Please be patient as the page loads
|
TXD11_MOUSE
|
||||||
| SwissProt Accessions | Q8K2W3, Q8BMR8, Q8VCK9 | Gene names | Txndc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 11. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TXD11_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.859562 (rank : 2) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q6PKC3, O95887, Q6PJA6, Q8N2Q4, Q96K45, Q96K53 | Gene names | TXNDC11, EFP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 11 (EF-hand-binding protein 1). | |||||
|
TXD11_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8K2W3, Q8BMR8, Q8VCK9 | Gene names | Txndc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 11. | |||||
|
PDIA3_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 3) | NC score | 0.421889 (rank : 3) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P27773 | Gene names | Pdia3, Erp, Erp60, Grp58 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A3 precursor (EC 5.3.4.1) (Disulfide isomerase ER-60) (ERp60) (58 kDa microsomal protein) (p58) (ERp57). | |||||
|
PDIA3_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 4) | NC score | 0.417199 (rank : 4) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P30101, Q13453, Q14255, Q8IYF8, Q9UMU7 | Gene names | PDIA3, ERP60, GRP58 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A3 precursor (EC 5.3.4.1) (Disulfide isomerase ER-60) (ERp60) (58 kDa microsomal protein) (p58) (ERp57) (58 kDa glucose-regulated protein). | |||||
|
PDIA4_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 5) | NC score | 0.390190 (rank : 5) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P13667 | Gene names | PDIA4, ERP70, ERP72 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A4 precursor (EC 5.3.4.1) (Protein ERp-72) (ERp72). | |||||
|
PDIA4_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 6) | NC score | 0.380424 (rank : 6) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P08003, P15841 | Gene names | Pdia4, Cai, Erp72 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A4 precursor (EC 5.3.4.1) (Protein ERp-72) (ERp72). | |||||
|
PDIA1_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 7) | NC score | 0.343049 (rank : 8) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P07237, P30037, P32079, Q15205, Q6LDE5 | Gene names | P4HB, PDI, PDIA1, PO4DB | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase precursor (EC 5.3.4.1) (PDI) (Prolyl 4- hydroxylase subunit beta) (Cellular thyroid hormone-binding protein) (p55). | |||||
|
PDIA1_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 8) | NC score | 0.339444 (rank : 9) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P09103 | Gene names | P4hb, Pdia1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase precursor (EC 5.3.4.1) (PDI) (Prolyl 4- hydroxylase subunit beta) (Cellular thyroid hormone-binding protein) (p55) (Erp59). | |||||
|
PDIA2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 9) | NC score | 0.343667 (rank : 7) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13087, Q96KJ6, Q9BW95 | Gene names | PDIA2, PDIP | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A2 precursor (EC 5.3.4.1) (PDIp). | |||||
|
PDIA6_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 10) | NC score | 0.313619 (rank : 10) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q922R8, Q8BK54 | Gene names | Pdia6, Txndc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein disulfide-isomerase A6 precursor (EC 5.3.4.1) (Thioredoxin domain-containing protein 7). | |||||
|
CP135_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 11) | NC score | 0.066228 (rank : 44) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
PDIA6_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 12) | NC score | 0.307022 (rank : 11) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15084, Q99778 | Gene names | PDIA6, TXNDC7 | |||
|
Domain Architecture |
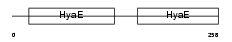
|
|||||
| Description | Protein disulfide-isomerase A6 precursor (EC 5.3.4.1) (Protein disulfide isomerase P5) (Thioredoxin domain-containing protein 7). | |||||
|
TXND4_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 13) | NC score | 0.283322 (rank : 13) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BS26, O60319, Q5VWZ7, Q6UW14, Q8WX67 | Gene names | TXNDC4, ERP44, KIAA0573 | |||
|
Domain Architecture |
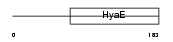
|
|||||
| Description | Thioredoxin domain-containing protein 4 precursor (Endoplasmic reticulum resident protein ERp44). | |||||
|
PDIA5_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 14) | NC score | 0.288263 (rank : 12) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q921X9 | Gene names | Pdia5, Pdir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein disulfide-isomerase A5 precursor (EC 5.3.4.1) (Protein disulfide isomerase-related protein). | |||||
|
TXND5_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 15) | NC score | 0.282748 (rank : 14) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q91W90, Q8R1I6 | Gene names | Txndc5, Tlp46 | |||
|
Domain Architecture |
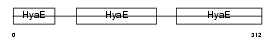
|
|||||
| Description | Thioredoxin domain-containing protein 5 precursor (Thioredoxin-like protein p46) (Endoplasmic reticulum protein ERp46) (Plasma cell- specific thioredoxin-related protein) (PC-TRP). | |||||
|
TXND4_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 16) | NC score | 0.278872 (rank : 15) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D1Q6, Q3TVI1, Q6A045 | Gene names | Txndc4, Erp44, Kiaa0573 | |||
|
Domain Architecture |
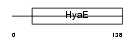
|
|||||
| Description | Thioredoxin domain-containing protein 4 precursor (Endoplasmic reticulum resident protein ERp44). | |||||
|
PDIA5_HUMAN
|
||||||
| θ value | 0.125558 (rank : 17) | NC score | 0.278449 (rank : 16) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q14554 | Gene names | PDIA5, PDIR | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A5 precursor (EC 5.3.4.1) (Protein disulfide isomerase-related protein). | |||||
|
TXND3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.118916 (rank : 28) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N427, Q9NZH1 | Gene names | TXNDC3, NME8, SPTRX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 3 (Spermatid-specific thioredoxin-2) (Sptrx-2) (NM23-H8). | |||||
|
A16L1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.035102 (rank : 97) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q676U5, Q6IPN1, Q6UXW4, Q6ZVZ5, Q8NCY2, Q96JV5, Q9H619 | Gene names | ATG16L1, APG16L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 16-1 (APG16-like 1). | |||||
|
KI21A_MOUSE
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.035873 (rank : 96) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
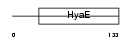
TXND1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.244529 (rank : 18) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H3N1, Q8N487, Q8NBN5, Q9Y4T6 | Gene names | TXNDC1, TMX, TXNDC | |||
|
Domain Architecture |
|
|||||
| Description | Thioredoxin domain-containing protein 1 precursor (Transmembrane Trx- related protein) (Thioredoxin-related transmembrane protein). | |||||
|
EP15R_MOUSE
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.054415 (rank : 71) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.032839 (rank : 98) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
ADIP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.082243 (rank : 38) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y2D8, Q6P2P8, Q6ULS1, Q7L168, Q9UIX0 | Gene names | SSX2IP, KIAA0923 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Afadin- and alpha-actinin-binding protein (ADIP) (Afadin DIL domain- interacting protein) (SSX2-interacting protein). | |||||
|
KI21A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.044045 (rank : 93) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
CCD41_MOUSE
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.051311 (rank : 83) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
MYOC_HUMAN
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.059960 (rank : 53) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99972, O00620, Q7Z6Q9 | Gene names | MYOC, GLC1A, TIGR | |||
|
Domain Architecture |

|
|||||
| Description | Myocilin precursor (Trabecular meshwork-induced glucocorticoid response protein). | |||||
|
K2C1B_MOUSE
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.057516 (rank : 57) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6IFZ6 | Gene names | Krt1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 1b (Type II keratin Kb39) (Embryonic type II keratin-1). | |||||
|
K2C6B_MOUSE
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.055035 (rank : 68) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z331 | Gene names | Krt6b, K6-beta, Krt2-6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 6B (Cytokeratin-6B) (CK 6B) (K6b keratin) (Keratin-6 beta) (mK6-beta). | |||||
|
TXD10_MOUSE
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.243612 (rank : 19) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BXZ1, Q3US84, Q6PGA1, Q6ZPH5, Q8BZB8 | Gene names | Txndc10, Kiaa1830 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein disulfide-isomerase TXNDC10 precursor (EC 5.3.4.1) (Thioredoxin domain-containing protein 10). | |||||
|
ADIP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.084845 (rank : 36) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VC66, Q8BG59, Q8C7X0, Q8K2F7 | Gene names | Ssx2ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Afadin- and alpha-actinin-binding protein (ADIP) (Afadin DIL domain- interacting protein). | |||||
|
CASP_HUMAN
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.066610 (rank : 43) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CASP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.068142 (rank : 39) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.065202 (rank : 45) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.067423 (rank : 41) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
TXND5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.272934 (rank : 17) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NBS9, Q8ND33, Q8TCT2, Q9BVH9 | Gene names | TXNDC5, TLP46 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin domain-containing protein 5 precursor (Thioredoxin-like protein p46) (Endoplasmic reticulum protein ERp46). | |||||
|
CP135_HUMAN
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.056403 (rank : 62) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
K2C6A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.055797 (rank : 65) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P02538, Q6NT67, Q96CL4 | Gene names | KRT6A, K6A | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6A (Cytokeratin-6A) (CK 6A) (K6a keratin). | |||||
|
K2C6A_MOUSE
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.053783 (rank : 72) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P50446, Q9Z332 | Gene names | Krt6a, Ker2, Krt2-6, Krt2-6a, Krt6 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6A (Cytokeratin-6A) (CK 6A) (K6a keratin) (Keratin-6 alpha) (mK6-alpha). | |||||
|
K2C6B_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.056552 (rank : 60) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P04259, P48669 | Gene names | KRT6B, K6B | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6B (Cytokeratin-6B) (CK 6B) (K6b keratin). | |||||
|
K2C6C_HUMAN
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.055907 (rank : 64) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P48666 | Gene names | KRT6C | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6C (Cytokeratin-6C) (CK 6C) (K6c keratin). | |||||
|
K2C6E_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.056768 (rank : 59) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P48668, Q7RTN9 | Gene names | KRT6E | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6E (Cytokeratin-6E) (CK 6E) (K6e keratin) (Keratin K6h). | |||||
|
RABE2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.058745 (rank : 55) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 598 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q91WG2, Q99KN3 | Gene names | Rabep2, Rabpt5b | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 2 (Rabaptin-5beta). | |||||
|
TRAIP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.060633 (rank : 52) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
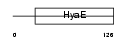
TXND1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | 0.223433 (rank : 21) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VBT0, Q3UCI8, Q9CSD5 | Gene names | Txndc1, Txndc | |||
|
Domain Architecture |
|
|||||
| Description | Thioredoxin domain-containing protein 1 precursor. | |||||
|
TXND3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.084248 (rank : 37) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q715T0, Q80W74 | Gene names | Txndc3, Sptrx2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 3 (Spermatid-specific thioredoxin-2) (Sptrx-2). | |||||
|
CING_HUMAN
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.056454 (rank : 61) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
FYCO1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.061030 (rank : 49) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
FYCO1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.060875 (rank : 51) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
LZTS1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.066666 (rank : 42) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P60853 | Gene names | Lzts1, Fez1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/Esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
MRCKB_MOUSE
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.030634 (rank : 99) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MRCKG_MOUSE
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.020118 (rank : 102) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1466 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q80UW5 | Gene names | Cdc42bpg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.049202 (rank : 91) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
K2C5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.054957 (rank : 69) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q922U2, Q920F2 | Gene names | Krt5, Krt2-5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 5 (Cytokeratin-5) (CK-5) (Keratin-5) (K5). | |||||
|
SPTN5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.036464 (rank : 94) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.007527 (rank : 112) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
TXD10_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.231673 (rank : 20) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96JJ7, Q8N5J0, Q9NWJ9 | Gene names | TXNDC10, KIAA1830, TMX3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein disulfide-isomerase TXNDC10 precursor (EC 5.3.4.1) (Thioredoxin domain-containing protein 10) (Thioredoxin-related transmembrane protein 3). | |||||
|
IBP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.019690 (rank : 103) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.054450 (rank : 70) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
K2C1B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.057274 (rank : 58) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7Z794, Q7RTS8 | Gene names | KRT1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 1b. | |||||
|
K2C5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.055946 (rank : 63) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P13647, Q6PI71, Q6UBJ0, Q8TA91 | Gene names | KRT5 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 5 (Cytokeratin-5) (CK-5) (Keratin-5) (K5) (58 kDa cytokeratin). | |||||
|
A16L1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.028303 (rank : 100) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 776 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8C0J2, Q6KAT7, Q80U97, Q80U98, Q80U99, Q80Y53, Q9DB63 | Gene names | Atg16l1, Apg16l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 16-1 (APG16-like 1). | |||||
|
ARSG_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.014017 (rank : 109) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96EG1, Q6UXF2, Q9Y2K4 | Gene names | ARSG, KIAA1001 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylsulfatase G precursor (EC 3.1.6.-) (ASG). | |||||
|
CMTA2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.017624 (rank : 105) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80Y50, Q3TFK0, Q5SX68, Q80TP1, Q8R0D9, Q8R2N5 | Gene names | Camta2, Kiaa0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
CCD13_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.052448 (rank : 78) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IYE1 | Gene names | CCDC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 13. | |||||
|
CNTN6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.004908 (rank : 113) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JMB8, Q8C6X1 | Gene names | Cntn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-6 precursor (Neural recognition molecule NB-3) (mNB-3). | |||||
|
ZN294_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.018158 (rank : 104) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6A009, Q6PIW6, Q8BZ44 | Gene names | Znf294, Kiaa0714, zfp294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 294 (Zfp-294). | |||||
|
CI093_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.050853 (rank : 85) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
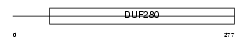
MAS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.007789 (rank : 111) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04201, Q6FG47 | Gene names | MAS1, MAS | |||
|
Domain Architecture |
|
|||||
| Description | MAS proto-oncogene. | |||||
|
PDCL3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.014249 (rank : 108) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BVF2, Q3TH06, Q99JX2, Q9D0W3 | Gene names | Pdcl3, Viaf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 3 (Viral IAP-associated factor 1) (VIAF-1). | |||||
|
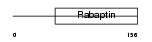
RABE2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.058180 (rank : 56) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 545 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H5N1 | Gene names | RABEP2, RABPT5B | |||
|
Domain Architecture |
|
|||||
| Description | Rab GTPase-binding effector protein 2 (Rabaptin-5beta). | |||||
|
RUFY1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.036030 (rank : 95) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
THIOM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.196024 (rank : 22) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99757, Q5JZA0, Q9UH29 | Gene names | TXN2, TRX2 | |||
|
Domain Architecture |
|
|||||
| Description | Thioredoxin, mitochondrial precursor (Mt-Trx) (MTRX) (Thioredoxin-2). | |||||
|
THIOM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.195438 (rank : 23) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97493, Q545D5 | Gene names | Txn2 | |||
|
Domain Architecture |
|
|||||
| Description | Thioredoxin, mitochondrial precursor (Mt-Trx) (MTRX) (Thioredoxin-2). | |||||
|
ABCBB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.003684 (rank : 115) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95342, Q9UNB2 | Gene names | ABCB11, BSEP | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt export pump (ATP-binding cassette sub-family B member 11). | |||||
|
DTNB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.013138 (rank : 110) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O70585, O70563, Q9CTZ1 | Gene names | Dtnb | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin beta (Beta-dystrobrevin) (DTN-B) (MDTN-B). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.046978 (rank : 92) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
MAST1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.003903 (rank : 114) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
SPTB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.025521 (rank : 101) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
TRPM7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.016981 (rank : 107) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
TRPM7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.017149 (rank : 106) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
CASQ1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.060889 (rank : 50) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P31415, Q8TBW7 | Gene names | CASQ1, CASQ | |||
|
Domain Architecture |

|
|||||
| Description | Calsequestrin-1 precursor (Calsequestrin, skeletal muscle isoform) (Calmitin). | |||||
|
CASQ1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.055234 (rank : 67) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O09165 | Gene names | Casq1 | |||
|
Domain Architecture |

|
|||||
| Description | Calsequestrin-1 precursor (Calsequestrin, skeletal muscle isoform). | |||||
|
CASQ2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.059272 (rank : 54) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14958, Q8TBW8 | Gene names | CASQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Calsequestrin-2 precursor (Calsequestrin, cardiac muscle isoform). | |||||
|
CASQ2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.052540 (rank : 77) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O09161, O88505 | Gene names | Casq2 | |||
|
Domain Architecture |

|
|||||
| Description | Calsequestrin-2 precursor (Calsequestrin, cardiac muscle isoform). | |||||
|
CING_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.051578 (rank : 80) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CP250_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.050146 (rank : 89) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
EP15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.064888 (rank : 46) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
GFAP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.050351 (rank : 87) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 725 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P03995, Q7TQ30, Q925K3 | Gene names | Gfap | |||
|
Domain Architecture |
|
|||||
| Description | Glial fibrillary acidic protein, astrocyte (GFAP). | |||||
|
K22O_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.051776 (rank : 79) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q01546, Q7Z795 | Gene names | KRT76, KRT2B, KRT2P | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 2 oral (Cytokeratin-2P) (K2P) (CK 2P) (Keratin 76). | |||||
|
K2C1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.050325 (rank : 88) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P04264, Q14720, Q6GSJ0, Q9H298 | Gene names | KRT1, KRTA | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin) (Hair alpha protein). | |||||
|
K2C3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.050044 (rank : 90) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P12035, Q701L8 | Gene names | KRT3 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cytoskeletal 3 (Cytokeratin-3) (CK-3) (Keratin-3) (K3) (65 kDa cytokeratin). | |||||
|
K2C4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.053008 (rank : 75) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P19013, Q6GTR8, Q96LA7, Q9BTL1 | Gene names | KRT4, CYK4 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 4 (Cytokeratin-4) (CK-4) (Keratin-4) (K4). | |||||
|
K2C4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.051323 (rank : 82) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P07744, Q6P3F5 | Gene names | Krt4, Krt2-4 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 4 (Cytokeratin-4) (CK-4) (Keratin-4) (K4) (Cytoskeletal 57 kDa keratin). | |||||
|
K2C6G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.051148 (rank : 84) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9R0H5, Q7TPF3, Q9D0X6 | Gene names | Krt6g, K6irs1, Krt2-6g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 6G (Cytokeratin-6G) (CK 6G) (K6g keratin) (Keratin-K6irs) (mK6irs1/Krt2-6g). | |||||
|
K2C7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.050581 (rank : 86) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P08729, Q92676, Q9BUD8, Q9Y3R7 | Gene names | KRT7, SCL | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cytoskeletal 7 (Cytokeratin-7) (CK-7) (Keratin-7) (K7) (Sarcolectin). | |||||
|
MACOI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.062866 (rank : 47) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8N5G2, Q2TLX5, Q2TLX6, Q9NVG6 | Gene names | TMEM57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57). | |||||
|
MACOI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.062593 (rank : 48) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 620 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q7TQE6 | Gene names | Tmem57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57) (Brain-specific adaptor protein C61). | |||||
|
MD1L1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.053225 (rank : 74) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
QSC6L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.101648 (rank : 33) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZRP7, Q5TB37, Q7Z7B6, Q86VV7, Q8N3G2 | Gene names | QSCN6L1, QSOX2, SOXN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfhydryl oxidase 2 precursor (EC 1.8.3.2) (Quiescin Q6-like protein 1) (Neuroblastoma-derived sulfhydryl oxidase). | |||||
|
QSC6L_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.092215 (rank : 34) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3TMX7, Q3TZA5, Q8C9I4, Q8K0M2 | Gene names | Qscn6l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfhydryl oxidase 2 precursor (EC 1.8.3.2) (Quiescin Q6-like protein 1). | |||||
|
QSCN6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.104830 (rank : 32) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00391, Q59G29, Q5T2X0, Q8TDL6, Q8WVP4 | Gene names | QSCN6, QSOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfhydryl oxidase 1 precursor (EC 1.8.3.2) (Quiescin Q6) (hQSOX). | |||||
|
QSCN6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.113164 (rank : 30) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BND5, Q3TDY9, Q3TE19, Q3TR29, Q3UEL4, Q8K041, Q9DBL6 | Gene names | Qscn6, Sox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfhydryl oxidase 1 precursor (EC 1.8.3.2) (Quiescin Q6) (Skin sulfhydryl oxidase) (mSOx). | |||||
|
RABE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.053346 (rank : 73) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
RABE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.052555 (rank : 76) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
THIO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.121453 (rank : 27) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P10599, Q96KI3 | Gene names | TXN, TRDX, TRX, TRX1 | |||
|
Domain Architecture |
|
|||||
| Description | Thioredoxin (Trx) (ATL-derived factor) (ADF) (Surface-associated sulphydryl protein) (SASP). | |||||
|
THIO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.111588 (rank : 31) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P10639, Q52KC4, Q9D8R0 | Gene names | Txn, Txn1 | |||
|
Domain Architecture |
|
|||||
| Description | Thioredoxin (Trx) (ATL-derived factor) (ADF). | |||||
|
TXD13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.134563 (rank : 26) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H1E5, Q8N4P7, Q8NCC1, Q9UJA1, Q9ULQ8 | Gene names | TXNDC13, KIAA1162 | |||
|
Domain Architecture |
|
|||||
| Description | Thioredoxin domain-containing protein 13 precursor. | |||||
|
TXD13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.135121 (rank : 25) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8C0L0, Q3UHC6, Q6ZPW7, Q80X49 | Gene names | Txndc13, D2Bwg1356e, Kiaa1162 | |||
|
Domain Architecture |
|
|||||
| Description | Thioredoxin domain-containing protein 13 precursor. | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.051473 (rank : 81) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
TXND6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.068126 (rank : 40) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86XW9, Q7Z4A8, Q8N1V7 | Gene names | TXNDC6, TXL2 | |||
|
Domain Architecture |
|
|||||
| Description | Thioredoxin domain-containing protein 6 (Thioredoxin-like protein 2) (Txl-2). | |||||
|
TXND8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.055723 (rank : 66) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6A555, Q5T934 | Gene names | TXNDC8, SPTRX3, TRX6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 8 (Spermatid-specific thioredoxin-3) (Sptrx-3) (Thioredoxin-6). | |||||
|
TXND8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.139446 (rank : 24) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q69AB2, Q78Y03, Q80YE0, Q9CQ96 | Gene names | Txndc8, Sptrx3, Trx6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 8 (Spermatid-specific thioredoxin-3) (Sptrx-3) (Thioredoxin-6). | |||||
|
TXNL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.117037 (rank : 29) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O76003, Q96CE0, Q9P1B0, Q9P1B1 | Gene names | TXNL2, PICOT | |||
|
Domain Architecture |
|
|||||
| Description | Thioredoxin-like protein 2 (PKC-interacting cousin of thioredoxin) (PKC-theta-interacting protein) (PKCq-interacting protein). | |||||
|
TXNL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.085012 (rank : 35) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9CQM9, Q9JLZ2 | Gene names | Txnl2, Picot | |||
|
Domain Architecture |
|
|||||
| Description | Thioredoxin-like protein 2 (PKC-interacting cousin of thioredoxin) (PKC-theta-interacting protein) (PKCq-interacting protein). | |||||
|
TXD11_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8K2W3, Q8BMR8, Q8VCK9 | Gene names | Txndc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 11. | |||||
|
TXD11_HUMAN
|
||||||
| NC score | 0.859562 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q6PKC3, O95887, Q6PJA6, Q8N2Q4, Q96K45, Q96K53 | Gene names | TXNDC11, EFP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 11 (EF-hand-binding protein 1). | |||||
|
PDIA3_MOUSE
|
||||||
| NC score | 0.421889 (rank : 3) | θ value | 5.62301e-10 (rank : 3) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P27773 | Gene names | Pdia3, Erp, Erp60, Grp58 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A3 precursor (EC 5.3.4.1) (Disulfide isomerase ER-60) (ERp60) (58 kDa microsomal protein) (p58) (ERp57). | |||||
|
PDIA3_HUMAN
|
||||||
| NC score | 0.417199 (rank : 4) | θ value | 3.64472e-09 (rank : 4) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P30101, Q13453, Q14255, Q8IYF8, Q9UMU7 | Gene names | PDIA3, ERP60, GRP58 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A3 precursor (EC 5.3.4.1) (Disulfide isomerase ER-60) (ERp60) (58 kDa microsomal protein) (p58) (ERp57) (58 kDa glucose-regulated protein). | |||||
|
PDIA4_HUMAN
|
||||||
| NC score | 0.390190 (rank : 5) | θ value | 6.87365e-08 (rank : 5) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P13667 | Gene names | PDIA4, ERP70, ERP72 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A4 precursor (EC 5.3.4.1) (Protein ERp-72) (ERp72). | |||||
|
PDIA4_MOUSE
|
||||||
| NC score | 0.380424 (rank : 6) | θ value | 1.29631e-06 (rank : 6) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P08003, P15841 | Gene names | Pdia4, Cai, Erp72 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A4 precursor (EC 5.3.4.1) (Protein ERp-72) (ERp72). | |||||
|
PDIA2_HUMAN
|
||||||
| NC score | 0.343667 (rank : 7) | θ value | 0.00298849 (rank : 9) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13087, Q96KJ6, Q9BW95 | Gene names | PDIA2, PDIP | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A2 precursor (EC 5.3.4.1) (PDIp). | |||||
|
PDIA1_HUMAN
|
||||||
| NC score | 0.343049 (rank : 8) | θ value | 0.00035302 (rank : 7) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P07237, P30037, P32079, Q15205, Q6LDE5 | Gene names | P4HB, PDI, PDIA1, PO4DB | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase precursor (EC 5.3.4.1) (PDI) (Prolyl 4- hydroxylase subunit beta) (Cellular thyroid hormone-binding protein) (p55). | |||||
|
PDIA1_MOUSE
|
||||||
| NC score | 0.339444 (rank : 9) | θ value | 0.000461057 (rank : 8) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P09103 | Gene names | P4hb, Pdia1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase precursor (EC 5.3.4.1) (PDI) (Prolyl 4- hydroxylase subunit beta) (Cellular thyroid hormone-binding protein) (p55) (Erp59). | |||||
|
PDIA6_MOUSE
|
||||||
| NC score | 0.313619 (rank : 10) | θ value | 0.00665767 (rank : 10) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q922R8, Q8BK54 | Gene names | Pdia6, Txndc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein disulfide-isomerase A6 precursor (EC 5.3.4.1) (Thioredoxin domain-containing protein 7). | |||||
|
PDIA6_HUMAN
|
||||||
| NC score | 0.307022 (rank : 11) | θ value | 0.0193708 (rank : 12) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15084, Q99778 | Gene names | PDIA6, TXNDC7 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A6 precursor (EC 5.3.4.1) (Protein disulfide isomerase P5) (Thioredoxin domain-containing protein 7). | |||||
|
PDIA5_MOUSE
|
||||||
| NC score | 0.288263 (rank : 12) | θ value | 0.0252991 (rank : 14) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q921X9 | Gene names | Pdia5, Pdir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein disulfide-isomerase A5 precursor (EC 5.3.4.1) (Protein disulfide isomerase-related protein). | |||||
|
TXND4_HUMAN
|
||||||
| NC score | 0.283322 (rank : 13) | θ value | 0.0193708 (rank : 13) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BS26, O60319, Q5VWZ7, Q6UW14, Q8WX67 | Gene names | TXNDC4, ERP44, KIAA0573 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin domain-containing protein 4 precursor (Endoplasmic reticulum resident protein ERp44). | |||||
|
TXND5_MOUSE
|
||||||
| NC score | 0.282748 (rank : 14) | θ value | 0.0431538 (rank : 15) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q91W90, Q8R1I6 | Gene names | Txndc5, Tlp46 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin domain-containing protein 5 precursor (Thioredoxin-like protein p46) (Endoplasmic reticulum protein ERp46) (Plasma cell- specific thioredoxin-related protein) (PC-TRP). | |||||
|
TXND4_MOUSE
|
||||||
| NC score | 0.278872 (rank : 15) | θ value | 0.0736092 (rank : 16) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9D1Q6, Q3TVI1, Q6A045 | Gene names | Txndc4, Erp44, Kiaa0573 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin domain-containing protein 4 precursor (Endoplasmic reticulum resident protein ERp44). | |||||
|
PDIA5_HUMAN
|
||||||
| NC score | 0.278449 (rank : 16) | θ value | 0.125558 (rank : 17) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q14554 | Gene names | PDIA5, PDIR | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A5 precursor (EC 5.3.4.1) (Protein disulfide isomerase-related protein). | |||||
|
TXND5_HUMAN
|
||||||
| NC score | 0.272934 (rank : 17) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8NBS9, Q8ND33, Q8TCT2, Q9BVH9 | Gene names | TXNDC5, TLP46 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin domain-containing protein 5 precursor (Thioredoxin-like protein p46) (Endoplasmic reticulum protein ERp46). | |||||
|
TXND1_HUMAN
|
||||||
| NC score | 0.244529 (rank : 18) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H3N1, Q8N487, Q8NBN5, Q9Y4T6 | Gene names | TXNDC1, TMX, TXNDC | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin domain-containing protein 1 precursor (Transmembrane Trx- related protein) (Thioredoxin-related transmembrane protein). | |||||
|
TXD10_MOUSE
|
||||||
| NC score | 0.243612 (rank : 19) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BXZ1, Q3US84, Q6PGA1, Q6ZPH5, Q8BZB8 | Gene names | Txndc10, Kiaa1830 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein disulfide-isomerase TXNDC10 precursor (EC 5.3.4.1) (Thioredoxin domain-containing protein 10). | |||||
|
TXD10_HUMAN
|
||||||
| NC score | 0.231673 (rank : 20) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96JJ7, Q8N5J0, Q9NWJ9 | Gene names | TXNDC10, KIAA1830, TMX3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein disulfide-isomerase TXNDC10 precursor (EC 5.3.4.1) (Thioredoxin domain-containing protein 10) (Thioredoxin-related transmembrane protein 3). | |||||
|
TXND1_MOUSE
|
||||||
| NC score | 0.223433 (rank : 21) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VBT0, Q3UCI8, Q9CSD5 | Gene names | Txndc1, Txndc | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin domain-containing protein 1 precursor. | |||||
|
THIOM_HUMAN
|
||||||
| NC score | 0.196024 (rank : 22) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99757, Q5JZA0, Q9UH29 | Gene names | TXN2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin, mitochondrial precursor (Mt-Trx) (MTRX) (Thioredoxin-2). | |||||
|
THIOM_MOUSE
|
||||||
| NC score | 0.195438 (rank : 23) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97493, Q545D5 | Gene names | Txn2 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin, mitochondrial precursor (Mt-Trx) (MTRX) (Thioredoxin-2). | |||||
|
TXND8_MOUSE
|
||||||
| NC score | 0.139446 (rank : 24) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q69AB2, Q78Y03, Q80YE0, Q9CQ96 | Gene names | Txndc8, Sptrx3, Trx6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 8 (Spermatid-specific thioredoxin-3) (Sptrx-3) (Thioredoxin-6). | |||||
|
TXD13_MOUSE
|
||||||
| NC score | 0.135121 (rank : 25) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8C0L0, Q3UHC6, Q6ZPW7, Q80X49 | Gene names | Txndc13, D2Bwg1356e, Kiaa1162 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin domain-containing protein 13 precursor. | |||||
|
TXD13_HUMAN
|
||||||
| NC score | 0.134563 (rank : 26) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H1E5, Q8N4P7, Q8NCC1, Q9UJA1, Q9ULQ8 | Gene names | TXNDC13, KIAA1162 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin domain-containing protein 13 precursor. | |||||
|
THIO_HUMAN
|
||||||
| NC score | 0.121453 (rank : 27) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P10599, Q96KI3 | Gene names | TXN, TRDX, TRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin (Trx) (ATL-derived factor) (ADF) (Surface-associated sulphydryl protein) (SASP). | |||||
|
TXND3_HUMAN
|
||||||
| NC score | 0.118916 (rank : 28) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N427, Q9NZH1 | Gene names | TXNDC3, NME8, SPTRX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 3 (Spermatid-specific thioredoxin-2) (Sptrx-2) (NM23-H8). | |||||
|
TXNL2_HUMAN
|
||||||
| NC score | 0.117037 (rank : 29) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O76003, Q96CE0, Q9P1B0, Q9P1B1 | Gene names | TXNL2, PICOT | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin-like protein 2 (PKC-interacting cousin of thioredoxin) (PKC-theta-interacting protein) (PKCq-interacting protein). | |||||
|
QSCN6_MOUSE
|
||||||
| NC score | 0.113164 (rank : 30) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BND5, Q3TDY9, Q3TE19, Q3TR29, Q3UEL4, Q8K041, Q9DBL6 | Gene names | Qscn6, Sox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfhydryl oxidase 1 precursor (EC 1.8.3.2) (Quiescin Q6) (Skin sulfhydryl oxidase) (mSOx). | |||||
|
THIO_MOUSE
|
||||||
| NC score | 0.111588 (rank : 31) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P10639, Q52KC4, Q9D8R0 | Gene names | Txn, Txn1 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin (Trx) (ATL-derived factor) (ADF). | |||||
|
QSCN6_HUMAN
|
||||||
| NC score | 0.104830 (rank : 32) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00391, Q59G29, Q5T2X0, Q8TDL6, Q8WVP4 | Gene names | QSCN6, QSOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfhydryl oxidase 1 precursor (EC 1.8.3.2) (Quiescin Q6) (hQSOX). | |||||
|
QSC6L_HUMAN
|
||||||
| NC score | 0.101648 (rank : 33) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZRP7, Q5TB37, Q7Z7B6, Q86VV7, Q8N3G2 | Gene names | QSCN6L1, QSOX2, SOXN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfhydryl oxidase 2 precursor (EC 1.8.3.2) (Quiescin Q6-like protein 1) (Neuroblastoma-derived sulfhydryl oxidase). | |||||
|
QSC6L_MOUSE
|
||||||
| NC score | 0.092215 (rank : 34) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3TMX7, Q3TZA5, Q8C9I4, Q8K0M2 | Gene names | Qscn6l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfhydryl oxidase 2 precursor (EC 1.8.3.2) (Quiescin Q6-like protein 1). | |||||
|
TXNL2_MOUSE
|
||||||
| NC score | 0.085012 (rank : 35) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9CQM9, Q9JLZ2 | Gene names | Txnl2, Picot | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin-like protein 2 (PKC-interacting cousin of thioredoxin) (PKC-theta-interacting protein) (PKCq-interacting protein). | |||||
|
ADIP_MOUSE
|
||||||
| NC score | 0.084845 (rank : 36) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VC66, Q8BG59, Q8C7X0, Q8K2F7 | Gene names | Ssx2ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Afadin- and alpha-actinin-binding protein (ADIP) (Afadin DIL domain- interacting protein). | |||||
|
TXND3_MOUSE
|
||||||
| NC score | 0.084248 (rank : 37) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q715T0, Q80W74 | Gene names | Txndc3, Sptrx2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 3 (Spermatid-specific thioredoxin-2) (Sptrx-2). | |||||
|
ADIP_HUMAN
|
||||||
| NC score | 0.082243 (rank : 38) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y2D8, Q6P2P8, Q6ULS1, Q7L168, Q9UIX0 | Gene names | SSX2IP, KIAA0923 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Afadin- and alpha-actinin-binding protein (ADIP) (Afadin DIL domain- interacting protein) (SSX2-interacting protein). | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.068142 (rank : 39) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
TXND6_HUMAN
|
||||||
| NC score | 0.068126 (rank : 40) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q86XW9, Q7Z4A8, Q8N1V7 | Gene names | TXNDC6, TXL2 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin domain-containing protein 6 (Thioredoxin-like protein 2) (Txl-2). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.067423 (rank : 41) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
LZTS1_MOUSE
|
||||||
| NC score | 0.066666 (rank : 42) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P60853 | Gene names | Lzts1, Fez1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/Esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
CASP_HUMAN
|
||||||
| NC score | 0.066610 (rank : 43) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.066228 (rank : 44) | θ value | 0.0148317 (rank : 11) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.065202 (rank : 45) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
EP15_HUMAN
|
||||||
| NC score | 0.064888 (rank : 46) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
MACOI_HUMAN
|
||||||
| NC score | 0.062866 (rank : 47) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8N5G2, Q2TLX5, Q2TLX6, Q9NVG6 | Gene names | TMEM57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57). | |||||
|
MACOI_MOUSE
|
||||||
| NC score | 0.062593 (rank : 48) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 620 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q7TQE6 | Gene names | Tmem57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57) (Brain-specific adaptor protein C61). | |||||
|
FYCO1_HUMAN
|
||||||
| NC score | 0.061030 (rank : 49) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
CASQ1_HUMAN
|
||||||
| NC score | 0.060889 (rank : 50) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P31415, Q8TBW7 | Gene names | CASQ1, CASQ | |||
|
Domain Architecture |

|
|||||
| Description | Calsequestrin-1 precursor (Calsequestrin, skeletal muscle isoform) (Calmitin). | |||||
|
FYCO1_MOUSE
|
||||||
| NC score | 0.060875 (rank : 51) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
TRAIP_MOUSE
|
||||||
| NC score | 0.060633 (rank : 52) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
MYOC_HUMAN
|
||||||
| NC score | 0.059960 (rank : 53) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99972, O00620, Q7Z6Q9 | Gene names | MYOC, GLC1A, TIGR | |||
|
Domain Architecture |

|
|||||
| Description | Myocilin precursor (Trabecular meshwork-induced glucocorticoid response protein). | |||||
|
CASQ2_HUMAN
|
||||||
| NC score | 0.059272 (rank : 54) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14958, Q8TBW8 | Gene names | CASQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Calsequestrin-2 precursor (Calsequestrin, cardiac muscle isoform). | |||||
|
RABE2_MOUSE
|
||||||
| NC score | 0.058745 (rank : 55) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 598 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q91WG2, Q99KN3 | Gene names | Rabep2, Rabpt5b | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 2 (Rabaptin-5beta). | |||||
|
RABE2_HUMAN
|
||||||
| NC score | 0.058180 (rank : 56) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 545 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H5N1 | Gene names | RABEP2, RABPT5B | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 2 (Rabaptin-5beta). | |||||
|
K2C1B_MOUSE
|
||||||
| NC score | 0.057516 (rank : 57) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6IFZ6 | Gene names | Krt1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 1b (Type II keratin Kb39) (Embryonic type II keratin-1). | |||||
|
K2C1B_HUMAN
|
||||||
| NC score | 0.057274 (rank : 58) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7Z794, Q7RTS8 | Gene names | KRT1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 1b. | |||||
|
K2C6E_HUMAN
|
||||||
| NC score | 0.056768 (rank : 59) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P48668, Q7RTN9 | Gene names | KRT6E | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6E (Cytokeratin-6E) (CK 6E) (K6e keratin) (Keratin K6h). | |||||
|
K2C6B_HUMAN
|
||||||
| NC score | 0.056552 (rank : 60) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P04259, P48669 | Gene names | KRT6B, K6B | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6B (Cytokeratin-6B) (CK 6B) (K6b keratin). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.056454 (rank : 61) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.056403 (rank : 62) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
K2C5_HUMAN
|
||||||
| NC score | 0.055946 (rank : 63) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P13647, Q6PI71, Q6UBJ0, Q8TA91 | Gene names | KRT5 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 5 (Cytokeratin-5) (CK-5) (Keratin-5) (K5) (58 kDa cytokeratin). | |||||
|
K2C6C_HUMAN
|
||||||
| NC score | 0.055907 (rank : 64) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P48666 | Gene names | KRT6C | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6C (Cytokeratin-6C) (CK 6C) (K6c keratin). | |||||
|
K2C6A_HUMAN
|
||||||
| NC score | 0.055797 (rank : 65) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P02538, Q6NT67, Q96CL4 | Gene names | KRT6A, K6A | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6A (Cytokeratin-6A) (CK 6A) (K6a keratin). | |||||
|
TXND8_HUMAN
|
||||||
| NC score | 0.055723 (rank : 66) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6A555, Q5T934 | Gene names | TXNDC8, SPTRX3, TRX6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 8 (Spermatid-specific thioredoxin-3) (Sptrx-3) (Thioredoxin-6). | |||||
|
CASQ1_MOUSE
|
||||||
| NC score | 0.055234 (rank : 67) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O09165 | Gene names | Casq1 | |||
|
Domain Architecture |

|
|||||
| Description | Calsequestrin-1 precursor (Calsequestrin, skeletal muscle isoform). | |||||
|
K2C6B_MOUSE
|
||||||
| NC score | 0.055035 (rank : 68) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z331 | Gene names | Krt6b, K6-beta, Krt2-6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 6B (Cytokeratin-6B) (CK 6B) (K6b keratin) (Keratin-6 beta) (mK6-beta). | |||||
|
K2C5_MOUSE
|
||||||
| NC score | 0.054957 (rank : 69) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q922U2, Q920F2 | Gene names | Krt5, Krt2-5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 5 (Cytokeratin-5) (CK-5) (Keratin-5) (K5). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.054450 (rank : 70) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
EP15R_MOUSE
|
||||||
| NC score | 0.054415 (rank : 71) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
K2C6A_MOUSE
|
||||||
| NC score | 0.053783 (rank : 72) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P50446, Q9Z332 | Gene names | Krt6a, Ker2, Krt2-6, Krt2-6a, Krt6 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 6A (Cytokeratin-6A) (CK 6A) (K6a keratin) (Keratin-6 alpha) (mK6-alpha). | |||||
|
RABE1_HUMAN
|
||||||
| NC score | 0.053346 (rank : 73) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
MD1L1_HUMAN
|
||||||
| NC score | 0.053225 (rank : 74) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
K2C4_HUMAN
|
||||||
| NC score | 0.053008 (rank : 75) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P19013, Q6GTR8, Q96LA7, Q9BTL1 | Gene names | KRT4, CYK4 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 4 (Cytokeratin-4) (CK-4) (Keratin-4) (K4). | |||||
|
RABE1_MOUSE
|
||||||
| NC score | 0.052555 (rank : 76) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
CASQ2_MOUSE
|
||||||
| NC score | 0.052540 (rank : 77) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O09161, O88505 | Gene names | Casq2 | |||
|
Domain Architecture |

|
|||||
| Description | Calsequestrin-2 precursor (Calsequestrin, cardiac muscle isoform). | |||||
|
CCD13_HUMAN
|
||||||
| NC score | 0.052448 (rank : 78) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IYE1 | Gene names | CCDC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 13. | |||||
|
K22O_HUMAN
|
||||||
| NC score | 0.051776 (rank : 79) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q01546, Q7Z795 | Gene names | KRT76, KRT2B, KRT2P | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 2 oral (Cytokeratin-2P) (K2P) (CK 2P) (Keratin 76). | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.051578 (rank : 80) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.051473 (rank : 81) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
K2C4_MOUSE
|
||||||
| NC score | 0.051323 (rank : 82) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P07744, Q6P3F5 | Gene names | Krt4, Krt2-4 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 4 (Cytokeratin-4) (CK-4) (Keratin-4) (K4) (Cytoskeletal 57 kDa keratin). | |||||
|
CCD41_MOUSE
|
||||||
| NC score | 0.051311 (rank : 83) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
K2C6G_MOUSE
|
||||||
| NC score | 0.051148 (rank : 84) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9R0H5, Q7TPF3, Q9D0X6 | Gene names | Krt6g, K6irs1, Krt2-6g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 6G (Cytokeratin-6G) (CK 6G) (K6g keratin) (Keratin-K6irs) (mK6irs1/Krt2-6g). | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.050853 (rank : 85) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
K2C7_HUMAN
|
||||||
| NC score | 0.050581 (rank : 86) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P08729, Q92676, Q9BUD8, Q9Y3R7 | Gene names | KRT7, SCL | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 7 (Cytokeratin-7) (CK-7) (Keratin-7) (K7) (Sarcolectin). | |||||
|
GFAP_MOUSE
|
||||||
| NC score | 0.050351 (rank : 87) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 725 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P03995, Q7TQ30, Q925K3 | Gene names | Gfap | |||
|
Domain Architecture |

|
|||||
| Description | Glial fibrillary acidic protein, astrocyte (GFAP). | |||||
|
K2C1_HUMAN
|
||||||
| NC score | 0.050325 (rank : 88) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P04264, Q14720, Q6GSJ0, Q9H298 | Gene names | KRT1, KRTA | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin) (Hair alpha protein). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.050146 (rank : 89) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
K2C3_HUMAN
|
||||||
| NC score | 0.050044 (rank : 90) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P12035, Q701L8 | Gene names | KRT3 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 3 (Cytokeratin-3) (CK-3) (Keratin-3) (K3) (65 kDa cytokeratin). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.049202 (rank : 91) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.046978 (rank : 92) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
KI21A_HUMAN
|
||||||
| NC score | 0.044045 (rank : 93) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
SPTN5_HUMAN
|
||||||
| NC score | 0.036464 (rank : 94) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
RUFY1_HUMAN
|
||||||
| NC score | 0.036030 (rank : 95) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
KI21A_MOUSE
|
||||||
| NC score | 0.035873 (rank : 96) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
A16L1_HUMAN
|
||||||
| NC score | 0.035102 (rank : 97) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q676U5, Q6IPN1, Q6UXW4, Q6ZVZ5, Q8NCY2, Q96JV5, Q9H619 | Gene names | ATG16L1, APG16L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 16-1 (APG16-like 1). | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.032839 (rank : 98) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MRCKB_MOUSE
|
||||||
| NC score | 0.030634 (rank : 99) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
A16L1_MOUSE
|
||||||
| NC score | 0.028303 (rank : 100) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 776 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8C0J2, Q6KAT7, Q80U97, Q80U98, Q80U99, Q80Y53, Q9DB63 | Gene names | Atg16l1, Apg16l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autophagy-related protein 16-1 (APG16-like 1). | |||||
|
SPTB1_MOUSE
|
||||||
| NC score | 0.025521 (rank : 101) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
MRCKG_MOUSE
|
||||||
| NC score | 0.020118 (rank : 102) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1466 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q80UW5 | Gene names | Cdc42bpg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
IBP1_MOUSE
|
||||||
| NC score | 0.019690 (rank : 103) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P47876, Q61732 | Gene names | Igfbp1, Igfbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 1 precursor (IGFBP-1) (IBP- 1) (IGF-binding protein 1). | |||||
|
ZN294_MOUSE
|
||||||
| NC score | 0.018158 (rank : 104) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6A009, Q6PIW6, Q8BZ44 | Gene names | Znf294, Kiaa0714, zfp294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 294 (Zfp-294). | |||||
|
CMTA2_MOUSE
|
||||||
| NC score | 0.017624 (rank : 105) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80Y50, Q3TFK0, Q5SX68, Q80TP1, Q8R0D9, Q8R2N5 | Gene names | Camta2, Kiaa0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
TRPM7_MOUSE
|
||||||
| NC score | 0.017149 (rank : 106) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
TRPM7_HUMAN
|
||||||
| NC score | 0.016981 (rank : 107) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
PDCL3_MOUSE
|
||||||
| NC score | 0.014249 (rank : 108) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BVF2, Q3TH06, Q99JX2, Q9D0W3 | Gene names | Pdcl3, Viaf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 3 (Viral IAP-associated factor 1) (VIAF-1). | |||||
|
ARSG_HUMAN
|
||||||
| NC score | 0.014017 (rank : 109) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96EG1, Q6UXF2, Q9Y2K4 | Gene names | ARSG, KIAA1001 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylsulfatase G precursor (EC 3.1.6.-) (ASG). | |||||
|
DTNB_MOUSE
|
||||||
| NC score | 0.013138 (rank : 110) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O70585, O70563, Q9CTZ1 | Gene names | Dtnb | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin beta (Beta-dystrobrevin) (DTN-B) (MDTN-B). | |||||
|
MAS_HUMAN
|
||||||
| NC score | 0.007789 (rank : 111) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04201, Q6FG47 | Gene names | MAS1, MAS | |||
|
Domain Architecture |

|
|||||
| Description | MAS proto-oncogene. | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.007527 (rank : 112) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
CNTN6_MOUSE
|
||||||
| NC score | 0.004908 (rank : 113) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JMB8, Q8C6X1 | Gene names | Cntn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-6 precursor (Neural recognition molecule NB-3) (mNB-3). | |||||
|
MAST1_HUMAN
|
||||||
| NC score | 0.003903 (rank : 114) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
ABCBB_HUMAN
|
||||||
| NC score | 0.003684 (rank : 115) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 81 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95342, Q9UNB2 | Gene names | ABCB11, BSEP | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt export pump (ATP-binding cassette sub-family B member 11). | |||||