Please be patient as the page loads
|
RGS1_MOUSE
|
||||||
| SwissProt Accessions | Q9JL25, Q3TBD1, Q3TC18, Q3TD56 | Gene names | Rgs1 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RGS1_MOUSE
|
||||||
| θ value | 1.0648e-109 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9JL25, Q3TBD1, Q3TC18, Q3TD56 | Gene names | Rgs1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1). | |||||
|
RGS1_HUMAN
|
||||||
| θ value | 9.6755e-95 (rank : 2) | NC score | 0.997874 (rank : 2) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q08116, Q07918, Q9H1W2 | Gene names | RGS1, 1R20, BL34 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1) (Early response protein 1R20) (B-cell activation protein BL34). | |||||
|
RGS18_HUMAN
|
||||||
| θ value | 2.26708e-35 (rank : 3) | NC score | 0.970640 (rank : 11) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NS28 | Gene names | RGS18, RGS13 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 18 (RGS18). | |||||
|
RGS18_MOUSE
|
||||||
| θ value | 1.12514e-34 (rank : 4) | NC score | 0.969580 (rank : 12) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99PG4 | Gene names | Rgs18 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 18 (RGS18). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 1.46948e-34 (rank : 5) | NC score | 0.619011 (rank : 40) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
RGS5_MOUSE
|
||||||
| θ value | 1.46948e-34 (rank : 6) | NC score | 0.977964 (rank : 3) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08850, Q543B1, Q9D0Z2 | Gene names | Rgs5 | |||
|
Domain Architecture |
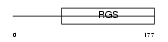
|
|||||
| Description | Regulator of G-protein signaling 5 (RGS5). | |||||
|
RGS8_HUMAN
|
||||||
| θ value | 7.29293e-34 (rank : 7) | NC score | 0.975895 (rank : 4) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P57771, Q3SYD2 | Gene names | RGS8 | |||
|
Domain Architecture |
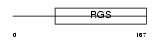
|
|||||
| Description | Regulator of G-protein signaling 8 (RGS8). | |||||
|
RGS8_MOUSE
|
||||||
| θ value | 7.29293e-34 (rank : 8) | NC score | 0.975638 (rank : 5) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BXT1 | Gene names | Rgs8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 8 (RGS8). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 1.24399e-33 (rank : 9) | NC score | 0.696225 (rank : 36) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
RGS4_HUMAN
|
||||||
| θ value | 2.77131e-33 (rank : 10) | NC score | 0.974273 (rank : 8) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P49798 | Gene names | RGS4 | |||
|
Domain Architecture |
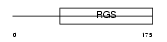
|
|||||
| Description | Regulator of G-protein signaling 4 (RGS4) (RGP4). | |||||
|
RGS5_HUMAN
|
||||||
| θ value | 1.0531e-32 (rank : 11) | NC score | 0.975062 (rank : 7) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O15539 | Gene names | RGS5 | |||
|
Domain Architecture |
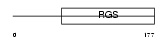
|
|||||
| Description | Regulator of G-protein signaling 5 (RGS5). | |||||
|
RGS4_MOUSE
|
||||||
| θ value | 2.34606e-32 (rank : 12) | NC score | 0.973141 (rank : 9) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08899, Q99L30 | Gene names | Rgs4 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 4 (RGS4). | |||||
|
RGS13_HUMAN
|
||||||
| θ value | 3.06405e-32 (rank : 13) | NC score | 0.975192 (rank : 6) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14921, Q6PGR2, Q8TD63, Q9BX45 | Gene names | RGS13 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 13 (RGS13). | |||||
|
RGS2_HUMAN
|
||||||
| θ value | 8.91499e-32 (rank : 14) | NC score | 0.967526 (rank : 13) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P41220 | Gene names | RGS2, G0S8 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 2 (RGS2) (G0/G1 switch regulatory protein 8). | |||||
|
RGS2_MOUSE
|
||||||
| θ value | 1.16434e-31 (rank : 15) | NC score | 0.966318 (rank : 14) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O08849, Q544S7, Q91WX1, Q9JL24 | Gene names | Rgs2 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 2 (RGS2). | |||||
|
RGS13_MOUSE
|
||||||
| θ value | 8.3442e-30 (rank : 16) | NC score | 0.971111 (rank : 10) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8K443 | Gene names | Rgs13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 13 (RGS13). | |||||
|
RGS16_HUMAN
|
||||||
| θ value | 1.2049e-28 (rank : 17) | NC score | 0.964660 (rank : 15) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O15492, Q99701 | Gene names | RGS16, RGSR | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 16 (RGS16) (Retinally abundant regulator of G-protein signaling) (RGS-R) (A28-RGS14P). | |||||
|
RGS16_MOUSE
|
||||||
| θ value | 3.87602e-27 (rank : 18) | NC score | 0.963902 (rank : 16) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P97428, O09091, P97420, Q80V16 | Gene names | Rgs16, Rgsr | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 16 (RGS16) (Retinally abundant regulator of G-protein signaling) (RGS-R) (A28-RGS14P). | |||||
|
RGS19_MOUSE
|
||||||
| θ value | 3.28125e-26 (rank : 19) | NC score | 0.954074 (rank : 17) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9CX84, Q99L50 | Gene names | Rgs19 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 19 (RGS19). | |||||
|
RGS20_HUMAN
|
||||||
| θ value | 1.62847e-25 (rank : 20) | NC score | 0.949198 (rank : 24) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O76081, Q96BG9 | Gene names | RGS20, RGSZ1, ZGAP1 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 20 (RGS20) (Regulator of Gz-selective protein signaling 1) (Gz-selective GTPase-activating protein) (G(z)GAP). | |||||
|
RGS17_HUMAN
|
||||||
| θ value | 2.12685e-25 (rank : 21) | NC score | 0.953134 (rank : 20) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UGC6, Q5TF49, Q8TD61, Q9UJS8 | Gene names | RGS17 | |||
|
Domain Architecture |
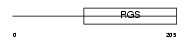
|
|||||
| Description | Regulator of G-protein signaling 17 (RGS17). | |||||
|
RGS19_HUMAN
|
||||||
| θ value | 2.77775e-25 (rank : 22) | NC score | 0.952798 (rank : 22) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P49795, Q53XN0, Q8TD60 | Gene names | RGS19, GAIP, GNAI3IP | |||
|
Domain Architecture |
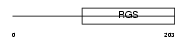
|
|||||
| Description | Regulator of G-protein signaling 19 (RGS19) (G-alpha-interacting protein) (GAIP protein). | |||||
|
RGS17_MOUSE
|
||||||
| θ value | 4.73814e-25 (rank : 23) | NC score | 0.953039 (rank : 21) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QZB0, Q543T9 | Gene names | Rgs17, Rgsz2 | |||
|
Domain Architecture |
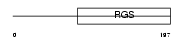
|
|||||
| Description | Regulator of G-protein signaling 17 (RGS17) (Regulator of Gz-selective protein signaling 2). | |||||
|
RGS20_MOUSE
|
||||||
| θ value | 1.05554e-24 (rank : 24) | NC score | 0.953884 (rank : 18) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QZB1, Q3TY63, Q9CUV8, Q9QZB2 | Gene names | Rgs20, Rgsz1 | |||
|
Domain Architecture |
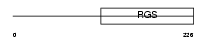
|
|||||
| Description | Regulator of G-protein signaling 20 (RGS20) (Regulator of G-protein signaling Z1). | |||||
|
RGS10_MOUSE
|
||||||
| θ value | 2.59989e-23 (rank : 25) | NC score | 0.953176 (rank : 19) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9CQE5, Q9D3L2 | Gene names | Rgs10 | |||
|
Domain Architecture |
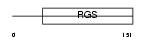
|
|||||
| Description | Regulator of G-protein signaling 10 (RGS10). | |||||
|
RGS6_MOUSE
|
||||||
| θ value | 5.79196e-23 (rank : 26) | NC score | 0.851559 (rank : 32) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Z2H2, Q8BGI8 | Gene names | Rgs6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 6 (RGS6). | |||||
|
RGS9_HUMAN
|
||||||
| θ value | 2.87452e-22 (rank : 27) | NC score | 0.853739 (rank : 30) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O75916, O75573, Q696R2, Q8TD64, Q8TD65, Q9HC32, Q9HC33 | Gene names | RGS9 | |||
|
Domain Architecture |
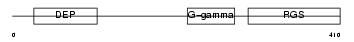
|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
RGS10_HUMAN
|
||||||
| θ value | 4.9032e-22 (rank : 28) | NC score | 0.950980 (rank : 23) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O43665 | Gene names | RGS10 | |||
|
Domain Architecture |
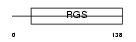
|
|||||
| Description | Regulator of G-protein signaling 10 (RGS10). | |||||
|
RGS9_MOUSE
|
||||||
| θ value | 8.36355e-22 (rank : 29) | NC score | 0.853077 (rank : 31) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O54828, Q9Z0S0 | Gene names | Rgs9 | |||
|
Domain Architecture |
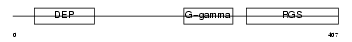
|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
RGS7_HUMAN
|
||||||
| θ value | 1.86321e-21 (rank : 30) | NC score | 0.857639 (rank : 28) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P49802, Q8TD66, Q8TD67, Q9UNU7, Q9Y6B9 | Gene names | RGS7 | |||
|
Domain Architecture |
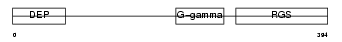
|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RGS7_MOUSE
|
||||||
| θ value | 5.42112e-21 (rank : 31) | NC score | 0.854807 (rank : 29) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O54829 | Gene names | Rgs7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RGS6_HUMAN
|
||||||
| θ value | 5.99374e-20 (rank : 32) | NC score | 0.827493 (rank : 33) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P49758, O75576, O75577, Q8TE13, Q8TE14, Q8TE15, Q8TE16, Q8TE17, Q8TE18, Q8TE19, Q8TE20, Q8TE21, Q8TE22, Q9Y245, Q9Y647 | Gene names | RGS6 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 6 (RGS6) (S914). | |||||
|
RGS11_HUMAN
|
||||||
| θ value | 2.5182e-18 (rank : 33) | NC score | 0.866421 (rank : 26) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O94810, O75883 | Gene names | RGS11 | |||
|
Domain Architecture |
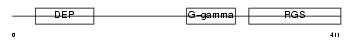
|
|||||
| Description | Regulator of G-protein signaling 11 (RGS11). | |||||
|
RGS14_MOUSE
|
||||||
| θ value | 3.28887e-18 (rank : 34) | NC score | 0.871004 (rank : 25) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P97492, Q9DCD1 | Gene names | Rgs14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14) (RAP1/RAP2-interacting protein). | |||||
|
RGS12_HUMAN
|
||||||
| θ value | 3.63628e-17 (rank : 35) | NC score | 0.771135 (rank : 34) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
RGS14_HUMAN
|
||||||
| θ value | 4.74913e-17 (rank : 36) | NC score | 0.865943 (rank : 27) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O43566, O43565, Q8TD62 | Gene names | RGS14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14). | |||||
|
AXN1_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 37) | NC score | 0.697665 (rank : 35) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35625 | Gene names | Axin1, Axin, Fu | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (Fused protein). | |||||
|
AXN1_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 38) | NC score | 0.684743 (rank : 37) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15169, Q4TT26, Q4TT27, Q86YA7, Q8WVW6, Q96S28 | Gene names | AXIN1, AXIN | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (hAxin). | |||||
|
AXN2_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 39) | NC score | 0.667241 (rank : 38) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y2T1, Q9H3M6, Q9UH84 | Gene names | AXIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
AXN2_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 40) | NC score | 0.656473 (rank : 39) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O88566, Q9QXJ6 | Gene names | Axin2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
SNX13_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 41) | NC score | 0.347097 (rank : 44) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y5W8, O94821, Q8WVZ2, Q8WXH8 | Gene names | SNX13, KIAA0713 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-13 (RGS domain- and PHOX domain-containing protein) (RGS-PX1). | |||||
|
SNX13_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 42) | NC score | 0.350693 (rank : 41) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6PHS6 | Gene names | Snx13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-13. | |||||
|
AKA10_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 43) | NC score | 0.349794 (rank : 42) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O43572, Q96AJ7 | Gene names | AKAP10 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
AKA10_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 44) | NC score | 0.348571 (rank : 43) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O88845, Q5SUB5, Q5SUB7, Q7TPE7, Q8BQL6 | Gene names | Akap10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
TRPM7_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 45) | NC score | 0.029209 (rank : 50) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
TRPM7_HUMAN
|
||||||
| θ value | 0.125558 (rank : 46) | NC score | 0.028088 (rank : 51) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
SNX14_HUMAN
|
||||||
| θ value | 0.279714 (rank : 47) | NC score | 0.099521 (rank : 47) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5W7, Q9BSD1 | Gene names | SNX14 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-14. | |||||
|
SNX14_MOUSE
|
||||||
| θ value | 0.279714 (rank : 48) | NC score | 0.105534 (rank : 46) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BHY8 | Gene names | Snx14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-14. | |||||
|
TRIP4_MOUSE
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.027264 (rank : 52) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QXN3, Q8CAD5 | Gene names | Trip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 (ASC-1) (Thyroid receptor-interacting protein 4) (Trip-4). | |||||
|
RADI_MOUSE
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.002916 (rank : 60) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26043, Q9QW27 | Gene names | Rdx | |||
|
Domain Architecture |

|
|||||
| Description | Radixin (ESP10). | |||||
|
RK_MOUSE
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | -0.000410 (rank : 63) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WVL4 | Gene names | Grk1, Rhok | |||
|
Domain Architecture |

|
|||||
| Description | Rhodopsin kinase precursor (EC 2.7.11.14) (RK) (G protein-coupled receptor kinase 1). | |||||
|
DYH5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.008665 (rank : 54) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
RK_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.000196 (rank : 62) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15835 | Gene names | GRK1, RHOK | |||
|
Domain Architecture |

|
|||||
| Description | Rhodopsin kinase precursor (EC 2.7.11.14) (RK) (G protein-coupled receptor kinase 1). | |||||
|
ARHGB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.025042 (rank : 53) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
RAD18_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.006553 (rank : 56) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QXK2, Q9CZB8 | Gene names | Rad18, Rad18sc | |||
|
Domain Architecture |

|
|||||
| Description | Postreplication repair protein RAD18 (mRAD18Sc). | |||||
|
RHG05_MOUSE
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.002937 (rank : 59) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
4ET_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.007592 (rank : 55) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EST3, Q9CSS3 | Gene names | Eif4enif1, Clast4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
NBEA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.004529 (rank : 57) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPN1, Q8C931, Q9EPM9, Q9EPN0, Q9WVM9 | Gene names | Nbea, Lyst2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein neurobeachin (Lysosomal-trafficking regulator 2). | |||||
|
RADI_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.001498 (rank : 61) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
SIAT9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.003209 (rank : 58) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UNP4, O94902, Q53QU1 | Gene names | ST3GAL5, SIAT9 | |||
|
Domain Architecture |
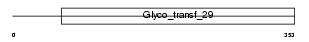
|
|||||
| Description | Lactosylceramide alpha-2,3-sialyltransferase (EC 2.4.99.9) (CMP- NeuAc:lactosylceramide alpha-2,3-sialyltransferase) (Ganglioside GM3 synthase) (ST3Gal V) (Sialyltransferase 9). | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.053986 (rank : 48) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
PS1C2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.051916 (rank : 49) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UIG4 | Gene names | PSORS1C2, C6orf17, SPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein precursor (SPR1 protein). | |||||
|
SNX25_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.163229 (rank : 45) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H3E2 | Gene names | SNX25 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-25. | |||||
|
RGS1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.0648e-109 (rank : 1) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9JL25, Q3TBD1, Q3TC18, Q3TD56 | Gene names | Rgs1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1). | |||||
|
RGS1_HUMAN
|
||||||
| NC score | 0.997874 (rank : 2) | θ value | 9.6755e-95 (rank : 2) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q08116, Q07918, Q9H1W2 | Gene names | RGS1, 1R20, BL34 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1) (Early response protein 1R20) (B-cell activation protein BL34). | |||||
|
RGS5_MOUSE
|
||||||
| NC score | 0.977964 (rank : 3) | θ value | 1.46948e-34 (rank : 6) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08850, Q543B1, Q9D0Z2 | Gene names | Rgs5 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 5 (RGS5). | |||||
|
RGS8_HUMAN
|
||||||
| NC score | 0.975895 (rank : 4) | θ value | 7.29293e-34 (rank : 7) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P57771, Q3SYD2 | Gene names | RGS8 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 8 (RGS8). | |||||
|
RGS8_MOUSE
|
||||||
| NC score | 0.975638 (rank : 5) | θ value | 7.29293e-34 (rank : 8) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BXT1 | Gene names | Rgs8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 8 (RGS8). | |||||
|
RGS13_HUMAN
|
||||||
| NC score | 0.975192 (rank : 6) | θ value | 3.06405e-32 (rank : 13) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14921, Q6PGR2, Q8TD63, Q9BX45 | Gene names | RGS13 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 13 (RGS13). | |||||
|
RGS5_HUMAN
|
||||||
| NC score | 0.975062 (rank : 7) | θ value | 1.0531e-32 (rank : 11) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O15539 | Gene names | RGS5 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 5 (RGS5). | |||||
|
RGS4_HUMAN
|
||||||
| NC score | 0.974273 (rank : 8) | θ value | 2.77131e-33 (rank : 10) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P49798 | Gene names | RGS4 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 4 (RGS4) (RGP4). | |||||
|
RGS4_MOUSE
|
||||||
| NC score | 0.973141 (rank : 9) | θ value | 2.34606e-32 (rank : 12) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08899, Q99L30 | Gene names | Rgs4 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 4 (RGS4). | |||||
|
RGS13_MOUSE
|
||||||
| NC score | 0.971111 (rank : 10) | θ value | 8.3442e-30 (rank : 16) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8K443 | Gene names | Rgs13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 13 (RGS13). | |||||
|
RGS18_HUMAN
|
||||||
| NC score | 0.970640 (rank : 11) | θ value | 2.26708e-35 (rank : 3) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NS28 | Gene names | RGS18, RGS13 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 18 (RGS18). | |||||
|
RGS18_MOUSE
|
||||||
| NC score | 0.969580 (rank : 12) | θ value | 1.12514e-34 (rank : 4) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q99PG4 | Gene names | Rgs18 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 18 (RGS18). | |||||
|
RGS2_HUMAN
|
||||||
| NC score | 0.967526 (rank : 13) | θ value | 8.91499e-32 (rank : 14) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P41220 | Gene names | RGS2, G0S8 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 2 (RGS2) (G0/G1 switch regulatory protein 8). | |||||
|
RGS2_MOUSE
|
||||||
| NC score | 0.966318 (rank : 14) | θ value | 1.16434e-31 (rank : 15) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O08849, Q544S7, Q91WX1, Q9JL24 | Gene names | Rgs2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 2 (RGS2). | |||||
|
RGS16_HUMAN
|
||||||
| NC score | 0.964660 (rank : 15) | θ value | 1.2049e-28 (rank : 17) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O15492, Q99701 | Gene names | RGS16, RGSR | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 16 (RGS16) (Retinally abundant regulator of G-protein signaling) (RGS-R) (A28-RGS14P). | |||||
|
RGS16_MOUSE
|
||||||
| NC score | 0.963902 (rank : 16) | θ value | 3.87602e-27 (rank : 18) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P97428, O09091, P97420, Q80V16 | Gene names | Rgs16, Rgsr | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 16 (RGS16) (Retinally abundant regulator of G-protein signaling) (RGS-R) (A28-RGS14P). | |||||
|
RGS19_MOUSE
|
||||||
| NC score | 0.954074 (rank : 17) | θ value | 3.28125e-26 (rank : 19) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9CX84, Q99L50 | Gene names | Rgs19 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 19 (RGS19). | |||||
|
RGS20_MOUSE
|
||||||
| NC score | 0.953884 (rank : 18) | θ value | 1.05554e-24 (rank : 24) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QZB1, Q3TY63, Q9CUV8, Q9QZB2 | Gene names | Rgs20, Rgsz1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 20 (RGS20) (Regulator of G-protein signaling Z1). | |||||
|
RGS10_MOUSE
|
||||||
| NC score | 0.953176 (rank : 19) | θ value | 2.59989e-23 (rank : 25) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9CQE5, Q9D3L2 | Gene names | Rgs10 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 10 (RGS10). | |||||
|
RGS17_HUMAN
|
||||||
| NC score | 0.953134 (rank : 20) | θ value | 2.12685e-25 (rank : 21) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UGC6, Q5TF49, Q8TD61, Q9UJS8 | Gene names | RGS17 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 17 (RGS17). | |||||
|
RGS17_MOUSE
|
||||||
| NC score | 0.953039 (rank : 21) | θ value | 4.73814e-25 (rank : 23) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QZB0, Q543T9 | Gene names | Rgs17, Rgsz2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 17 (RGS17) (Regulator of Gz-selective protein signaling 2). | |||||
|
RGS19_HUMAN
|
||||||
| NC score | 0.952798 (rank : 22) | θ value | 2.77775e-25 (rank : 22) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P49795, Q53XN0, Q8TD60 | Gene names | RGS19, GAIP, GNAI3IP | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 19 (RGS19) (G-alpha-interacting protein) (GAIP protein). | |||||
|
RGS10_HUMAN
|
||||||
| NC score | 0.950980 (rank : 23) | θ value | 4.9032e-22 (rank : 28) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O43665 | Gene names | RGS10 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 10 (RGS10). | |||||
|
RGS20_HUMAN
|
||||||
| NC score | 0.949198 (rank : 24) | θ value | 1.62847e-25 (rank : 20) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O76081, Q96BG9 | Gene names | RGS20, RGSZ1, ZGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 20 (RGS20) (Regulator of Gz-selective protein signaling 1) (Gz-selective GTPase-activating protein) (G(z)GAP). | |||||
|
RGS14_MOUSE
|
||||||
| NC score | 0.871004 (rank : 25) | θ value | 3.28887e-18 (rank : 34) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P97492, Q9DCD1 | Gene names | Rgs14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14) (RAP1/RAP2-interacting protein). | |||||
|
RGS11_HUMAN
|
||||||
| NC score | 0.866421 (rank : 26) | θ value | 2.5182e-18 (rank : 33) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O94810, O75883 | Gene names | RGS11 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 11 (RGS11). | |||||
|
RGS14_HUMAN
|
||||||
| NC score | 0.865943 (rank : 27) | θ value | 4.74913e-17 (rank : 36) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O43566, O43565, Q8TD62 | Gene names | RGS14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14). | |||||
|
RGS7_HUMAN
|
||||||
| NC score | 0.857639 (rank : 28) | θ value | 1.86321e-21 (rank : 30) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P49802, Q8TD66, Q8TD67, Q9UNU7, Q9Y6B9 | Gene names | RGS7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RGS7_MOUSE
|
||||||
| NC score | 0.854807 (rank : 29) | θ value | 5.42112e-21 (rank : 31) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O54829 | Gene names | Rgs7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RGS9_HUMAN
|
||||||
| NC score | 0.853739 (rank : 30) | θ value | 2.87452e-22 (rank : 27) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O75916, O75573, Q696R2, Q8TD64, Q8TD65, Q9HC32, Q9HC33 | Gene names | RGS9 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
RGS9_MOUSE
|
||||||
| NC score | 0.853077 (rank : 31) | θ value | 8.36355e-22 (rank : 29) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O54828, Q9Z0S0 | Gene names | Rgs9 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
RGS6_MOUSE
|
||||||
| NC score | 0.851559 (rank : 32) | θ value | 5.79196e-23 (rank : 26) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Z2H2, Q8BGI8 | Gene names | Rgs6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 6 (RGS6). | |||||
|
RGS6_HUMAN
|
||||||
| NC score | 0.827493 (rank : 33) | θ value | 5.99374e-20 (rank : 32) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P49758, O75576, O75577, Q8TE13, Q8TE14, Q8TE15, Q8TE16, Q8TE17, Q8TE18, Q8TE19, Q8TE20, Q8TE21, Q8TE22, Q9Y245, Q9Y647 | Gene names | RGS6 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 6 (RGS6) (S914). | |||||
|
RGS12_HUMAN
|
||||||
| NC score | 0.771135 (rank : 34) | θ value | 3.63628e-17 (rank : 35) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
AXN1_MOUSE
|
||||||
| NC score | 0.697665 (rank : 35) | θ value | 3.52202e-12 (rank : 37) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O35625 | Gene names | Axin1, Axin, Fu | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (Fused protein). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.696225 (rank : 36) | θ value | 1.24399e-33 (rank : 9) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
AXN1_HUMAN
|
||||||
| NC score | 0.684743 (rank : 37) | θ value | 2.28291e-11 (rank : 38) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15169, Q4TT26, Q4TT27, Q86YA7, Q8WVW6, Q96S28 | Gene names | AXIN1, AXIN | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (hAxin). | |||||
|
AXN2_HUMAN
|
||||||
| NC score | 0.667241 (rank : 38) | θ value | 3.89403e-11 (rank : 39) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y2T1, Q9H3M6, Q9UH84 | Gene names | AXIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
AXN2_MOUSE
|
||||||
| NC score | 0.656473 (rank : 39) | θ value | 1.47974e-10 (rank : 40) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O88566, Q9QXJ6 | Gene names | Axin2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.619011 (rank : 40) | θ value | 1.46948e-34 (rank : 5) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SNX13_MOUSE
|
||||||
| NC score | 0.350693 (rank : 41) | θ value | 2.88788e-06 (rank : 42) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6PHS6 | Gene names | Snx13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-13. | |||||
|
AKA10_HUMAN
|
||||||
| NC score | 0.349794 (rank : 42) | θ value | 0.00020696 (rank : 43) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O43572, Q96AJ7 | Gene names | AKAP10 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
AKA10_MOUSE
|
||||||
| NC score | 0.348571 (rank : 43) | θ value | 0.00020696 (rank : 44) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O88845, Q5SUB5, Q5SUB7, Q7TPE7, Q8BQL6 | Gene names | Akap10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
SNX13_HUMAN
|
||||||
| NC score | 0.347097 (rank : 44) | θ value | 2.88788e-06 (rank : 41) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y5W8, O94821, Q8WVZ2, Q8WXH8 | Gene names | SNX13, KIAA0713 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-13 (RGS domain- and PHOX domain-containing protein) (RGS-PX1). | |||||
|
SNX25_HUMAN
|
||||||
| NC score | 0.163229 (rank : 45) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H3E2 | Gene names | SNX25 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-25. | |||||
|
SNX14_MOUSE
|
||||||
| NC score | 0.105534 (rank : 46) | θ value | 0.279714 (rank : 48) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BHY8 | Gene names | Snx14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-14. | |||||
|
SNX14_HUMAN
|
||||||
| NC score | 0.099521 (rank : 47) | θ value | 0.279714 (rank : 47) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5W7, Q9BSD1 | Gene names | SNX14 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-14. | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.053986 (rank : 48) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
PS1C2_HUMAN
|
||||||
| NC score | 0.051916 (rank : 49) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UIG4 | Gene names | PSORS1C2, C6orf17, SPR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein precursor (SPR1 protein). | |||||
|
TRPM7_MOUSE
|
||||||
| NC score | 0.029209 (rank : 50) | θ value | 0.0563607 (rank : 45) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q923J1, Q8C7S7, Q8CE54, Q921Y5, Q925B2, Q9CUT2, Q9JLQ1 | Gene names | Trpm7, Chak, Ltrpc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1) (Transient receptor potential-phospholipase C- interacting kinase) (TRP-PLIK). | |||||
|
TRPM7_HUMAN
|
||||||
| NC score | 0.028088 (rank : 51) | θ value | 0.125558 (rank : 46) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96QT4, Q6ZMF5, Q86VJ4, Q8NBW2, Q9BXB2, Q9NXQ2 | Gene names | TRPM7, CHAK1, LTRPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily M member 7 (EC 2.7.11.1) (Long transient receptor potential channel 7) (LTrpC7) (Channel-kinase 1). | |||||
|
TRIP4_MOUSE
|
||||||
| NC score | 0.027264 (rank : 52) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QXN3, Q8CAD5 | Gene names | Trip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 (ASC-1) (Thyroid receptor-interacting protein 4) (Trip-4). | |||||
|
ARHGB_HUMAN
|
||||||
| NC score | 0.025042 (rank : 53) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
DYH5_HUMAN
|
||||||
| NC score | 0.008665 (rank : 54) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
4ET_MOUSE
|
||||||
| NC score | 0.007592 (rank : 55) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EST3, Q9CSS3 | Gene names | Eif4enif1, Clast4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
RAD18_MOUSE
|
||||||
| NC score | 0.006553 (rank : 56) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QXK2, Q9CZB8 | Gene names | Rad18, Rad18sc | |||
|
Domain Architecture |

|
|||||
| Description | Postreplication repair protein RAD18 (mRAD18Sc). | |||||
|
NBEA_MOUSE
|
||||||
| NC score | 0.004529 (rank : 57) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPN1, Q8C931, Q9EPM9, Q9EPN0, Q9WVM9 | Gene names | Nbea, Lyst2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein neurobeachin (Lysosomal-trafficking regulator 2). | |||||
|
SIAT9_HUMAN
|
||||||
| NC score | 0.003209 (rank : 58) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UNP4, O94902, Q53QU1 | Gene names | ST3GAL5, SIAT9 | |||
|
Domain Architecture |
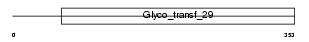
|
|||||
| Description | Lactosylceramide alpha-2,3-sialyltransferase (EC 2.4.99.9) (CMP- NeuAc:lactosylceramide alpha-2,3-sialyltransferase) (Ganglioside GM3 synthase) (ST3Gal V) (Sialyltransferase 9). | |||||
|
RHG05_MOUSE
|
||||||
| NC score | 0.002937 (rank : 59) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97393 | Gene names | Arhgap5, Rhogap5 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 5 (p190-B). | |||||
|
RADI_MOUSE
|
||||||
| NC score | 0.002916 (rank : 60) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26043, Q9QW27 | Gene names | Rdx | |||
|
Domain Architecture |

|
|||||
| Description | Radixin (ESP10). | |||||
|
RADI_HUMAN
|
||||||
| NC score | 0.001498 (rank : 61) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 741 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35241, Q86Y61 | Gene names | RDX | |||
|
Domain Architecture |

|
|||||
| Description | Radixin. | |||||
|
RK_HUMAN
|
||||||
| NC score | 0.000196 (rank : 62) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15835 | Gene names | GRK1, RHOK | |||
|
Domain Architecture |

|
|||||
| Description | Rhodopsin kinase precursor (EC 2.7.11.14) (RK) (G protein-coupled receptor kinase 1). | |||||
|
RK_MOUSE
|
||||||
| NC score | -0.000410 (rank : 63) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WVL4 | Gene names | Grk1, Rhok | |||
|
Domain Architecture |

|
|||||
| Description | Rhodopsin kinase precursor (EC 2.7.11.14) (RK) (G protein-coupled receptor kinase 1). | |||||