Please be patient as the page loads
|
TRPM7_HUMAN
|
||||||
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TRPM3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.961433 (rank : 5) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9HCF6, Q5VW02, Q5VW03, Q5VW04, Q5W5T7, Q86SH0, Q86SH6, Q86UL0, Q86WK1, Q86WK2, Q86WK3, Q86WK4, Q86YZ9, Q86Z00, Q86Z01 | Gene names | TRPM3, KIAA1616, LTRPC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 3 (Long transient receptor potential channel 3) (LTrpC3) (Melastatin-2) (MLSN2). | |||||
|
TRPM6_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.975906 (rank : 4) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9BX84, Q6VPR8, Q6VPR9, Q6VPS0, Q6VPS1, Q6VPS2 | Gene names | TRPM6, CHAK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM6_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.977476 (rank : 3) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CIR4 | Gene names | Trpm6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM7_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
TRPM7_MOUSE
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.995423 (rank : 2) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
TRPM2_HUMAN
|
||||||
| θ value | 3.27947e-143 (rank : 6) | NC score | 0.858745 (rank : 7) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O94759, Q5KTC2, Q6J3P5, Q96KN6, Q96Q93 | Gene names | TRPM2, EREG1, KNP3, LTRPC2, TRPC7 | |||
|
Domain Architecture |

|
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7) (Estrogen- responsive element-associated gene 1 protein). | |||||
|
TRPM2_MOUSE
|
||||||
| θ value | 1.52337e-140 (rank : 7) | NC score | 0.861586 (rank : 6) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91YD4 | Gene names | Trpm2, Ltrpc2, Trpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7). | |||||
|
TRPM8_HUMAN
|
||||||
| θ value | 3.35642e-63 (rank : 8) | NC score | 0.845135 (rank : 8) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z2W7, Q6QNH9, Q8TAC3, Q8TDX8, Q9BVK1 | Gene names | TRPM8, LTRPC6, TRPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8 (Transient receptor potential-p8) (Trp-p8) (Long transient receptor potential channel 6) (LTrpC6). | |||||
|
TRPM8_MOUSE
|
||||||
| θ value | 5.0155e-59 (rank : 9) | NC score | 0.841301 (rank : 9) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8R4D5 | Gene names | Trpm8, Trpp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8. | |||||
|
TRPC4_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 10) | NC score | 0.494015 (rank : 10) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UBN4, Q15721, Q9UIB0, Q9UIB1, Q9UIB2 | Gene names | TRPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (trp-related protein 4) (hTrp-4) (hTrp4). | |||||
|
TRPC4_MOUSE
|
||||||
| θ value | 5.25075e-16 (rank : 11) | NC score | 0.492805 (rank : 11) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QUQ5, Q62350, Q9QUQ9, Q9QZC0 | Gene names | Trpc4, Trrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (Receptor- activated cation channel TRP4) (Capacitative calcium entry channel Trp4). | |||||
|
TRPC5_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 12) | NC score | 0.475208 (rank : 13) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UL62, O75233, Q5JXY8, Q9Y514 | Gene names | TRPC5, TRP5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Htrp-5) (Htrp5). | |||||
|
TRPC5_MOUSE
|
||||||
| θ value | 1.86753e-13 (rank : 13) | NC score | 0.475440 (rank : 12) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QX29, Q61059, Q9QWT1, Q9R0D4 | Gene names | Trpc5, Trp5, Trrp5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Transient receptor protein 5) (Mtrp5) (trp-related protein 5) (Capacitative calcium entry channel 2) (CCE2). | |||||
|
TRPC2_MOUSE
|
||||||
| θ value | 6.64225e-11 (rank : 14) | NC score | 0.441360 (rank : 14) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9R244, Q9ES59, Q9ES60, Q9R243 | Gene names | Trpc2, Trp2, Trrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 2 (TrpC2) (mTrp2). | |||||
|
EF2K_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 15) | NC score | 0.326661 (rank : 23) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08796 | Gene names | Eef2k | |||
|
Domain Architecture |
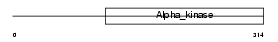
|
|||||
| Description | Elongation factor 2 kinase (EC 2.7.11.20) (eEF-2 kinase) (eEF-2K) (Calcium/calmodulin-dependent eukaryotic elongation factor 2 kinase). | |||||
|
TRPC3_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 16) | NC score | 0.421239 (rank : 15) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QZC1, Q61058 | Gene names | Trpc3, Trp3, Trrp3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Transient receptor protein 3) (Mtrp3) (Receptor-activated cation channel TRP3) (trp-related protein 3). | |||||
|
EF2K_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 17) | NC score | 0.318571 (rank : 24) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00418 | Gene names | EEF2K | |||
|
Domain Architecture |
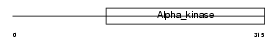
|
|||||
| Description | Elongation factor 2 kinase (EC 2.7.11.20) (eEF-2 kinase) (eEF-2K) (Calcium/calmodulin-dependent eukaryotic elongation factor 2 kinase). | |||||
|
TRPC6_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 18) | NC score | 0.388170 (rank : 18) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y210, Q9HCW3, Q9NQA8, Q9NQA9 | Gene names | TRPC6, TRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6). | |||||
|
TRPC6_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 19) | NC score | 0.388692 (rank : 17) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61143, Q9Z2J1 | Gene names | Trpc6, Trp6, Trrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6) (Calcium entry channel). | |||||
|
TRPC7_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 20) | NC score | 0.378681 (rank : 20) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HCX4 | Gene names | TRPC7, TRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (TRP7 protein). | |||||
|
TRPC7_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 21) | NC score | 0.379377 (rank : 19) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WVC5 | Gene names | Trpc7, Trp7, Trrp8 | |||
|
Domain Architecture |
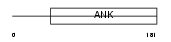
|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (mTRP7). | |||||
|
TRPC3_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 22) | NC score | 0.411242 (rank : 16) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13507, O00593, Q15660 | Gene names | TRPC3, TRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Htrp-3) (Htrp3). | |||||
|
PK2L1_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 23) | NC score | 0.138072 (rank : 31) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
TRPC1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 24) | NC score | 0.353861 (rank : 22) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P48995 | Gene names | TRPC1, TRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (TRP-1 protein). | |||||
|
TRPC1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 25) | NC score | 0.354114 (rank : 21) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61056, O35722 | Gene names | Trpc1, Trp1, Trrp1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (Transient receptor protein 1) (Mtrp1) (Trp-related protein 1). | |||||
|
PK2L2_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 26) | NC score | 0.116111 (rank : 39) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PCNT_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 27) | NC score | 0.043550 (rank : 46) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
LUZP1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 28) | NC score | 0.036778 (rank : 48) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
TRPV3_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 29) | NC score | 0.212980 (rank : 25) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 30) | NC score | 0.044629 (rank : 45) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
PK2L2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 31) | NC score | 0.099600 (rank : 40) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
TRPV3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 32) | NC score | 0.212401 (rank : 26) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
TRPV6_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 33) | NC score | 0.147908 (rank : 29) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H1D0, Q8TDL3, Q8WXR8, Q96LC5, Q9H1D1, Q9H296 | Gene names | TRPV6, ECAC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1) (CaT-like) (CaT-L). | |||||
|
RGS1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 34) | NC score | 0.028088 (rank : 52) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JL25, Q3TBD1, Q3TC18, Q3TD56 | Gene names | Rgs1 | |||
|
Domain Architecture |
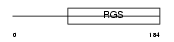
|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 0.163984 (rank : 35) | NC score | 0.036876 (rank : 47) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CNGA1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 36) | NC score | 0.019098 (rank : 67) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
STAT1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 37) | NC score | 0.014686 (rank : 80) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P42224 | Gene names | STAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 1-alpha/beta (Transcription factor ISGF-3 components p91/p84). | |||||
|
TRPV6_MOUSE
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.132679 (rank : 33) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91WD2 | Gene names | Trpv6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1). | |||||
|
LAMA2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 39) | NC score | 0.009671 (rank : 95) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
SM1L2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.019433 (rank : 65) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
TRPV5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.129181 (rank : 35) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NQA5, Q8NDW5, Q8NDX7, Q8NDX8, Q96PM6 | Gene names | TRPV5, ECAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
TRPV1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.142636 (rank : 30) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8NER1, Q9H0G9, Q9H303, Q9H304, Q9NQ74, Q9NY22 | Gene names | TRPV1, VR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1) (Capsaicin receptor). | |||||
|
CING_HUMAN
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.018458 (rank : 69) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
LMNA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.025491 (rank : 58) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 653 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P02545, P02546, Q969I8, Q96JA2 | Gene names | LMNA, LMN1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-A/C (70 kDa lamin) (NY-REN-32 antigen). | |||||
|
LMNA_MOUSE
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.025629 (rank : 56) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P48678, P11516, P97859, Q3TIH0, Q3TTS8, Q3UCA0, Q3UCJ8, Q91WF2, Q9DC21 | Gene names | Lmna, Lmn1 | |||
|
Domain Architecture |
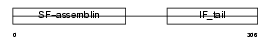
|
|||||
| Description | Lamin-A/C. | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.012118 (rank : 88) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
RBM25_HUMAN
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.020222 (rank : 64) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
TRPV2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.097371 (rank : 41) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WTR1, Q99K71 | Gene names | Trpv2, Grc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Growth factor-regulated calcium channel) (GRC). | |||||
|
UBXD2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.025577 (rank : 57) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92575, Q8IYM5 | Gene names | UBXD2, KIAA0242, UBXDC1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
KIF3B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.014283 (rank : 81) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O15066 | Gene names | KIF3B, KIAA0359 | |||
|
Domain Architecture |
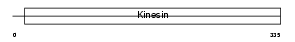
|
|||||
| Description | Kinesin-like protein KIF3B (Microtubule plus end-directed kinesin motor 3B) (HH0048). | |||||
|
KIF3B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.013096 (rank : 83) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61771 | Gene names | Kif3b | |||
|
Domain Architecture |
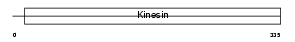
|
|||||
| Description | Kinesin-like protein KIF3B (Microtubule plus end-directed kinesin motor 3B). | |||||
|
LUZP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.028145 (rank : 51) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.021867 (rank : 61) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
TRAK2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.027667 (rank : 53) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60296, Q8WVH7, Q96NS2, Q9C0K5, Q9C0K6 | Gene names | TRAK2, ALS2CR3, KIAA0549 | |||
|
Domain Architecture |
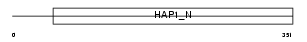
|
|||||
| Description | Trafficking kinesin-binding protein 2 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 3 protein). | |||||
|
TRPV1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.136525 (rank : 32) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q704Y3, Q5SSE1, Q5SSE2, Q5SSE4, Q5WPV5, Q68SW0 | Gene names | Trpv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1). | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.026828 (rank : 54) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
LMAN1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.026817 (rank : 55) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D0F3 | Gene names | Lman1, Ergic53 | |||
|
Domain Architecture |
|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (p58). | |||||
|
PHF12_MOUSE
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.012915 (rank : 84) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.016304 (rank : 76) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.018289 (rank : 70) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
TRPV5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.124194 (rank : 36) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P69744 | Gene names | Trpv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
DEMA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.010956 (rank : 90) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WV69 | Gene names | Epb49, Epb4.9 | |||
|
Domain Architecture |

|
|||||
| Description | Dematin (Erythrocyte membrane protein band 4.9). | |||||
|
PROM1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.020697 (rank : 62) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O54990, O35408 | Gene names | Prom1, Prom, Proml1 | |||
|
Domain Architecture |

|
|||||
| Description | Prominin-1 precursor (Prominin-like protein 1) (Antigen AC133 homolog). | |||||
|
SENP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.010822 (rank : 91) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59110 | Gene names | Senp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 1 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP1). | |||||
|
SH3G1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.007916 (rank : 102) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99961, Q99668 | Gene names | SH3GL1, CNSA1, SH3D2B | |||
|
Domain Architecture |

|
|||||
| Description | SH3-containing GRB2-like protein 1 (Endophilin-2) (Endophilin-A2) (SH3 domain protein 2B) (Extra eleven-nineteen leukemia fusion gene) (EEN) (EEN fusion partner of MLL). | |||||
|
TXD11_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.022219 (rank : 60) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PKC3, O95887, Q6PJA6, Q8N2Q4, Q96K45, Q96K53 | Gene names | TXNDC11, EFP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 11 (EF-hand-binding protein 1). | |||||
|
AT1B1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.009160 (rank : 97) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P05026, Q5TGZ3 | Gene names | ATP1B1, ATP1B | |||
|
Domain Architecture |
|
|||||
| Description | Sodium/potassium-transporting ATPase subunit beta-1 (Sodium/potassium- dependent ATPase beta-1 subunit). | |||||
|
BRD8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.023460 (rank : 59) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
CC47_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.029192 (rank : 50) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
CCD47_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.032749 (rank : 49) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96A33, Q96D00, Q96JZ7, Q9H3E4, Q9NRG3 | Gene names | CCDC47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor. | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.012868 (rank : 85) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
HOOK1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.012234 (rank : 87) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.009879 (rank : 94) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
RCOR2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.010116 (rank : 93) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C796, Q9JMK4 | Gene names | Rcor2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2 (M-CoREST). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.014763 (rank : 78) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
DIAP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.008408 (rank : 101) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O70566, Q8C2G8 | Gene names | Diaph2, Diap2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2) (mDia3). | |||||
|
P73_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.008586 (rank : 100) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15350, O15351, Q5TBV5, Q5TBV6, Q8NHW9, Q8TDY5, Q8TDY6, Q9NTK8 | Gene names | TP73, P73 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein p73 (p53-like transcription factor) (p53-related protein). | |||||
|
RAI14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.019084 (rank : 68) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
TRPV4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.132186 (rank : 34) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
UBXD2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.020330 (rank : 63) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VCH8, Q8BW17 | Gene names | Ubxd2, Ubxdc1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
3BP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.003908 (rank : 107) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
CL011_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.010354 (rank : 92) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8QZV7, Q99JT8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C12orf11 homolog. | |||||
|
CP7B1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.006899 (rank : 105) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60991 | Gene names | Cyp7b1 | |||
|
Domain Architecture |
|
|||||
| Description | Cytochrome P450 7B1 (EC 1.14.13.100) (25-hydroxycholesterol 7-alpha- hydroxylase) (Oxysterol 7-alpha-hydroxylase) (HCT-1). | |||||
|
DIAP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.011665 (rank : 89) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NSV4, Q5T2S7, Q5XKF6, Q6MZF0, Q6NUP0, Q86VS4, Q8NAV4 | Gene names | DIAPH3, DIAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein diaphanous homolog 3 (Diaphanous-related formin-3) (DRF3). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.012823 (rank : 86) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
LIPS_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.009212 (rank : 96) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54310, P97866, Q3TE34, Q6GU16 | Gene names | Lipe | |||
|
Domain Architecture |
|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
LMAN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.019395 (rank : 66) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49257, Q12895, Q8N5I7, Q9UQG1, Q9UQG2, Q9UQG3, Q9UQG4, Q9UQG5, Q9UQG6, Q9UQG7, Q9UQG8, Q9UQG9, Q9UQH0, Q9UQH1, Q9UQH2 | Gene names | LMAN1, ERGIC53, F5F8D | |||
|
Domain Architecture |
|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (Gp58) (Intracellular mannose- specific lectin MR60). | |||||
|
NOVA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.007160 (rank : 104) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UNW9, O43267, Q9UEA1 | Gene names | NOVA2, ANOVA, NOVA3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-2 (Neuro-oncological ventral antigen 2) (Astrocytic NOVA1-like RNA-binding protein). | |||||
|
OR2H1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | -0.003080 (rank : 108) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9GZK4, O43629, O43661, O43662, Q9GZK9 | Gene names | OR2H1, OR2H6, OR2H8 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 2H1 (Hs6M1-16) (Olfactory receptor 6-2) (OR6-2) (OLFR42A-9004.14/9026.2). | |||||
|
RCOR2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.008748 (rank : 99) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IZ40, Q96FP3 | Gene names | RCOR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2. | |||||
|
SH3G1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.006309 (rank : 106) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62419 | Gene names | Sh3gl1, Sh3d2b | |||
|
Domain Architecture |

|
|||||
| Description | SH3-containing GRB2-like protein 1 (Endophilin-2) (Endophilin-A2) (SH3 domain protein 2B) (SH3p8). | |||||
|
TRPV4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.117652 (rank : 38) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9EPK8, Q91XR5, Q9EQZ4, Q9ERZ7, Q9ES76 | Gene names | Trpv4, Trp12, Vrl2, Vroac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (Vanilloid receptor- related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.017844 (rank : 72) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
CCDC6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.014693 (rank : 79) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q16204, Q15250 | Gene names | CCDC6, D10S170 | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil domain-containing protein 6 (H4 protein) (Papillary thyroid carcinoma-encoded protein). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.014790 (rank : 77) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CKAP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.016312 (rank : 74) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q07065, Q504S5, Q53ES6 | Gene names | CKAP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.013222 (rank : 82) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.018090 (rank : 71) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MCLN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.016310 (rank : 75) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZK6, Q8N9R3 | Gene names | MCOLN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
S4A11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.007717 (rank : 103) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NBS3, Q9BXF4, Q9NTW9 | Gene names | SLC4A11, BTR1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium bicarbonate transporter-like protein 11 (Bicarbonate transporter-related protein 1) (Solute carrier family 4 member 11). | |||||
|
TRPV2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.076152 (rank : 42) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y5S1, Q9Y670 | Gene names | TRPV2, VRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Vanilloid receptor-like protein 1) (VRL-1). | |||||
|
TXD11_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.016981 (rank : 73) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K2W3, Q8BMR8, Q8VCK9 | Gene names | Txndc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 11. | |||||
|
UBP8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.009159 (rank : 98) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
ZN548_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | -0.003664 (rank : 109) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NEK5, Q96M05 | Gene names | ZNF548 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 548. | |||||
|
NUD9P_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.117929 (rank : 37) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N2Z3 | Gene names | NUDT9P1, C10orf98 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative nucleoside diphosphate-linked moiety X motif 9P1. | |||||
|
NUDT9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.154602 (rank : 28) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BW91, Q8NBN1, Q8NCB9, Q8NG25 | Gene names | NUDT9, NUDT10 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribose pyrophosphatase, mitochondrial precursor (EC 3.6.1.13) (ADP-ribose diphosphatase) (Adenosine diphosphoribose pyrophosphatase) (ADPR-PPase) (ADP-ribose phosphohydrolase) (Nucleoside diphosphate- linked moiety X motif 9) (Nudix motif 9). | |||||
|
NUDT9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.155964 (rank : 27) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BVU5, Q3TZ68, Q8K1J4 | Gene names | Nudt9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribose pyrophosphatase, mitochondrial precursor (EC 3.6.1.13) (ADP-ribose diphosphatase) (Adenosine diphosphoribose pyrophosphatase) (ADPR-PPase) (ADP-ribose phosphohydrolase) (Nucleoside diphosphate- linked moiety X motif 9) (Nudix motif 9). | |||||
|
PKD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.073989 (rank : 43) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
PKD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.068312 (rank : 44) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
TRPM7_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
TRPM7_MOUSE
|
||||||
| NC score | 0.995423 (rank : 2) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
TRPM6_MOUSE
|
||||||
| NC score | 0.977476 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CIR4 | Gene names | Trpm6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM6_HUMAN
|
||||||
| NC score | 0.975906 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9BX84, Q6VPR8, Q6VPR9, Q6VPS0, Q6VPS1, Q6VPS2 | Gene names | TRPM6, CHAK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 6 (EC 2.7.11.1) (Channel kinase 2) (Melastatin-related TRP cation channel 6). | |||||
|
TRPM3_HUMAN
|
||||||
| NC score | 0.961433 (rank : 5) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9HCF6, Q5VW02, Q5VW03, Q5VW04, Q5W5T7, Q86SH0, Q86SH6, Q86UL0, Q86WK1, Q86WK2, Q86WK3, Q86WK4, Q86YZ9, Q86Z00, Q86Z01 | Gene names | TRPM3, KIAA1616, LTRPC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 3 (Long transient receptor potential channel 3) (LTrpC3) (Melastatin-2) (MLSN2). | |||||
|
TRPM2_MOUSE
|
||||||
| NC score | 0.861586 (rank : 6) | θ value | 1.52337e-140 (rank : 7) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q91YD4 | Gene names | Trpm2, Ltrpc2, Trpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7). | |||||
|
TRPM2_HUMAN
|
||||||
| NC score | 0.858745 (rank : 7) | θ value | 3.27947e-143 (rank : 6) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O94759, Q5KTC2, Q6J3P5, Q96KN6, Q96Q93 | Gene names | TRPM2, EREG1, KNP3, LTRPC2, TRPC7 | |||
|
Domain Architecture |

|
|||||
| Description | Transient receptor potential cation channel subfamily M member 2 (EC 3.6.1.13) (Long transient receptor potential channel 2) (LTrpC2) (LTrpC-2) (Transient receptor potential channel 7) (TrpC7) (Estrogen- responsive element-associated gene 1 protein). | |||||
|
TRPM8_HUMAN
|
||||||
| NC score | 0.845135 (rank : 8) | θ value | 3.35642e-63 (rank : 8) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z2W7, Q6QNH9, Q8TAC3, Q8TDX8, Q9BVK1 | Gene names | TRPM8, LTRPC6, TRPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8 (Transient receptor potential-p8) (Trp-p8) (Long transient receptor potential channel 6) (LTrpC6). | |||||
|
TRPM8_MOUSE
|
||||||
| NC score | 0.841301 (rank : 9) | θ value | 5.0155e-59 (rank : 9) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8R4D5 | Gene names | Trpm8, Trpp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 8. | |||||
|
TRPC4_HUMAN
|
||||||
| NC score | 0.494015 (rank : 10) | θ value | 4.02038e-16 (rank : 10) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UBN4, Q15721, Q9UIB0, Q9UIB1, Q9UIB2 | Gene names | TRPC4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (trp-related protein 4) (hTrp-4) (hTrp4). | |||||
|
TRPC4_MOUSE
|
||||||
| NC score | 0.492805 (rank : 11) | θ value | 5.25075e-16 (rank : 11) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9QUQ5, Q62350, Q9QUQ9, Q9QZC0 | Gene names | Trpc4, Trrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 4 (TrpC4) (Receptor- activated cation channel TRP4) (Capacitative calcium entry channel Trp4). | |||||
|
TRPC5_MOUSE
|
||||||
| NC score | 0.475440 (rank : 12) | θ value | 1.86753e-13 (rank : 13) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QX29, Q61059, Q9QWT1, Q9R0D4 | Gene names | Trpc5, Trp5, Trrp5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Transient receptor protein 5) (Mtrp5) (trp-related protein 5) (Capacitative calcium entry channel 2) (CCE2). | |||||
|
TRPC5_HUMAN
|
||||||
| NC score | 0.475208 (rank : 13) | θ value | 1.86753e-13 (rank : 12) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UL62, O75233, Q5JXY8, Q9Y514 | Gene names | TRPC5, TRP5 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 5 (TrpC5) (Htrp-5) (Htrp5). | |||||
|
TRPC2_MOUSE
|
||||||
| NC score | 0.441360 (rank : 14) | θ value | 6.64225e-11 (rank : 14) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9R244, Q9ES59, Q9ES60, Q9R243 | Gene names | Trpc2, Trp2, Trrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 2 (TrpC2) (mTrp2). | |||||
|
TRPC3_MOUSE
|
||||||
| NC score | 0.421239 (rank : 15) | θ value | 2.52405e-10 (rank : 16) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QZC1, Q61058 | Gene names | Trpc3, Trp3, Trrp3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Transient receptor protein 3) (Mtrp3) (Receptor-activated cation channel TRP3) (trp-related protein 3). | |||||
|
TRPC3_HUMAN
|
||||||
| NC score | 0.411242 (rank : 16) | θ value | 6.21693e-09 (rank : 22) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13507, O00593, Q15660 | Gene names | TRPC3, TRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 3 (TrpC3) (Htrp-3) (Htrp3). | |||||
|
TRPC6_MOUSE
|
||||||
| NC score | 0.388692 (rank : 17) | θ value | 2.79066e-09 (rank : 19) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61143, Q9Z2J1 | Gene names | Trpc6, Trp6, Trrp6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6) (Calcium entry channel). | |||||
|
TRPC6_HUMAN
|
||||||
| NC score | 0.388170 (rank : 18) | θ value | 1.63604e-09 (rank : 18) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y210, Q9HCW3, Q9NQA8, Q9NQA9 | Gene names | TRPC6, TRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 6 (TrpC6). | |||||
|
TRPC7_MOUSE
|
||||||
| NC score | 0.379377 (rank : 19) | θ value | 3.64472e-09 (rank : 21) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9WVC5 | Gene names | Trpc7, Trp7, Trrp8 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (mTRP7). | |||||
|
TRPC7_HUMAN
|
||||||
| NC score | 0.378681 (rank : 20) | θ value | 3.64472e-09 (rank : 20) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HCX4 | Gene names | TRPC7, TRP7 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 7 (TrpC7) (TRP7 protein). | |||||
|
TRPC1_MOUSE
|
||||||
| NC score | 0.354114 (rank : 21) | θ value | 0.00134147 (rank : 25) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61056, O35722 | Gene names | Trpc1, Trp1, Trrp1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (Transient receptor protein 1) (Mtrp1) (Trp-related protein 1). | |||||
|
TRPC1_HUMAN
|
||||||
| NC score | 0.353861 (rank : 22) | θ value | 0.00134147 (rank : 24) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P48995 | Gene names | TRPC1, TRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Short transient receptor potential channel 1 (TrpC1) (TRP-1 protein). | |||||
|
EF2K_MOUSE
|
||||||
| NC score | 0.326661 (rank : 23) | θ value | 1.47974e-10 (rank : 15) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08796 | Gene names | Eef2k | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 2 kinase (EC 2.7.11.20) (eEF-2 kinase) (eEF-2K) (Calcium/calmodulin-dependent eukaryotic elongation factor 2 kinase). | |||||
|
EF2K_HUMAN
|
||||||
| NC score | 0.318571 (rank : 24) | θ value | 9.59137e-10 (rank : 17) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00418 | Gene names | EEF2K | |||
|
Domain Architecture |

|
|||||
| Description | Elongation factor 2 kinase (EC 2.7.11.20) (eEF-2 kinase) (eEF-2K) (Calcium/calmodulin-dependent eukaryotic elongation factor 2 kinase). | |||||
|
TRPV3_MOUSE
|
||||||
| NC score | 0.212980 (rank : 25) | θ value | 0.0193708 (rank : 29) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8K424, Q5SV08 | Gene names | Trpv3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3). | |||||
|
TRPV3_HUMAN
|
||||||
| NC score | 0.212401 (rank : 26) | θ value | 0.0431538 (rank : 32) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
NUDT9_MOUSE
|
||||||
| NC score | 0.155964 (rank : 27) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BVU5, Q3TZ68, Q8K1J4 | Gene names | Nudt9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribose pyrophosphatase, mitochondrial precursor (EC 3.6.1.13) (ADP-ribose diphosphatase) (Adenosine diphosphoribose pyrophosphatase) (ADPR-PPase) (ADP-ribose phosphohydrolase) (Nucleoside diphosphate- linked moiety X motif 9) (Nudix motif 9). | |||||
|
NUDT9_HUMAN
|
||||||
| NC score | 0.154602 (rank : 28) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BW91, Q8NBN1, Q8NCB9, Q8NG25 | Gene names | NUDT9, NUDT10 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribose pyrophosphatase, mitochondrial precursor (EC 3.6.1.13) (ADP-ribose diphosphatase) (Adenosine diphosphoribose pyrophosphatase) (ADPR-PPase) (ADP-ribose phosphohydrolase) (Nucleoside diphosphate- linked moiety X motif 9) (Nudix motif 9). | |||||
|
TRPV6_HUMAN
|
||||||
| NC score | 0.147908 (rank : 29) | θ value | 0.0736092 (rank : 33) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H1D0, Q8TDL3, Q8WXR8, Q96LC5, Q9H1D1, Q9H296 | Gene names | TRPV6, ECAC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1) (CaT-like) (CaT-L). | |||||
|
TRPV1_HUMAN
|
||||||
| NC score | 0.142636 (rank : 30) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8NER1, Q9H0G9, Q9H303, Q9H304, Q9NQ74, Q9NY22 | Gene names | TRPV1, VR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1) (Capsaicin receptor). | |||||
|
PK2L1_HUMAN
|
||||||
| NC score | 0.138072 (rank : 31) | θ value | 0.000786445 (rank : 23) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
TRPV1_MOUSE
|
||||||
| NC score | 0.136525 (rank : 32) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q704Y3, Q5SSE1, Q5SSE2, Q5SSE4, Q5WPV5, Q68SW0 | Gene names | Trpv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 1 (TrpV1) (osm-9-like TRP channel 1) (OTRPC1) (Vanilloid receptor 1). | |||||
|
TRPV6_MOUSE
|
||||||
| NC score | 0.132679 (rank : 33) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q91WD2 | Gene names | Trpv6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 6 (TrpV6) (Epithelial calcium channel 2) (ECaC2) (Calcium transport protein 1) (CaT1). | |||||
|
TRPV4_HUMAN
|
||||||
| NC score | 0.132186 (rank : 34) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
TRPV5_HUMAN
|
||||||
| NC score | 0.129181 (rank : 35) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NQA5, Q8NDW5, Q8NDX7, Q8NDX8, Q96PM6 | Gene names | TRPV5, ECAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
TRPV5_MOUSE
|
||||||
| NC score | 0.124194 (rank : 36) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P69744 | Gene names | Trpv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
NUD9P_HUMAN
|
||||||
| NC score | 0.117929 (rank : 37) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N2Z3 | Gene names | NUDT9P1, C10orf98 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative nucleoside diphosphate-linked moiety X motif 9P1. | |||||
|
TRPV4_MOUSE
|
||||||
| NC score | 0.117652 (rank : 38) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9EPK8, Q91XR5, Q9EQZ4, Q9ERZ7, Q9ES76 | Gene names | Trpv4, Trp12, Vrl2, Vroac | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (Vanilloid receptor- related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
PK2L2_MOUSE
|
||||||
| NC score | 0.116111 (rank : 39) | θ value | 0.00228821 (rank : 26) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L2_HUMAN
|
||||||
| NC score | 0.099600 (rank : 40) | θ value | 0.0431538 (rank : 31) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
TRPV2_MOUSE
|
||||||
| NC score | 0.097371 (rank : 41) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9WTR1, Q99K71 | Gene names | Trpv2, Grc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Growth factor-regulated calcium channel) (GRC). | |||||
|
TRPV2_HUMAN
|
||||||
| NC score | 0.076152 (rank : 42) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y5S1, Q9Y670 | Gene names | TRPV2, VRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 2 (TrpV2) (osm-9-like TRP channel 2) (OTRPC2) (Vanilloid receptor-like protein 1) (VRL-1). | |||||
|
PKD2_HUMAN
|
||||||
| NC score | 0.073989 (rank : 43) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
PKD2_MOUSE
|
||||||
| NC score | 0.068312 (rank : 44) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.044629 (rank : 45) | θ value | 0.0252991 (rank : 30) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.043550 (rank : 46) | θ value | 0.00869519 (rank : 27) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.036876 (rank : 47) | θ value | 0.163984 (rank : 35) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
LUZP1_MOUSE
|
||||||
| NC score | 0.036778 (rank : 48) | θ value | 0.0193708 (rank : 28) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
CCD47_HUMAN
|
||||||
| NC score | 0.032749 (rank : 49) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96A33, Q96D00, Q96JZ7, Q9H3E4, Q9NRG3 | Gene names | CCDC47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor. | |||||
|
CC47_MOUSE
|
||||||
| NC score | 0.029192 (rank : 50) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
LUZP1_HUMAN
|
||||||
| NC score | 0.028145 (rank : 51) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
RGS1_MOUSE
|
||||||
| NC score | 0.028088 (rank : 52) | θ value | 0.125558 (rank : 34) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JL25, Q3TBD1, Q3TC18, Q3TD56 | Gene names | Rgs1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1). | |||||
|
TRAK2_HUMAN
|
||||||
| NC score | 0.027667 (rank : 53) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60296, Q8WVH7, Q96NS2, Q9C0K5, Q9C0K6 | Gene names | TRAK2, ALS2CR3, KIAA0549 | |||
|
Domain Architecture |

|
|||||
| Description | Trafficking kinesin-binding protein 2 (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 3 protein). | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.026828 (rank : 54) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
LMAN1_MOUSE
|
||||||
| NC score | 0.026817 (rank : 55) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D0F3 | Gene names | Lman1, Ergic53 | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (p58). | |||||
|
LMNA_MOUSE
|
||||||
| NC score | 0.025629 (rank : 56) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P48678, P11516, P97859, Q3TIH0, Q3TTS8, Q3UCA0, Q3UCJ8, Q91WF2, Q9DC21 | Gene names | Lmna, Lmn1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-A/C. | |||||
|
UBXD2_HUMAN
|
||||||
| NC score | 0.025577 (rank : 57) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92575, Q8IYM5 | Gene names | UBXD2, KIAA0242, UBXDC1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
LMNA_HUMAN
|
||||||
| NC score | 0.025491 (rank : 58) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 653 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P02545, P02546, Q969I8, Q96JA2 | Gene names | LMNA, LMN1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-A/C (70 kDa lamin) (NY-REN-32 antigen). | |||||
|
BRD8_HUMAN
|
||||||
| NC score | 0.023460 (rank : 59) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
TXD11_HUMAN
|
||||||
| NC score | 0.022219 (rank : 60) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PKC3, O95887, Q6PJA6, Q8N2Q4, Q96K45, Q96K53 | Gene names | TXNDC11, EFP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 11 (EF-hand-binding protein 1). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.021867 (rank : 61) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
PROM1_MOUSE
|
||||||
| NC score | 0.020697 (rank : 62) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O54990, O35408 | Gene names | Prom1, Prom, Proml1 | |||
|
Domain Architecture |

|
|||||
| Description | Prominin-1 precursor (Prominin-like protein 1) (Antigen AC133 homolog). | |||||
|
UBXD2_MOUSE
|
||||||
| NC score | 0.020330 (rank : 63) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VCH8, Q8BW17 | Gene names | Ubxd2, Ubxdc1 | |||
|
Domain Architecture |

|
|||||
| Description | UBX domain-containing protein 2. | |||||
|
RBM25_HUMAN
|
||||||
| NC score | 0.020222 (rank : 64) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
SM1L2_HUMAN
|
||||||
| NC score | 0.019433 (rank : 65) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
LMAN1_HUMAN
|
||||||
| NC score | 0.019395 (rank : 66) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49257, Q12895, Q8N5I7, Q9UQG1, Q9UQG2, Q9UQG3, Q9UQG4, Q9UQG5, Q9UQG6, Q9UQG7, Q9UQG8, Q9UQG9, Q9UQH0, Q9UQH1, Q9UQH2 | Gene names | LMAN1, ERGIC53, F5F8D | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (Gp58) (Intracellular mannose- specific lectin MR60). | |||||
|
CNGA1_HUMAN
|
||||||
| NC score | 0.019098 (rank : 67) | θ value | 0.163984 (rank : 36) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
RAI14_HUMAN
|
||||||
| NC score | 0.019084 (rank : 68) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.018458 (rank : 69) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.018289 (rank : 70) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.018090 (rank : 71) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.017844 (rank : 72) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
TXD11_MOUSE
|
||||||
| NC score | 0.016981 (rank : 73) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K2W3, Q8BMR8, Q8VCK9 | Gene names | Txndc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 11. | |||||
|
CKAP4_HUMAN
|
||||||
| NC score | 0.016312 (rank : 74) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q07065, Q504S5, Q53ES6 | Gene names | CKAP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
MCLN2_HUMAN
|
||||||
| NC score | 0.016310 (rank : 75) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZK6, Q8N9R3 | Gene names | MCOLN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.016304 (rank : 76) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.014790 (rank : 77) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.014763 (rank : 78) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
CCDC6_HUMAN
|
||||||
| NC score | 0.014693 (rank : 79) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q16204, Q15250 | Gene names | CCDC6, D10S170 | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil domain-containing protein 6 (H4 protein) (Papillary thyroid carcinoma-encoded protein). | |||||
|
STAT1_HUMAN
|
||||||
| NC score | 0.014686 (rank : 80) | θ value | 0.365318 (rank : 37) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P42224 | Gene names | STAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 1-alpha/beta (Transcription factor ISGF-3 components p91/p84). | |||||
|
KIF3B_HUMAN
|
||||||
| NC score | 0.014283 (rank : 81) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 540 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O15066 | Gene names | KIF3B, KIAA0359 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3B (Microtubule plus end-directed kinesin motor 3B) (HH0048). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.013222 (rank : 82) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
KIF3B_MOUSE
|
||||||
| NC score | 0.013096 (rank : 83) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61771 | Gene names | Kif3b | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3B (Microtubule plus end-directed kinesin motor 3B). | |||||
|
PHF12_MOUSE
|
||||||
| NC score | 0.012915 (rank : 84) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.012868 (rank : 85) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.012823 (rank : 86) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
HOOK1_HUMAN
|
||||||
| NC score | 0.012234 (rank : 87) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.012118 (rank : 88) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
DIAP3_HUMAN
|
||||||
| NC score | 0.011665 (rank : 89) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NSV4, Q5T2S7, Q5XKF6, Q6MZF0, Q6NUP0, Q86VS4, Q8NAV4 | Gene names | DIAPH3, DIAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein diaphanous homolog 3 (Diaphanous-related formin-3) (DRF3). | |||||
|
DEMA_MOUSE
|
||||||
| NC score | 0.010956 (rank : 90) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WV69 | Gene names | Epb49, Epb4.9 | |||
|
Domain Architecture |

|
|||||
| Description | Dematin (Erythrocyte membrane protein band 4.9). | |||||
|
SENP1_MOUSE
|
||||||
| NC score | 0.010822 (rank : 91) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59110 | Gene names | Senp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 1 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP1). | |||||
|
CL011_MOUSE
|
||||||
| NC score | 0.010354 (rank : 92) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8QZV7, Q99JT8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C12orf11 homolog. | |||||
|
RCOR2_MOUSE
|
||||||
| NC score | 0.010116 (rank : 93) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C796, Q9JMK4 | Gene names | Rcor2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2 (M-CoREST). | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.009879 (rank : 94) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
LAMA2_MOUSE
|
||||||
| NC score | 0.009671 (rank : 95) | θ value | 0.62314 (rank : 39) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
LIPS_MOUSE
|
||||||
| NC score | 0.009212 (rank : 96) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54310, P97866, Q3TE34, Q6GU16 | Gene names | Lipe | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
AT1B1_HUMAN
|
||||||
| NC score | 0.009160 (rank : 97) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P05026, Q5TGZ3 | Gene names | ATP1B1, ATP1B | |||
|
Domain Architecture |
|
|||||
| Description | Sodium/potassium-transporting ATPase subunit beta-1 (Sodium/potassium- dependent ATPase beta-1 subunit). | |||||
|
UBP8_HUMAN
|
||||||
| NC score | 0.009159 (rank : 98) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P40818, Q7Z3U2, Q86VA0, Q8IWI7 | Gene names | USP8, KIAA0055, UBPY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 8 (EC 3.1.2.15) (Ubiquitin thioesterase 8) (Ubiquitin-specific-processing protease 8) (Deubiquitinating enzyme 8) (hUBPy). | |||||
|
RCOR2_HUMAN
|
||||||
| NC score | 0.008748 (rank : 99) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IZ40, Q96FP3 | Gene names | RCOR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2. | |||||
|
P73_HUMAN
|
||||||
| NC score | 0.008586 (rank : 100) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15350, O15351, Q5TBV5, Q5TBV6, Q8NHW9, Q8TDY5, Q8TDY6, Q9NTK8 | Gene names | TP73, P73 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein p73 (p53-like transcription factor) (p53-related protein). | |||||
|
DIAP2_MOUSE
|
||||||
| NC score | 0.008408 (rank : 101) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O70566, Q8C2G8 | Gene names | Diaph2, Diap2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2) (mDia3). | |||||
|
SH3G1_HUMAN
|
||||||
| NC score | 0.007916 (rank : 102) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99961, Q99668 | Gene names | SH3GL1, CNSA1, SH3D2B | |||
|
Domain Architecture |

|
|||||
| Description | SH3-containing GRB2-like protein 1 (Endophilin-2) (Endophilin-A2) (SH3 domain protein 2B) (Extra eleven-nineteen leukemia fusion gene) (EEN) (EEN fusion partner of MLL). | |||||
|
S4A11_HUMAN
|
||||||
| NC score | 0.007717 (rank : 103) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NBS3, Q9BXF4, Q9NTW9 | Gene names | SLC4A11, BTR1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium bicarbonate transporter-like protein 11 (Bicarbonate transporter-related protein 1) (Solute carrier family 4 member 11). | |||||
|
NOVA2_HUMAN
|
||||||
| NC score | 0.007160 (rank : 104) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UNW9, O43267, Q9UEA1 | Gene names | NOVA2, ANOVA, NOVA3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein Nova-2 (Neuro-oncological ventral antigen 2) (Astrocytic NOVA1-like RNA-binding protein). | |||||
|
CP7B1_MOUSE
|
||||||
| NC score | 0.006899 (rank : 105) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60991 | Gene names | Cyp7b1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 7B1 (EC 1.14.13.100) (25-hydroxycholesterol 7-alpha- hydroxylase) (Oxysterol 7-alpha-hydroxylase) (HCT-1). | |||||
|
SH3G1_MOUSE
|
||||||
| NC score | 0.006309 (rank : 106) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62419 | Gene names | Sh3gl1, Sh3d2b | |||
|
Domain Architecture |

|
|||||
| Description | SH3-containing GRB2-like protein 1 (Endophilin-2) (Endophilin-A2) (SH3 domain protein 2B) (SH3p8). | |||||
|
3BP1_MOUSE
|
||||||
| NC score | 0.003908 (rank : 107) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55194, Q99KK8 | Gene names | Sh3bp1, 3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
OR2H1_HUMAN
|
||||||
| NC score | -0.003080 (rank : 108) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9GZK4, O43629, O43661, O43662, Q9GZK9 | Gene names | OR2H1, OR2H6, OR2H8 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 2H1 (Hs6M1-16) (Olfactory receptor 6-2) (OR6-2) (OLFR42A-9004.14/9026.2). | |||||
|
ZN548_HUMAN
|
||||||
| NC score | -0.003664 (rank : 109) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 104 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NEK5, Q96M05 | Gene names | ZNF548 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 548. | |||||