P10493, Q8BQI3, Q8C3U8, Q8C9P6 | Gene names | Nid1, Ent | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
SPRC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.064570 (rank : 68) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P09486 | Gene names | SPARC, ON | |||
|
Domain Architecture |
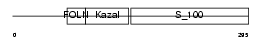
|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
SPRC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.063200 (rank : 69) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P07214 | Gene names | Sparc | |||
|
Domain Architecture |
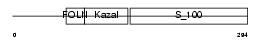
|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
SPRL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.054302 (rank : 74) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
TACD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.206747 (rank : 36) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P16422, P18180, Q96C47 | Gene names | TACSTD1, GA733-2, M1S2, TROP1 | |||
|
Domain Architecture |
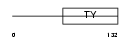
|
|||||
| Description | Tumor-associated calcium signal transducer 1 precursor (Major gastrointestinal tumor-associated protein GA733-2) (Epithelial cell surface antigen) (Epithelial glycoprotein) (EGP) (Adenocarcinoma- associated antigen) (KSA) (KS 1/4 antigen) (Cell surface glycoprotein Trop-1) (CD326 antigen). | |||||
|
TACD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.178327 (rank : 40) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P09758, Q15658, Q96QD2 | Gene names | TACSTD2, GA733-1, M1S1, TROP2 | |||
|
Domain Architecture |
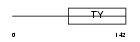
|
|||||
| Description | Tumor-associated calcium signal transducer 2 precursor (Pancreatic carcinoma marker protein GA733-1) (Cell surface glycoprotein Trop-2). | |||||
|
TEFF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.054327 (rank : 73) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UIK5, Q4ZFW4, Q53H90, Q53RE1, Q8N2R5, Q9NR15, Q9NSS5, Q9P2Y9, Q9UK65 | Gene names | TMEFF2, HPP1, TENB2, TPEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Transmembrane protein containing epidermal growth factor and follistatin domains) (Tomoregulin) (TR) (Hyperplastic polyposis protein 1). | |||||
|
TEFF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.055168 (rank : 72) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QYM9, Q3UY20, Q8CDH1, Q9JJE3 | Gene names | Tmeff2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Tomoregulin) (TR). | |||||
|
IBP1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 4.70271e-166 (rank : 1) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
|
IBP1_HUMAN
|
||||||
| NC score | 0.982076 (rank : 2) | θ value | 4.1872e-106 (rank : 2) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P08833, Q8IYP5 | Gene names | IGFBP1, IBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1) (Placental protein 12) (PP12). | |||||
|
IBP4_HUMAN
|
||||||
| NC score | 0.918668 (rank : 3) | θ value | 8.89434e-40 (rank : 3) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P22692 | Gene names | IGFBP4, IBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
IBP4_MOUSE
|
||||||
| NC score | 0.917393 (rank : 4) | θ value | 1.98146e-39 (rank : 4) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P47879, O35666 | Gene names | Igfbp4, Igfbp-4 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 4 precursor (IGFBP-4) (IBP- 4) (IGF-binding protein 4). | |||||
|
IBP2_MOUSE
|
||||||
| NC score | 0.914044 (rank : 5) | θ value | 2.42216e-37 (rank : 5) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P47877, Q61722 | Gene names | Igfbp2, Igfbp-2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
IBP2_HUMAN
|
||||||
| NC score | 0.913710 (rank : 6) | θ value | 3.4976e-36 (rank : 6) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P18065, Q14619, Q9UCL3 | Gene names | IGFBP2, BP2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 2 precursor (IGFBP-2) (IBP- 2) (IGF-binding protein 2). | |||||
|
IBP5_HUMAN
|
||||||
| NC score | 0.904623 (rank : 7) | θ value | 6.59618e-35 (rank : 7) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P24593 | Gene names | IGFBP5, IBP5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP5_MOUSE
|
||||||
| NC score | 0.903815 (rank : 8) | θ value | 1.46948e-34 (rank : 8) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q07079 | Gene names | Igfbp5, Igfbp-5 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 5 precursor (IGFBP-5) (IBP- 5) (IGF-binding protein 5). | |||||
|
IBP3_HUMAN
|
||||||
| NC score | 0.896221 (rank : 9) | θ value | 5.77852e-31 (rank : 9) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P17936, Q2V509, Q6P1M6 | Gene names | IGFBP3, IBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
IBP3_MOUSE
|
||||||
| NC score | 0.885102 (rank : 10) | θ value | 4.14116e-29 (rank : 10) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P47878 | Gene names | Igfbp3, Igfbp-3 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 3 precursor (IGFBP-3) (IBP- 3) (IGF-binding protein 3). | |||||
|
IBP6_HUMAN
|
||||||
| NC score | 0.839644 (rank : 11) | θ value | 1.47631e-18 (rank : 11) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P24592, Q14492 | Gene names | IGFBP6, IBP6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
IBP6_MOUSE
|
||||||
| NC score | 0.826219 (rank : 12) | θ value | 3.40345e-15 (rank : 12) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P47880 | Gene names | Igfbp6, Igfbp-6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
TICN1_MOUSE
|
||||||
| NC score | 0.382433 (rank : 13) | θ value | 0.000461057 (rank : 21) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q62288 | Gene names | Spock1, Spock, Ticn1 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
TICN1_HUMAN
|
||||||
| NC score | 0.380532 (rank : 14) | θ value | 0.000602161 (rank : 22) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q08629 | Gene names | SPOCK1, SPOCK, TIC1, TICN1 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-1 precursor (Protein SPOCK). | |||||
|
HG2A_MOUSE
|
||||||
| NC score | 0.380054 (rank : 15) | θ value | 0.00228821 (rank : 23) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P04441, O19452 | Gene names | Cd74, Ii | |||
|
Domain Architecture |

|
|||||
| Description | H-2 class II histocompatibility antigen gamma chain (MHC class II- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (CD74 antigen). | |||||
|
SMOC2_MOUSE
|
||||||
| NC score | 0.379833 (rank : 16) | θ value | 8.40245e-06 (rank : 15) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8CD91, Q8VCM1, Q9D9K2, Q9ER95 | Gene names | Smoc2 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2). | |||||
|
TICN2_HUMAN
|
||||||
| NC score | 0.379598 (rank : 17) | θ value | 0.000121331 (rank : 19) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q92563, Q6UW87 | Gene names | SPOCK2, KIAA0275, TICN2 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
TICN2_MOUSE
|
||||||
| NC score | 0.376294 (rank : 18) | θ value | 0.00390308 (rank : 28) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9ER58 | Gene names | Spock2, Ticn2 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-2 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2). | |||||
|
SMOC2_HUMAN
|
||||||
| NC score | 0.374843 (rank : 19) | θ value | 7.1131e-05 (rank : 16) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9H3U7, Q5TAT7, Q5TAT8, Q86VV9, Q96SF3, Q9H1L3, Q9H1L4, Q9H3U0, Q9H4F7, Q9HCV2 | Gene names | SMOC2, SMAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 2 precursor (Secreted modular calcium-binding protein 2) (SMOC-2) (Smooth muscle-associated protein 2) (SMAP-2). | |||||
|
TICN3_HUMAN
|
||||||
| NC score | 0.353041 (rank : 20) | θ value | 0.00869519 (rank : 30) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9BQ16, O75705, Q6UW53, Q96Q26 | Gene names | SPOCK3, TICN3 | |||
|
Domain Architecture |

|
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
SMOC1_MOUSE
|
||||||
| NC score | 0.350417 (rank : 21) | θ value | 0.00390308 (rank : 27) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BLY1, Q9WVN9 | Gene names | Smoc1, Srg | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1) (SPARC-related gene protein). | |||||
|
SMOC1_HUMAN
|
||||||
| NC score | 0.349846 (rank : 22) | θ value | 0.00298849 (rank : 26) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9H4F8, Q96F78 | Gene names | SMOC1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-related modular calcium-binding protein 1 precursor (Secreted modular calcium-binding protein 1) (SMOC-1). | |||||
|
HTRA1_HUMAN
|
||||||
| NC score | 0.349144 (rank : 23) | θ value | 5.26297e-08 (rank : 13) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92743, Q9UNS5 | Gene names | HTRA1, HTRA, PRSS11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-) (L56). | |||||
|
HG2A_HUMAN
|
||||||
| NC score | 0.346452 (rank : 24) | θ value | 0.00298849 (rank : 25) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P04233, Q14597, Q29832, Q5U0J8, Q8WLP6 | Gene names | CD74, DHLAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HLA class II histocompatibility antigen gamma chain (HLA-DR antigens- associated invariant chain) (Ia antigen-associated invariant chain) (Ii) (p33) (CD74 antigen). | |||||
|
TICN3_MOUSE
|
||||||
| NC score | 0.346390 (rank : 25) | θ value | 0.0431538 (rank : 35) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BKV0, Q9ER59 | Gene names | Spock3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testican-3 precursor (SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 3). | |||||
|
HTRA1_MOUSE
|
||||||
| NC score | 0.339042 (rank : 26) | θ value | 2.21117e-06 (rank : 14) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9R118 | Gene names | Htra1, Htra, Prss11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-). | |||||
|
IBP7_MOUSE
|
||||||
| NC score | 0.256872 (rank : 27) | θ value | 9.29e-05 (rank : 17) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61581, O88812, Q9EQW0 | Gene names | Igfbp7, Mac25 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein). | |||||
|
IBP7_HUMAN
|
||||||
| NC score | 0.255629 (rank : 28) | θ value | 0.00228821 (rank : 24) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q16270, Q07822 | Gene names | IGFBP7, MAC25, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 7 precursor (IGFBP-7) (IBP- 7) (IGF-binding protein 7) (MAC25 protein) (Prostacyclin-stimulating factor) (PGI2-stimulating factor) (IGFBP-rP1). | |||||
|
ESM1_HUMAN
|
||||||
| NC score | 0.244929 (rank : 29) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NQ30, Q15330, Q96ES3 | Gene names | ESM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-specific molecule 1 precursor (ESM-1 secretory protein) (ESM-1). | |||||
|
ESM1_MOUSE
|
||||||
| NC score | 0.240439 (rank : 30) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QYY7 | Gene names | Esm1 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial cell-specific molecule 1 precursor (ESM-1 secretory protein) (ESM-1). | |||||
|
THYG_HUMAN
|
||||||
| NC score | 0.227822 (rank : 31) | θ value | 0.000158464 (rank : 20) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P01266, O15274, O43899, Q15593, Q15948, Q9NYR1, Q9NYR2, Q9UMZ0, Q9UNY3 | Gene names | TG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
KAZD1_MOUSE
|
||||||
| NC score | 0.224861 (rank : 32) | θ value | 0.125558 (rank : 37) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BJ66 | Gene names | Kazald1, Bono1, Igfbprp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor (IGFBP-related protein 10) (IGFBP-rP10) (Bone and odontoblast expressed protein 1). | |||||
|
KAZD1_HUMAN
|
||||||
| NC score | 0.220726 (rank : 33) | θ value | 0.21417 (rank : 41) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96I82, Q6ZMB1, Q9BQ73 | Gene names | KAZALD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kazal-type serine protease inhibitor domain-containing protein 1 precursor. | |||||
|
THYG_MOUSE
|
||||||
| NC score | 0.213767 (rank : 34) | θ value | 9.29e-05 (rank : 18) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O08710, O88590, Q9QWY7 | Gene names | Tg, Tgn | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
LCE4A_HUMAN
|
||||||
| NC score | 0.207713 (rank : 35) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5TA78 | Gene names | LCE4A, LEP8, SPRL4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 4A (Late envelope protein 8) (Small proline-rich-like epidermal differentiation complex protein 4A). | |||||
|
TACD1_HUMAN
|
||||||
| NC score | 0.206747 (rank : 36) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P16422, P18180, Q96C47 | Gene names | TACSTD1, GA733-2, M1S2, TROP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor-associated calcium signal transducer 1 precursor (Major gastrointestinal tumor-associated protein GA733-2) (Epithelial cell surface antigen) (Epithelial glycoprotein) (EGP) (Adenocarcinoma- associated antigen) (KSA) (KS 1/4 antigen) (Cell surface glycoprotein Trop-1) (CD326 antigen). | |||||
|
WISP2_HUMAN
|
||||||
| NC score | 0.202412 (rank : 37) | θ value | 0.0113563 (rank : 32) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O76076 | Gene names | WISP2, CCN5, CT58, CTGFL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WNT1-inducible signaling pathway protein 2 precursor (WISP-2) (Connective tissue growth factor-like protein) (CTGF-L) (Connective tissue growth factor-related protein 58). | |||||
|
HTRA3_MOUSE
|
||||||
| NC score | 0.202340 (rank : 38) | θ value | 0.365318 (rank : 43) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9D236, Q6WLC5, Q6YDR0 | Gene names | Htra3, Prsp, Tasp | |||
|
Domain Architecture |

|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease) (Toll-associated serine protease). | |||||
|
HTRA3_HUMAN
|
||||||
| NC score | 0.201438 (rank : 39) | θ value | 0.47712 (rank : 44) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P83110, Q7Z7A2 | Gene names | HTRA3, PRSP | |||
|
Domain Architecture |

|
|||||
| Description | Probable serine protease HTRA3 precursor (EC 3.4.21.-) (High- temperature requirement factor A3) (Pregnancy-related serine protease). | |||||
|
TACD2_HUMAN
|
||||||
| NC score | 0.178327 (rank : 40) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P09758, Q15658, Q96QD2 | Gene names | TACSTD2, GA733-1, M1S1, TROP2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor-associated calcium signal transducer 2 precursor (Pancreatic carcinoma marker protein GA733-1) (Cell surface glycoprotein Trop-2). | |||||
|
HTRA4_HUMAN
|
||||||
| NC score | 0.176540 (rank : 41) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P83105, Q542Z4, Q6PF13 | Gene names | HTRA4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable serine protease HTRA4 precursor (EC 3.4.21.-). | |||||
|
WISP2_MOUSE
|
||||||
| NC score | 0.164022 (rank : 42) | θ value | 0.125558 (rank : 39) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z0G4, Q8CIC8 | Gene names | Wisp2, Ccn5, Ctgfl | |||
|
Domain Architecture |

|
|||||
| Description | WNT1-inducible signaling pathway protein 2 precursor (WISP-2) (Connective tissue growth factor-like protein) (CTGF-L). | |||||
|
CTGF_HUMAN
|
||||||
| NC score | 0.154986 (rank : 43) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P29279, Q96QX2 | Gene names | CTGF, CCN2, HCS24 | |||
|
Domain Architecture |

|
|||||
| Description | Connective tissue growth factor precursor (Hypertrophic chondrocyte- specific protein 24). | |||||
|
NID2_HUMAN
|
||||||
| NC score | 0.149760 (rank : 44) | θ value | 0.0113563 (rank : 31) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
NID2_MOUSE
|
||||||
| NC score | 0.146473 (rank : 45) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O88322 | Gene names | Nid2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Entactin-2). | |||||
|
NOV_HUMAN
|
||||||
| NC score | 0.142155 (rank : 46) | θ value | 0.62314 (rank : 48) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P48745, Q96BY5 | Gene names | NOV, CCN3, IGFBP9, NOVH | |||
|
Domain Architecture |

|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
CRIM1_HUMAN
|
||||||
| NC score | 0.141521 (rank : 47) | θ value | 0.125558 (rank : 36) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
NOV_MOUSE
|
||||||
| NC score | 0.134578 (rank : 48) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q64299, Q8CA67 | Gene names | Nov, Ccn3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein NOV homolog precursor (NovH) (Nephroblastoma overexpressed gene protein homolog). | |||||
|
NID1_HUMAN
|
||||||
| NC score | 0.134528 (rank : 49) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P14543, Q14942, Q59FL2, Q5TAF2, Q5TAF3, Q86XD7 | Gene names | NID1, NID | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
CYR61_MOUSE
|
||||||
| NC score | 0.129947 (rank : 50) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P18406 | Gene names | Cyr61, Ccn1, Igfbp10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CYR61 precursor (Cysteine-rich, angiogenic inducer, 61) (Insulin-like growth factor-binding protein 10) (3CH61). | |||||
|
CYR61_HUMAN
|
||||||
| NC score | 0.129767 (rank : 51) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O00622, O14934, O43775, Q9BZL7 | Gene names | CYR61, CCN1, GIG1, IGFBP10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CYR61 precursor (Cysteine-rich, angiogenic inducer, 61) (Insulin-like growth factor-binding protein 10) (Protein GIG1). | |||||
|
NID1_MOUSE
|
||||||
| NC score | 0.122672 (rank : 52) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P10493, Q8BQI3, Q8C3U8, Q8C9P6 | Gene names | Nid1, Ent | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
WISP3_HUMAN
|
||||||
| NC score | 0.121985 (rank : 53) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95389, Q6UXH6 | Gene names | WISP3, CCN6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WNT1-inducible signaling pathway protein 3 precursor (WISP-3). | |||||
|
LCE2D_HUMAN
|
||||||
| NC score | 0.112299 (rank : 54) | θ value | 0.125558 (rank : 38) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5TA82 | Gene names | LCE2D, LEP12, SPRL1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2D (Late envelope protein 12) (Small proline-rich-like epidermal differentiation complex protein 1A). | |||||
|
LCE1B_HUMAN
|
||||||
| NC score | 0.109804 (rank : 55) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5T7P3 | Gene names | LCE1B, LEP2, SPRL2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1B (Late envelope protein 2) (Small proline-rich-like epidermal differentiation complex protein 2A). | |||||
|
CTGF_MOUSE
|
||||||
| NC score | 0.109392 (rank : 56) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P29268, Q922U0 | Gene names | Ctgf, Ccn2, Fisp-12, Fisp12, Hcs24 | |||
|
Domain Architecture |

|
|||||
| Description | Connective tissue growth factor precursor (Protein FISP-12) (Hypertrophic chondrocyte-specific protein 24). | |||||
|
LCE1D_HUMAN
|
||||||
| NC score | 0.095572 (rank : 57) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
HTRA2_MOUSE
|
||||||
| NC score | 0.094675 (rank : 58) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JIY5, Q9R108 | Gene names | Htra2, Omi, Prss25 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA2, mitochondrial precursor (EC 3.4.21.-) (High temperature requirement protein A2) (HtrA2) (Omi stress-regulated endoprotease) (Serine proteinase OMI). | |||||
|
LCE1C_HUMAN
|
||||||
| NC score | 0.094624 (rank : 59) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5T751 | Gene names | LCE1C, LEP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1C (Late envelope protein 3). | |||||
|
HTRA2_HUMAN
|
||||||
| NC score | 0.094363 (rank : 60) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43464, Q9HBZ4, Q9P0Y3, Q9P0Y4 | Gene names | HTRA2, OMI, PRSS25 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA2, mitochondrial precursor (EC 3.4.21.-) (High temperature requirement protein A2) (HtrA2) (Omi stress-regulated endoprotease) (Serine proteinase OMI). | |||||
|
WISP1_HUMAN
|
||||||
| NC score | 0.093165 (rank : 61) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95388, Q9HCS3 | Gene names | WISP1, CCN4 | |||
|
Domain Architecture |

|
|||||
| Description | WNT1-inducible signaling pathway protein 1 precursor (WISP-1) (Wnt-1- induced secreted protein). | |||||
|
LCE2C_HUMAN
|
||||||
| NC score | 0.090901 (rank : 62) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5TA81 | Gene names | LCE2C, LEP11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2C (Late envelope protein 11). | |||||
|
WISP1_MOUSE
|
||||||
| NC score | 0.089415 (rank : 63) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O54775, Q80ZL1 | Gene names | Wisp1, Ccn4, Elm1 | |||
|
Domain Architecture |

|
|||||
| Description | WNT1-inducible signaling pathway protein 1 precursor (WISP-1) (ELM-1). | |||||
|
LCE1E_HUMAN
|
||||||
| NC score | 0.087865 (rank : 64) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5T753 | Gene names | LCE1E, LEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1E (Late envelope protein 5). | |||||
|
MCSP_MOUSE
|
||||||
| NC score | 0.077956 (rank : 65) | θ value | 0.0193708 (rank : 33) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P15265, O70613, Q6P8N3 | Gene names | Smcp, Mcs, Mcsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm mitochondrial-associated cysteine-rich protein. | |||||
|
FSTL3_HUMAN
|
||||||
| NC score | 0.070975 (rank : 66) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95633 | Gene names | FSTL3, FLRG | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
FSTL3_MOUSE
|
||||||
| NC score | 0.066505 (rank : 67) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9EQC7 | Gene names | Fstl3, Flrg | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
SPRC_HUMAN
|
||||||
| NC score | 0.064570 (rank : 68) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P09486 | Gene names | SPARC, ON | |||
|
Domain Architecture |

|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
SPRC_MOUSE
|
||||||
| NC score | 0.063200 (rank : 69) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P07214 | Gene names | Sparc | |||
|
Domain Architecture |

|
|||||
| Description | SPARC precursor (Secreted protein acidic and rich in cysteine) (Osteonectin) (ON) (Basement-membrane protein 40) (BM-40). | |||||
|
FST_HUMAN
|
||||||
| NC score | 0.058513 (rank : 70) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P19883, Q9BTH0 | Gene names | FST | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
FST_MOUSE
|
||||||
| NC score | 0.056725 (rank : 71) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P47931 | Gene names | Fst | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin precursor (FS) (Activin-binding protein). | |||||
|
TEFF2_MOUSE
|
||||||
| NC score | 0.055168 (rank : 72) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QYM9, Q3UY20, Q8CDH1, Q9JJE3 | Gene names | Tmeff2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Tomoregulin) (TR). | |||||
|
TEFF2_HUMAN
|
||||||
| NC score | 0.054327 (rank : 73) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UIK5, Q4ZFW4, Q53H90, Q53RE1, Q8N2R5, Q9NR15, Q9NSS5, Q9P2Y9, Q9UK65 | Gene names | TMEFF2, HPP1, TENB2, TPEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein with EGF-like and two follistatin-like domains 2 precursor (Transmembrane protein containing epidermal growth factor and follistatin domains) (Tomoregulin) (TR) (Hyperplastic polyposis protein 1). | |||||
|
SPRL1_HUMAN
|
||||||
| NC score | 0.054302 (rank : 74) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.052029 (rank : 75) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
FSTL1_MOUSE
|
||||||
| NC score | 0.051977 (rank : 76) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62356, Q99JI9 | Gene names | Fstl1, Frp, Fstl, Tsc36 | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1) (TGF- beta-inducible protein TSC-36). | |||||
|
FSTL1_HUMAN
|
||||||
| NC score | 0.051893 (rank : 77) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q12841 | Gene names | FSTL1, FRP | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 1 precursor (Follistatin-like 1). | |||||
|
RRMJ3_MOUSE
|
||||||
| NC score | 0.043954 (rank : 78) | θ value | 0.279714 (rank : 42) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBE9, Q3ULI1, Q921I7 | Gene names | Ftsj3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative rRNA methyltransferase 3 (EC 2.1.1.-) (rRNA (uridine-2'-O-)- methyltransferase 3). | |||||
|
RRMJ3_HUMAN
|
||||||
| NC score | 0.037251 (rank : 79) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IY81, Q8N3A3, Q8WXX1, Q9BWM4, Q9NXT6 | Gene names | FTSJ3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative rRNA methyltransferase 3 (EC 2.1.1.-) (rRNA (uridine-2'-O-)- methyltransferase 3). | |||||
|
GRN_MOUSE
|
||||||
| NC score | 0.034723 (rank : 80) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P28798 | Gene names | Grn | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) (PC cell-derived growth factor) (PCDGF) [Contains: Acrogranin; Granulin-1; Granulin-2; Granulin-3; Granulin-4; Granulin-5; Granulin-6; Granulin-7]. | |||||
|
KR103_HUMAN
|
||||||
| NC score | 0.032025 (rank : 81) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P60369, Q70LJ4 | Gene names | KRTAP10-3, KAP10.3, KAP18-3, KRTAP10.3, KRTAP18-3, KRTAP18.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-3 (Keratin-associated protein 10.3) (High sulfur keratin-associated protein 10.3) (Keratin-associated protein 18-3) (Keratin-associated protein 18.3). | |||||
|
MEGF6_HUMAN
|
||||||
| NC score | 0.029762 (rank : 82) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
NOTC3_HUMAN
|
||||||
| NC score | 0.027519 (rank : 83) | θ value | 0.00869519 (rank : 29) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
GSCL_HUMAN
|
||||||
| NC score | 0.023342 (rank : 84) | θ value | 0.0330416 (rank : 34) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15499 | Gene names | GSCL | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein goosecoid-like (GSC-2). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.021512 (rank : 85) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
TNR14_HUMAN
|
||||||
| NC score | 0.020939 (rank : 86) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92956, Q8WXR1, Q96J31, Q9UM65 | Gene names | TNFRSF14, HVEA, HVEM | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 14 precursor (Herpesvirus entry mediator A) (Tumor necrosis factor receptor-like 2) (TR2). | |||||
|
DLL3_HUMAN
|
||||||
| NC score | 0.020656 (rank : 87) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NYJ7 | Gene names | DLL3 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3). | |||||
|
NOTC3_MOUSE
|
||||||
| NC score | 0.019941 (rank : 88) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
TXD11_MOUSE
|
||||||
| NC score | 0.019690 (rank : 89) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K2W3, Q8BMR8, Q8VCK9 | Gene names | Txndc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 11. | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.019354 (rank : 90) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
LTBP4_MOUSE
|
||||||
| NC score | 0.018896 (rank : 91) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
CXXC6_HUMAN
|
||||||
| NC score | 0.018240 (rank : 92) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
|
ZNHI4_MOUSE
|
||||||
| NC score | 0.017330 (rank : 93) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99PT3, Q9CY38 | Gene names | Znhit4, Hmga1l4, Papa1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger HIT domain-containing protein 4 (PAP-1-associated protein 1) (PAPA-1) (High mobility group AT-hook 1-like 4). | |||||
|
I10R2_MOUSE
|
||||||
| NC score | 0.015485 (rank : 94) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61190 | Gene names | Il10rb, Crfb4 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-10 receptor beta chain precursor (IL-10R-B) (IL-10R2) (Cytokine receptor family 2 member 4) (Cytokine receptor class-II member 4) (CRF2-4) (CDw210b antigen). | |||||
|
MGR4_HUMAN
|
||||||
| NC score | 0.014928 (rank : 95) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14833, Q5SZ84 | Gene names | GRM4, GPRC1D, MGLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 4 precursor (mGluR4). | |||||
|
XPC_MOUSE
|
||||||
| NC score | 0.014426 (rank : 96) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51612, P54732, Q3TKI2, Q920M1, Q9DBW7 | Gene names | Xpc | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-C cells homolog (Xeroderma pigmentosum group C-complementing protein homolog) (p125). | |||||
|
PHX2B_HUMAN
|
||||||
| NC score | 0.014406 (rank : 97) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99453, Q6PJD9 | Gene names | PHOX2B, PMX2B | |||
|
Domain Architecture |

|
|||||
| Description | Paired mesoderm homeobox protein 2B (Paired-like homeobox 2B) (PHOX2B homeodomain protein) (Neuroblastoma Phox) (NBPhox). | |||||
|
PHX2B_MOUSE
|
||||||
| NC score | 0.014406 (rank : 98) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35690 | Gene names | Phox2b, Pmx2b | |||
|
Domain Architecture |

|
|||||
| Description | Paired mesoderm homeobox protein 2B (Paired-like homeobox 2B) (PHOX2B homeodomain protein) (Neuroblastoma Phox) (NBPhox). | |||||
|
I10R2_HUMAN
|
||||||
| NC score | 0.014215 (rank : 99) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08334, Q9BUU4 | Gene names | IL10RB, CRFB4 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-10 receptor beta chain precursor (IL-10R-B) (IL-10R2) (Cytokine receptor family 2 member 4) (Cytokine receptor class-II member 4) (CRF2-4) (CDw210b antigen). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.013679 (rank : 100) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
AMOT_HUMAN
|
||||||
| NC score | 0.012805 (rank : 101) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
MGR6_MOUSE
|
||||||
| NC score | 0.012425 (rank : 102) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5NCH9 | Gene names | Grm6, Gprc1f, Mglur6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metabotropic glutamate receptor 6 precursor (mGluR6). | |||||
|
MGR6_HUMAN
|
||||||
| NC score | 0.012353 (rank : 103) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15303 | Gene names | GRM6, GPRC1F, MGLUR6 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 6 precursor (mGluR6). | |||||
|
CABL1_HUMAN
|
||||||
| NC score | 0.012117 (rank : 104) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TDN4, Q8N3Y8, Q8NA22, Q9BTG1 | Gene names | CABLES1, CABLES | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
|
RNF31_HUMAN
|
||||||
| NC score | 0.011963 (rank : 105) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96EP0, Q86VI2, Q8TEI0, Q96GB4, Q96NF1, Q9H5F1, Q9NWD2 | Gene names | RNF31, ZIBRA | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 31 (Zinc in-between-RING-finger ubiquitin- associated domain protein). | |||||
|
IGHG3_HUMAN
|
||||||
| NC score | 0.010228 (rank : 106) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01860 | Gene names | IGHG3 | |||
|
Domain Architecture |

|
|||||
| Description | Ig gamma-3 chain C region (Heavy chain disease protein) (HDC). | |||||
|
EDA_MOUSE
|
||||||
| NC score | 0.009684 (rank : 107) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O54693, O35705, Q9QWJ8, Q9QZ01, Q9QZ02 | Gene names | Eda, Ed1, Ta | |||
|
Domain Architecture |

|
|||||
| Description | Ectodysplasin-A (EDA protein homolog) (Tabby protein) [Contains: Ectodysplasin-A, membrane form; Ectodysplasin-A, secreted form]. | |||||
|
ADA11_HUMAN
|
||||||
| NC score | 0.008413 (rank : 108) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75078, Q14808, Q14809, Q14810 | Gene names | ADAM11, MDC | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 11 precursor (A disintegrin and metalloproteinase domain 11) (Metalloproteinase-like, disintegrin-like, and cysteine-rich protein) (MDC). | |||||
|
ADA11_MOUSE
|
||||||
| NC score | 0.008332 (rank : 109) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R1V4 | Gene names | Adam11, Mdc | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 11 precursor (A disintegrin and metalloproteinase domain 11) (Metalloproteinase-like, disintegrin-like, and cysteine-rich protein) (MDC). | |||||
|
TRIM7_HUMAN
|
||||||
| NC score | 0.008100 (rank : 110) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9C029, Q969F5, Q96F67, Q96J89, Q96J90 | Gene names | TRIM7, GNIP, RNF90 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 7 (Glycogenin-interacting protein) (RING finger protein 90). | |||||
|
CELR3_HUMAN
|
||||||
| NC score | 0.005738 (rank : 111) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||
|
BRE1B_MOUSE
|
||||||
| NC score | 0.005611 (rank : 112) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
ABI2_HUMAN
|
||||||
| NC score | 0.005568 (rank : 113) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NYB9, Q13147, Q13249, Q13801, Q9BV70 | Gene names | ABI2, ARGBPIA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 2 (Abelson interactor 2) (Abi-2) (Abl-binding protein 3) (AblBP3) (Arg-binding protein 1) (ArgBP1). | |||||
|
ZN639_HUMAN
|
||||||
| NC score | 0.004930 (rank : 114) | θ value | 0.125558 (rank : 40) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UID6 | Gene names | ZNF639, ZASC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 639 (Zinc finger protein ZASC1) (Zinc finger protein ANC_2H01). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.004175 (rank : 115) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.002235 (rank : 116) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
ZBTB5_HUMAN
|
||||||
| NC score | 0.001252 (rank : 117) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15062 | Gene names | ZBTB5, KIAA0354 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 5. | |||||
|
ZBTB5_MOUSE
|
||||||
| NC score | 0.001210 (rank : 118) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 801 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TQG0, Q3TVR7, Q6GV09, Q8BIB5 | Gene names | Zbtb5, Kiaa0354 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 5 (Transcription factor ZNF-POZ). | |||||
|
DMPK_HUMAN
|
||||||
| NC score | -0.002901 (rank : 119) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q09013, Q16205 | Gene names | DMPK, MDPK | |||
|
Domain Architecture |

|
|||||
| Description | Myotonin-protein kinase (EC 2.7.11.1) (Myotonic dystrophy protein kinase) (MDPK) (DM-kinase) (DMK) (DMPK) (MT-PK). | |||||