Please be patient as the page loads
|
RGS9_HUMAN
|
||||||
| SwissProt Accessions | O75916, O75573, Q696R2, Q8TD64, Q8TD65, Q9HC32, Q9HC33 | Gene names | RGS9 | |||
|
Domain Architecture |
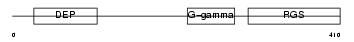
|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RGS9_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O75916, O75573, Q696R2, Q8TD64, Q8TD65, Q9HC32, Q9HC33 | Gene names | RGS9 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
RGS9_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998780 (rank : 2) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O54828, Q9Z0S0 | Gene names | Rgs9 | |||
|
Domain Architecture |
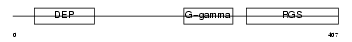
|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
RGS11_HUMAN
|
||||||
| θ value | 3.28708e-135 (rank : 3) | NC score | 0.981321 (rank : 3) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O94810, O75883 | Gene names | RGS11 | |||
|
Domain Architecture |
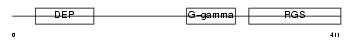
|
|||||
| Description | Regulator of G-protein signaling 11 (RGS11). | |||||
|
RGS7_MOUSE
|
||||||
| θ value | 8.77142e-88 (rank : 4) | NC score | 0.956928 (rank : 5) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O54829 | Gene names | Rgs7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RGS7_HUMAN
|
||||||
| θ value | 2.5521e-87 (rank : 5) | NC score | 0.956949 (rank : 4) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P49802, Q8TD66, Q8TD67, Q9UNU7, Q9Y6B9 | Gene names | RGS7 | |||
|
Domain Architecture |
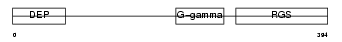
|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RGS6_MOUSE
|
||||||
| θ value | 3.11973e-85 (rank : 6) | NC score | 0.955169 (rank : 6) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Z2H2, Q8BGI8 | Gene names | Rgs6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 6 (RGS6). | |||||
|
RGS6_HUMAN
|
||||||
| θ value | 7.19216e-82 (rank : 7) | NC score | 0.947428 (rank : 7) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P49758, O75576, O75577, Q8TE13, Q8TE14, Q8TE15, Q8TE16, Q8TE17, Q8TE18, Q8TE19, Q8TE20, Q8TE21, Q8TE22, Q9Y245, Q9Y647 | Gene names | RGS6 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 6 (RGS6) (S914). | |||||
|
RGS1_HUMAN
|
||||||
| θ value | 5.79196e-23 (rank : 8) | NC score | 0.854982 (rank : 10) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q08116, Q07918, Q9H1W2 | Gene names | RGS1, 1R20, BL34 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1) (Early response protein 1R20) (B-cell activation protein BL34). | |||||
|
RGS10_MOUSE
|
||||||
| θ value | 2.87452e-22 (rank : 9) | NC score | 0.861267 (rank : 8) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9CQE5, Q9D3L2 | Gene names | Rgs10 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 10 (RGS10). | |||||
|
RGS1_MOUSE
|
||||||
| θ value | 2.87452e-22 (rank : 10) | NC score | 0.853739 (rank : 11) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9JL25, Q3TBD1, Q3TC18, Q3TD56 | Gene names | Rgs1 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1). | |||||
|
RGS20_MOUSE
|
||||||
| θ value | 6.40375e-22 (rank : 11) | NC score | 0.851958 (rank : 13) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QZB1, Q3TY63, Q9CUV8, Q9QZB2 | Gene names | Rgs20, Rgsz1 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 20 (RGS20) (Regulator of G-protein signaling Z1). | |||||
|
RGS20_HUMAN
|
||||||
| θ value | 8.36355e-22 (rank : 12) | NC score | 0.851608 (rank : 14) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O76081, Q96BG9 | Gene names | RGS20, RGSZ1, ZGAP1 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 20 (RGS20) (Regulator of Gz-selective protein signaling 1) (Gz-selective GTPase-activating protein) (G(z)GAP). | |||||
|
RGS10_HUMAN
|
||||||
| θ value | 2.43343e-21 (rank : 13) | NC score | 0.860613 (rank : 9) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O43665 | Gene names | RGS10 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 10 (RGS10). | |||||
|
RGS5_MOUSE
|
||||||
| θ value | 7.0802e-21 (rank : 14) | NC score | 0.852493 (rank : 12) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O08850, Q543B1, Q9D0Z2 | Gene names | Rgs5 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 5 (RGS5). | |||||
|
RGS18_HUMAN
|
||||||
| θ value | 2.69047e-20 (rank : 15) | NC score | 0.845992 (rank : 20) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NS28 | Gene names | RGS18, RGS13 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 18 (RGS18). | |||||
|
RGS17_HUMAN
|
||||||
| θ value | 3.51386e-20 (rank : 16) | NC score | 0.850040 (rank : 17) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UGC6, Q5TF49, Q8TD61, Q9UJS8 | Gene names | RGS17 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 17 (RGS17). | |||||
|
RGS5_HUMAN
|
||||||
| θ value | 4.58923e-20 (rank : 17) | NC score | 0.846356 (rank : 19) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15539 | Gene names | RGS5 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 5 (RGS5). | |||||
|
RGS19_MOUSE
|
||||||
| θ value | 5.99374e-20 (rank : 18) | NC score | 0.850285 (rank : 16) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9CX84, Q99L50 | Gene names | Rgs19 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 19 (RGS19). | |||||
|
RGS17_MOUSE
|
||||||
| θ value | 7.82807e-20 (rank : 19) | NC score | 0.848826 (rank : 18) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QZB0, Q543T9 | Gene names | Rgs17, Rgsz2 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 17 (RGS17) (Regulator of Gz-selective protein signaling 2). | |||||
|
RGS19_HUMAN
|
||||||
| θ value | 7.82807e-20 (rank : 20) | NC score | 0.851113 (rank : 15) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P49795, Q53XN0, Q8TD60 | Gene names | RGS19, GAIP, GNAI3IP | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 19 (RGS19) (G-alpha-interacting protein) (GAIP protein). | |||||
|
RGS12_HUMAN
|
||||||
| θ value | 1.02238e-19 (rank : 21) | NC score | 0.722612 (rank : 34) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 1.33526e-19 (rank : 22) | NC score | 0.531903 (rank : 38) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
RGS8_HUMAN
|
||||||
| θ value | 1.33526e-19 (rank : 23) | NC score | 0.842099 (rank : 21) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P57771, Q3SYD2 | Gene names | RGS8 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 8 (RGS8). | |||||
|
RGS8_MOUSE
|
||||||
| θ value | 2.97466e-19 (rank : 24) | NC score | 0.841049 (rank : 22) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BXT1 | Gene names | Rgs8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 8 (RGS8). | |||||
|
RGS14_MOUSE
|
||||||
| θ value | 3.88503e-19 (rank : 25) | NC score | 0.808692 (rank : 32) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97492, Q9DCD1 | Gene names | Rgs14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14) (RAP1/RAP2-interacting protein). | |||||
|
RGS14_HUMAN
|
||||||
| θ value | 5.07402e-19 (rank : 26) | NC score | 0.804666 (rank : 33) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O43566, O43565, Q8TD62 | Gene names | RGS14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14). | |||||
|
RGS16_HUMAN
|
||||||
| θ value | 6.62687e-19 (rank : 27) | NC score | 0.839190 (rank : 25) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15492, Q99701 | Gene names | RGS16, RGSR | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 16 (RGS16) (Retinally abundant regulator of G-protein signaling) (RGS-R) (A28-RGS14P). | |||||
|
RGS18_MOUSE
|
||||||
| θ value | 6.62687e-19 (rank : 28) | NC score | 0.840165 (rank : 23) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99PG4 | Gene names | Rgs18 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 18 (RGS18). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 1.13037e-18 (rank : 29) | NC score | 0.594965 (rank : 35) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
RGS2_MOUSE
|
||||||
| θ value | 2.5182e-18 (rank : 30) | NC score | 0.832887 (rank : 31) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O08849, Q544S7, Q91WX1, Q9JL24 | Gene names | Rgs2 | |||
|
Domain Architecture |
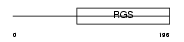
|
|||||
| Description | Regulator of G-protein signaling 2 (RGS2). | |||||
|
RGS4_HUMAN
|
||||||
| θ value | 2.5182e-18 (rank : 31) | NC score | 0.835039 (rank : 28) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P49798 | Gene names | RGS4 | |||
|
Domain Architecture |
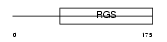
|
|||||
| Description | Regulator of G-protein signaling 4 (RGS4) (RGP4). | |||||
|
RGS2_HUMAN
|
||||||
| θ value | 7.32683e-18 (rank : 32) | NC score | 0.833880 (rank : 29) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P41220 | Gene names | RGS2, G0S8 | |||
|
Domain Architecture |
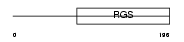
|
|||||
| Description | Regulator of G-protein signaling 2 (RGS2) (G0/G1 switch regulatory protein 8). | |||||
|
RGS4_MOUSE
|
||||||
| θ value | 7.32683e-18 (rank : 33) | NC score | 0.836952 (rank : 27) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O08899, Q99L30 | Gene names | Rgs4 | |||
|
Domain Architecture |
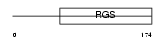
|
|||||
| Description | Regulator of G-protein signaling 4 (RGS4). | |||||
|
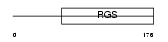
RGS16_MOUSE
|
||||||
| θ value | 1.63225e-17 (rank : 34) | NC score | 0.833130 (rank : 30) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97428, O09091, P97420, Q80V16 | Gene names | Rgs16, Rgsr | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 16 (RGS16) (Retinally abundant regulator of G-protein signaling) (RGS-R) (A28-RGS14P). | |||||
|
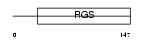
RGS13_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 35) | NC score | 0.839549 (rank : 24) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O14921, Q6PGR2, Q8TD63, Q9BX45 | Gene names | RGS13 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 13 (RGS13). | |||||
|
RGS13_MOUSE
|
||||||
| θ value | 6.85773e-16 (rank : 36) | NC score | 0.838113 (rank : 26) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8K443 | Gene names | Rgs13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 13 (RGS13). | |||||
|
AXN2_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 37) | NC score | 0.536527 (rank : 37) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y2T1, Q9H3M6, Q9UH84 | Gene names | AXIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
AXN1_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 38) | NC score | 0.545452 (rank : 36) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O35625 | Gene names | Axin1, Axin, Fu | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (Fused protein). | |||||
|
AXN2_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 39) | NC score | 0.530641 (rank : 39) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O88566, Q9QXJ6 | Gene names | Axin2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
AXN1_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 40) | NC score | 0.529709 (rank : 40) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O15169, Q4TT26, Q4TT27, Q86YA7, Q8WVW6, Q96S28 | Gene names | AXIN1, AXIN | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (hAxin). | |||||
|
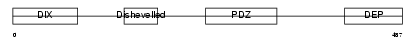
DVL3_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 41) | NC score | 0.123410 (rank : 47) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q92997, O14642, Q13531, Q8N5E9, Q92607 | Gene names | DVL3, KIAA0208 | |||
|
Domain Architecture |
|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
DVL3_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 42) | NC score | 0.123494 (rank : 46) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61062 | Gene names | Dvl3 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
DVL1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 43) | NC score | 0.097616 (rank : 51) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14640 | Gene names | DVL1 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
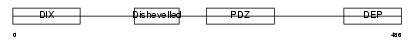
DVL1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 44) | NC score | 0.097759 (rank : 50) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P51141, Q60868 | Gene names | Dvl1, Dvl | |||
|
Domain Architecture |
|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
PSD7_MOUSE
|
||||||
| θ value | 0.163984 (rank : 45) | NC score | 0.051662 (rank : 54) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26516, Q3TW61, Q8C0I8 | Gene names | Psmd7, Mov-34, Mov34 | |||
|
Domain Architecture |
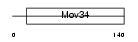
|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 7 (26S proteasome regulatory subunit rpn8) (26S proteasome regulatory subunit S12) (Proteasome subunit p40) (Mov34 protein). | |||||
|
DVL2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 46) | NC score | 0.099600 (rank : 48) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
DVL2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 47) | NC score | 0.098784 (rank : 49) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q60838 | Gene names | Dvl2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
BTBDC_HUMAN
|
||||||
| θ value | 0.365318 (rank : 48) | NC score | 0.032488 (rank : 58) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
CBPZ_MOUSE
|
||||||
| θ value | 0.365318 (rank : 49) | NC score | 0.019324 (rank : 62) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R4V4 | Gene names | Cpz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase Z precursor (EC 3.4.17.-) (CPZ). | |||||
|
DVL1L_HUMAN
|
||||||
| θ value | 0.47712 (rank : 50) | NC score | 0.097000 (rank : 52) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P54792 | Gene names | DVL1L1, DVL, DVL1 | |||
|
Domain Architecture |
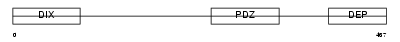
|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1-like (Dishevelled- 1-like) (DSH homolog 1-like). | |||||
|
PLEK_MOUSE
|
||||||
| θ value | 0.47712 (rank : 51) | NC score | 0.077325 (rank : 53) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JHK5, Q9ERI9 | Gene names | Plek | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin. | |||||
|
SFTPD_MOUSE
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.021099 (rank : 61) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P50404 | Gene names | Sftpd, Sftp4 | |||
|
Domain Architecture |

|
|||||
| Description | Pulmonary surfactant-associated protein D precursor (SP-D) (PSP-D). | |||||
|
HESX1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.005109 (rank : 66) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61658, Q61047 | Gene names | Hesx1, Hes-1, Rpx | |||
|
Domain Architecture |
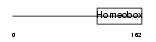
|
|||||
| Description | Homeobox expressed in ES cells 1 (Homeobox protein ANF) (Anterior- restricted homeobox protein) (Rathke pouch homeo box). | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.017056 (rank : 63) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
JHD2C_HUMAN
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.013257 (rank : 64) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15652, Q5SQZ8, Q5SQZ9, Q5SR00, Q7Z3E7, Q8N3U0, Q96KB9, Q9P2G7 | Gene names | JMJD1C, JHDM2C, KIAA1380, TRIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C) (Thyroid receptor-interacting protein 8) (TRIP-8). | |||||
|
PREX1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.038182 (rank : 56) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TCU6, Q5JS95, Q5JS96, Q69YL2, Q7Z2L9, Q9BQH0, Q9BX55, Q9H4Q6, Q9P2D2, Q9UGQ4 | Gene names | PREX1, KIAA1415 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (P-Rex1 protein). | |||||
|
SPTB1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.004371 (rank : 67) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
PSD7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.036927 (rank : 57) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51665, Q6PKI2, Q96E97 | Gene names | PSMD7, MOV34L | |||
|
Domain Architecture |
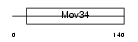
|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 7 (26S proteasome regulatory subunit rpn8) (26S proteasome regulatory subunit S12) (Proteasome subunit p40) (Mov34 protein homolog). | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.031027 (rank : 59) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
SNX25_HUMAN
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.168342 (rank : 45) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H3E2 | Gene names | SNX25 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-25. | |||||
|
PLEK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.030138 (rank : 60) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NYT0, Q96JT0 | Gene names | PLEK2 | |||
|
Domain Architecture |
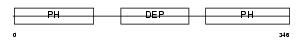
|
|||||
| Description | Pleckstrin-2. | |||||
|
PXDC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.010430 (rank : 65) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DC11, Q6PET5, Q91ZV6 | Gene names | Plxdc2, Tem7r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 2 precursor (Tumor endothelial marker 7-related protein). | |||||
|
BCL3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | -0.000792 (rank : 68) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2F6 | Gene names | Bcl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 3-encoded protein homolog (Protein Bcl-3). | |||||
|
CNTN6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | -0.001222 (rank : 69) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UQ52 | Gene names | CNTN6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-6 precursor (Neural recognition molecule NB-3) (hNB-3). | |||||
|
AKA10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.191485 (rank : 43) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O43572, Q96AJ7 | Gene names | AKAP10 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
AKA10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.191131 (rank : 44) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O88845, Q5SUB5, Q5SUB7, Q7TPE7, Q8BQL6 | Gene names | Akap10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
PLEK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.050734 (rank : 55) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
SNX13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.223122 (rank : 42) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y5W8, O94821, Q8WVZ2, Q8WXH8 | Gene names | SNX13, KIAA0713 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-13 (RGS domain- and PHOX domain-containing protein) (RGS-PX1). | |||||
|
SNX13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.224698 (rank : 41) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6PHS6 | Gene names | Snx13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-13. | |||||
|
RGS9_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O75916, O75573, Q696R2, Q8TD64, Q8TD65, Q9HC32, Q9HC33 | Gene names | RGS9 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
RGS9_MOUSE
|
||||||
| NC score | 0.998780 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O54828, Q9Z0S0 | Gene names | Rgs9 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
RGS11_HUMAN
|
||||||
| NC score | 0.981321 (rank : 3) | θ value | 3.28708e-135 (rank : 3) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O94810, O75883 | Gene names | RGS11 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 11 (RGS11). | |||||
|
RGS7_HUMAN
|
||||||
| NC score | 0.956949 (rank : 4) | θ value | 2.5521e-87 (rank : 5) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P49802, Q8TD66, Q8TD67, Q9UNU7, Q9Y6B9 | Gene names | RGS7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RGS7_MOUSE
|
||||||
| NC score | 0.956928 (rank : 5) | θ value | 8.77142e-88 (rank : 4) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O54829 | Gene names | Rgs7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RGS6_MOUSE
|
||||||
| NC score | 0.955169 (rank : 6) | θ value | 3.11973e-85 (rank : 6) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Z2H2, Q8BGI8 | Gene names | Rgs6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 6 (RGS6). | |||||
|
RGS6_HUMAN
|
||||||
| NC score | 0.947428 (rank : 7) | θ value | 7.19216e-82 (rank : 7) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P49758, O75576, O75577, Q8TE13, Q8TE14, Q8TE15, Q8TE16, Q8TE17, Q8TE18, Q8TE19, Q8TE20, Q8TE21, Q8TE22, Q9Y245, Q9Y647 | Gene names | RGS6 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 6 (RGS6) (S914). | |||||
|
RGS10_MOUSE
|
||||||
| NC score | 0.861267 (rank : 8) | θ value | 2.87452e-22 (rank : 9) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9CQE5, Q9D3L2 | Gene names | Rgs10 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 10 (RGS10). | |||||
|
RGS10_HUMAN
|
||||||
| NC score | 0.860613 (rank : 9) | θ value | 2.43343e-21 (rank : 13) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O43665 | Gene names | RGS10 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 10 (RGS10). | |||||
|
RGS1_HUMAN
|
||||||
| NC score | 0.854982 (rank : 10) | θ value | 5.79196e-23 (rank : 8) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q08116, Q07918, Q9H1W2 | Gene names | RGS1, 1R20, BL34 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1) (Early response protein 1R20) (B-cell activation protein BL34). | |||||
|
RGS1_MOUSE
|
||||||
| NC score | 0.853739 (rank : 11) | θ value | 2.87452e-22 (rank : 10) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9JL25, Q3TBD1, Q3TC18, Q3TD56 | Gene names | Rgs1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1). | |||||
|
RGS5_MOUSE
|
||||||
| NC score | 0.852493 (rank : 12) | θ value | 7.0802e-21 (rank : 14) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O08850, Q543B1, Q9D0Z2 | Gene names | Rgs5 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 5 (RGS5). | |||||
|
RGS20_MOUSE
|
||||||
| NC score | 0.851958 (rank : 13) | θ value | 6.40375e-22 (rank : 11) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QZB1, Q3TY63, Q9CUV8, Q9QZB2 | Gene names | Rgs20, Rgsz1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 20 (RGS20) (Regulator of G-protein signaling Z1). | |||||
|
RGS20_HUMAN
|
||||||
| NC score | 0.851608 (rank : 14) | θ value | 8.36355e-22 (rank : 12) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O76081, Q96BG9 | Gene names | RGS20, RGSZ1, ZGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 20 (RGS20) (Regulator of Gz-selective protein signaling 1) (Gz-selective GTPase-activating protein) (G(z)GAP). | |||||
|
RGS19_HUMAN
|
||||||
| NC score | 0.851113 (rank : 15) | θ value | 7.82807e-20 (rank : 20) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P49795, Q53XN0, Q8TD60 | Gene names | RGS19, GAIP, GNAI3IP | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 19 (RGS19) (G-alpha-interacting protein) (GAIP protein). | |||||
|
RGS19_MOUSE
|
||||||
| NC score | 0.850285 (rank : 16) | θ value | 5.99374e-20 (rank : 18) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9CX84, Q99L50 | Gene names | Rgs19 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 19 (RGS19). | |||||
|
RGS17_HUMAN
|
||||||
| NC score | 0.850040 (rank : 17) | θ value | 3.51386e-20 (rank : 16) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UGC6, Q5TF49, Q8TD61, Q9UJS8 | Gene names | RGS17 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 17 (RGS17). | |||||
|
RGS17_MOUSE
|
||||||
| NC score | 0.848826 (rank : 18) | θ value | 7.82807e-20 (rank : 19) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QZB0, Q543T9 | Gene names | Rgs17, Rgsz2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 17 (RGS17) (Regulator of Gz-selective protein signaling 2). | |||||
|
RGS5_HUMAN
|
||||||
| NC score | 0.846356 (rank : 19) | θ value | 4.58923e-20 (rank : 17) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15539 | Gene names | RGS5 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 5 (RGS5). | |||||
|
RGS18_HUMAN
|
||||||
| NC score | 0.845992 (rank : 20) | θ value | 2.69047e-20 (rank : 15) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NS28 | Gene names | RGS18, RGS13 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 18 (RGS18). | |||||
|
RGS8_HUMAN
|
||||||
| NC score | 0.842099 (rank : 21) | θ value | 1.33526e-19 (rank : 23) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P57771, Q3SYD2 | Gene names | RGS8 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 8 (RGS8). | |||||
|
RGS8_MOUSE
|
||||||
| NC score | 0.841049 (rank : 22) | θ value | 2.97466e-19 (rank : 24) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BXT1 | Gene names | Rgs8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 8 (RGS8). | |||||
|
RGS18_MOUSE
|
||||||
| NC score | 0.840165 (rank : 23) | θ value | 6.62687e-19 (rank : 28) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99PG4 | Gene names | Rgs18 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 18 (RGS18). | |||||
|
RGS13_HUMAN
|
||||||
| NC score | 0.839549 (rank : 24) | θ value | 1.38178e-16 (rank : 35) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O14921, Q6PGR2, Q8TD63, Q9BX45 | Gene names | RGS13 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 13 (RGS13). | |||||
|
RGS16_HUMAN
|
||||||
| NC score | 0.839190 (rank : 25) | θ value | 6.62687e-19 (rank : 27) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15492, Q99701 | Gene names | RGS16, RGSR | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 16 (RGS16) (Retinally abundant regulator of G-protein signaling) (RGS-R) (A28-RGS14P). | |||||
|
RGS13_MOUSE
|
||||||
| NC score | 0.838113 (rank : 26) | θ value | 6.85773e-16 (rank : 36) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8K443 | Gene names | Rgs13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 13 (RGS13). | |||||
|
RGS4_MOUSE
|
||||||
| NC score | 0.836952 (rank : 27) | θ value | 7.32683e-18 (rank : 33) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O08899, Q99L30 | Gene names | Rgs4 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 4 (RGS4). | |||||
|
RGS4_HUMAN
|
||||||
| NC score | 0.835039 (rank : 28) | θ value | 2.5182e-18 (rank : 31) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P49798 | Gene names | RGS4 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 4 (RGS4) (RGP4). | |||||
|
RGS2_HUMAN
|
||||||
| NC score | 0.833880 (rank : 29) | θ value | 7.32683e-18 (rank : 32) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P41220 | Gene names | RGS2, G0S8 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 2 (RGS2) (G0/G1 switch regulatory protein 8). | |||||
|
RGS16_MOUSE
|
||||||
| NC score | 0.833130 (rank : 30) | θ value | 1.63225e-17 (rank : 34) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97428, O09091, P97420, Q80V16 | Gene names | Rgs16, Rgsr | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 16 (RGS16) (Retinally abundant regulator of G-protein signaling) (RGS-R) (A28-RGS14P). | |||||
|
RGS2_MOUSE
|
||||||
| NC score | 0.832887 (rank : 31) | θ value | 2.5182e-18 (rank : 30) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O08849, Q544S7, Q91WX1, Q9JL24 | Gene names | Rgs2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 2 (RGS2). | |||||
|
RGS14_MOUSE
|
||||||
| NC score | 0.808692 (rank : 32) | θ value | 3.88503e-19 (rank : 25) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97492, Q9DCD1 | Gene names | Rgs14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14) (RAP1/RAP2-interacting protein). | |||||
|
RGS14_HUMAN
|
||||||
| NC score | 0.804666 (rank : 33) | θ value | 5.07402e-19 (rank : 26) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O43566, O43565, Q8TD62 | Gene names | RGS14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14). | |||||
|
RGS12_HUMAN
|
||||||
| NC score | 0.722612 (rank : 34) | θ value | 1.02238e-19 (rank : 21) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.594965 (rank : 35) | θ value | 1.13037e-18 (rank : 29) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
AXN1_MOUSE
|
||||||
| NC score | 0.545452 (rank : 36) | θ value | 2.44474e-05 (rank : 38) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O35625 | Gene names | Axin1, Axin, Fu | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (Fused protein). | |||||
|
AXN2_HUMAN
|
||||||
| NC score | 0.536527 (rank : 37) | θ value | 1.43324e-05 (rank : 37) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y2T1, Q9H3M6, Q9UH84 | Gene names | AXIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.531903 (rank : 38) | θ value | 1.33526e-19 (rank : 22) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
AXN2_MOUSE
|
||||||
| NC score | 0.530641 (rank : 39) | θ value | 2.44474e-05 (rank : 39) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O88566, Q9QXJ6 | Gene names | Axin2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
AXN1_HUMAN
|
||||||
| NC score | 0.529709 (rank : 40) | θ value | 0.000158464 (rank : 40) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O15169, Q4TT26, Q4TT27, Q86YA7, Q8WVW6, Q96S28 | Gene names | AXIN1, AXIN | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (hAxin). | |||||
|
SNX13_MOUSE
|
||||||
| NC score | 0.224698 (rank : 41) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6PHS6 | Gene names | Snx13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-13. | |||||
|
SNX13_HUMAN
|
||||||
| NC score | 0.223122 (rank : 42) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y5W8, O94821, Q8WVZ2, Q8WXH8 | Gene names | SNX13, KIAA0713 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-13 (RGS domain- and PHOX domain-containing protein) (RGS-PX1). | |||||
|
AKA10_HUMAN
|
||||||
| NC score | 0.191485 (rank : 43) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O43572, Q96AJ7 | Gene names | AKAP10 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
AKA10_MOUSE
|
||||||
| NC score | 0.191131 (rank : 44) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O88845, Q5SUB5, Q5SUB7, Q7TPE7, Q8BQL6 | Gene names | Akap10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
SNX25_HUMAN
|
||||||
| NC score | 0.168342 (rank : 45) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H3E2 | Gene names | SNX25 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-25. | |||||
|
DVL3_MOUSE
|
||||||
| NC score | 0.123494 (rank : 46) | θ value | 0.00298849 (rank : 42) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61062 | Gene names | Dvl3 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
DVL3_HUMAN
|
||||||
| NC score | 0.123410 (rank : 47) | θ value | 0.00298849 (rank : 41) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q92997, O14642, Q13531, Q8N5E9, Q92607 | Gene names | DVL3, KIAA0208 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
DVL2_HUMAN
|
||||||
| NC score | 0.099600 (rank : 48) | θ value | 0.279714 (rank : 46) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
DVL2_MOUSE
|
||||||
| NC score | 0.098784 (rank : 49) | θ value | 0.279714 (rank : 47) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q60838 | Gene names | Dvl2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
DVL1_MOUSE
|
||||||
| NC score | 0.097759 (rank : 50) | θ value | 0.163984 (rank : 44) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P51141, Q60868 | Gene names | Dvl1, Dvl | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
DVL1_HUMAN
|
||||||
| NC score | 0.097616 (rank : 51) | θ value | 0.163984 (rank : 43) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14640 | Gene names | DVL1 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1 (Dishevelled-1) (DSH homolog 1). | |||||
|
DVL1L_HUMAN
|
||||||
| NC score | 0.097000 (rank : 52) | θ value | 0.47712 (rank : 50) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P54792 | Gene names | DVL1L1, DVL, DVL1 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-1-like (Dishevelled- 1-like) (DSH homolog 1-like). | |||||
|
PLEK_MOUSE
|
||||||
| NC score | 0.077325 (rank : 53) | θ value | 0.47712 (rank : 51) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JHK5, Q9ERI9 | Gene names | Plek | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin. | |||||
|
PSD7_MOUSE
|
||||||
| NC score | 0.051662 (rank : 54) | θ value | 0.163984 (rank : 45) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26516, Q3TW61, Q8C0I8 | Gene names | Psmd7, Mov-34, Mov34 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 7 (26S proteasome regulatory subunit rpn8) (26S proteasome regulatory subunit S12) (Proteasome subunit p40) (Mov34 protein). | |||||
|
PLEK_HUMAN
|
||||||
| NC score | 0.050734 (rank : 55) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P08567, Q53SU8, Q6FGM8, Q8WV81 | Gene names | PLEK, P47 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin (Platelet p47 protein). | |||||
|
PREX1_HUMAN
|
||||||
| NC score | 0.038182 (rank : 56) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TCU6, Q5JS95, Q5JS96, Q69YL2, Q7Z2L9, Q9BQH0, Q9BX55, Q9H4Q6, Q9P2D2, Q9UGQ4 | Gene names | PREX1, KIAA1415 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein (P-Rex1 protein). | |||||
|
PSD7_HUMAN
|
||||||
| NC score | 0.036927 (rank : 57) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51665, Q6PKI2, Q96E97 | Gene names | PSMD7, MOV34L | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 7 (26S proteasome regulatory subunit rpn8) (26S proteasome regulatory subunit S12) (Proteasome subunit p40) (Mov34 protein homolog). | |||||
|
BTBDC_HUMAN
|
||||||
| NC score | 0.032488 (rank : 58) | θ value | 0.365318 (rank : 48) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.031027 (rank : 59) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
PLEK2_HUMAN
|
||||||
| NC score | 0.030138 (rank : 60) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NYT0, Q96JT0 | Gene names | PLEK2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin-2. | |||||
|
SFTPD_MOUSE
|
||||||
| NC score | 0.021099 (rank : 61) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P50404 | Gene names | Sftpd, Sftp4 | |||
|
Domain Architecture |

|
|||||
| Description | Pulmonary surfactant-associated protein D precursor (SP-D) (PSP-D). | |||||
|
CBPZ_MOUSE
|
||||||
| NC score | 0.019324 (rank : 62) | θ value | 0.365318 (rank : 49) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R4V4 | Gene names | Cpz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxypeptidase Z precursor (EC 3.4.17.-) (CPZ). | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.017056 (rank : 63) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
JHD2C_HUMAN
|
||||||
| NC score | 0.013257 (rank : 64) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15652, Q5SQZ8, Q5SQZ9, Q5SR00, Q7Z3E7, Q8N3U0, Q96KB9, Q9P2G7 | Gene names | JMJD1C, JHDM2C, KIAA1380, TRIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C) (Thyroid receptor-interacting protein 8) (TRIP-8). | |||||
|
PXDC2_MOUSE
|
||||||
| NC score | 0.010430 (rank : 65) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DC11, Q6PET5, Q91ZV6 | Gene names | Plxdc2, Tem7r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 2 precursor (Tumor endothelial marker 7-related protein). | |||||
|
HESX1_MOUSE
|
||||||
| NC score | 0.005109 (rank : 66) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61658, Q61047 | Gene names | Hesx1, Hes-1, Rpx | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox expressed in ES cells 1 (Homeobox protein ANF) (Anterior- restricted homeobox protein) (Rathke pouch homeo box). | |||||
|
SPTB1_MOUSE
|
||||||
| NC score | 0.004371 (rank : 67) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
BCL3_MOUSE
|
||||||
| NC score | -0.000792 (rank : 68) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2F6 | Gene names | Bcl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 3-encoded protein homolog (Protein Bcl-3). | |||||
|
CNTN6_HUMAN
|
||||||
| NC score | -0.001222 (rank : 69) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UQ52 | Gene names | CNTN6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-6 precursor (Neural recognition molecule NB-3) (hNB-3). | |||||