Please be patient as the page loads
|
RGS14_HUMAN
|
||||||
| SwissProt Accessions | O43566, O43565, Q8TD62 | Gene names | RGS14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RGS14_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O43566, O43565, Q8TD62 | Gene names | RGS14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14). | |||||
|
RGS14_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.994594 (rank : 2) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P97492, Q9DCD1 | Gene names | Rgs14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14) (RAP1/RAP2-interacting protein). | |||||
|
RGS12_HUMAN
|
||||||
| θ value | 6.27148e-94 (rank : 3) | NC score | 0.859969 (rank : 23) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
RGS10_HUMAN
|
||||||
| θ value | 4.42448e-31 (rank : 4) | NC score | 0.923426 (rank : 3) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O43665 | Gene names | RGS10 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 10 (RGS10). | |||||
|
RGS10_MOUSE
|
||||||
| θ value | 5.77852e-31 (rank : 5) | NC score | 0.921864 (rank : 4) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9CQE5, Q9D3L2 | Gene names | Rgs10 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 10 (RGS10). | |||||
|
RGS11_HUMAN
|
||||||
| θ value | 5.99374e-20 (rank : 6) | NC score | 0.817982 (rank : 28) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O94810, O75883 | Gene names | RGS11 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 11 (RGS11). | |||||
|
RGS5_MOUSE
|
||||||
| θ value | 5.99374e-20 (rank : 7) | NC score | 0.872379 (rank : 5) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08850, Q543B1, Q9D0Z2 | Gene names | Rgs5 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 5 (RGS5). | |||||
|
RGS19_MOUSE
|
||||||
| θ value | 1.02238e-19 (rank : 8) | NC score | 0.868310 (rank : 6) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9CX84, Q99L50 | Gene names | Rgs19 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 19 (RGS19). | |||||
|
RGS20_HUMAN
|
||||||
| θ value | 1.02238e-19 (rank : 9) | NC score | 0.863381 (rank : 16) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O76081, Q96BG9 | Gene names | RGS20, RGSZ1, ZGAP1 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 20 (RGS20) (Regulator of Gz-selective protein signaling 1) (Gz-selective GTPase-activating protein) (G(z)GAP). | |||||
|
RGS20_MOUSE
|
||||||
| θ value | 1.02238e-19 (rank : 10) | NC score | 0.867559 (rank : 8) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QZB1, Q3TY63, Q9CUV8, Q9QZB2 | Gene names | Rgs20, Rgsz1 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 20 (RGS20) (Regulator of G-protein signaling Z1). | |||||
|
RGS9_MOUSE
|
||||||
| θ value | 2.97466e-19 (rank : 11) | NC score | 0.805591 (rank : 29) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O54828, Q9Z0S0 | Gene names | Rgs9 | |||
|
Domain Architecture |
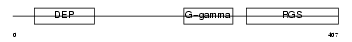
|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
RGS19_HUMAN
|
||||||
| θ value | 3.88503e-19 (rank : 12) | NC score | 0.864745 (rank : 13) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P49795, Q53XN0, Q8TD60 | Gene names | RGS19, GAIP, GNAI3IP | |||
|
Domain Architecture |
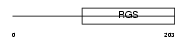
|
|||||
| Description | Regulator of G-protein signaling 19 (RGS19) (G-alpha-interacting protein) (GAIP protein). | |||||
|
RGS9_HUMAN
|
||||||
| θ value | 5.07402e-19 (rank : 13) | NC score | 0.804666 (rank : 30) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O75916, O75573, Q696R2, Q8TD64, Q8TD65, Q9HC32, Q9HC33 | Gene names | RGS9 | |||
|
Domain Architecture |
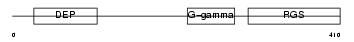
|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
RGS4_HUMAN
|
||||||
| θ value | 8.65492e-19 (rank : 14) | NC score | 0.867696 (rank : 7) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P49798 | Gene names | RGS4 | |||
|
Domain Architecture |
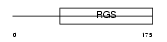
|
|||||
| Description | Regulator of G-protein signaling 4 (RGS4) (RGP4). | |||||
|
RGS18_MOUSE
|
||||||
| θ value | 1.13037e-18 (rank : 15) | NC score | 0.861383 (rank : 22) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99PG4 | Gene names | Rgs18 | |||
|
Domain Architecture |
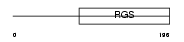
|
|||||
| Description | Regulator of G-protein signaling 18 (RGS18). | |||||
|
RGS7_MOUSE
|
||||||
| θ value | 1.47631e-18 (rank : 16) | NC score | 0.796584 (rank : 31) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O54829 | Gene names | Rgs7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RGS18_HUMAN
|
||||||
| θ value | 1.92812e-18 (rank : 17) | NC score | 0.862274 (rank : 17) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NS28 | Gene names | RGS18, RGS13 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 18 (RGS18). | |||||
|
RGS4_MOUSE
|
||||||
| θ value | 1.92812e-18 (rank : 18) | NC score | 0.866938 (rank : 9) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08899, Q99L30 | Gene names | Rgs4 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 4 (RGS4). | |||||
|
RGS6_MOUSE
|
||||||
| θ value | 1.92812e-18 (rank : 19) | NC score | 0.794371 (rank : 33) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Z2H2, Q8BGI8 | Gene names | Rgs6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 6 (RGS6). | |||||
|
RGS5_HUMAN
|
||||||
| θ value | 3.28887e-18 (rank : 20) | NC score | 0.863928 (rank : 15) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15539 | Gene names | RGS5 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 5 (RGS5). | |||||
|
RGS16_MOUSE
|
||||||
| θ value | 9.56915e-18 (rank : 21) | NC score | 0.861573 (rank : 19) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P97428, O09091, P97420, Q80V16 | Gene names | Rgs16, Rgsr | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 16 (RGS16) (Retinally abundant regulator of G-protein signaling) (RGS-R) (A28-RGS14P). | |||||
|
RGS7_HUMAN
|
||||||
| θ value | 9.56915e-18 (rank : 22) | NC score | 0.796460 (rank : 32) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49802, Q8TD66, Q8TD67, Q9UNU7, Q9Y6B9 | Gene names | RGS7 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RGS16_HUMAN
|
||||||
| θ value | 1.24977e-17 (rank : 23) | NC score | 0.861507 (rank : 20) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O15492, Q99701 | Gene names | RGS16, RGSR | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 16 (RGS16) (Retinally abundant regulator of G-protein signaling) (RGS-R) (A28-RGS14P). | |||||
|
RGS17_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 24) | NC score | 0.864983 (rank : 11) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UGC6, Q5TF49, Q8TD61, Q9UJS8 | Gene names | RGS17 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 17 (RGS17). | |||||
|
RGS2_MOUSE
|
||||||
| θ value | 3.63628e-17 (rank : 25) | NC score | 0.852976 (rank : 25) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O08849, Q544S7, Q91WX1, Q9JL24 | Gene names | Rgs2 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 2 (RGS2). | |||||
|
RGS1_MOUSE
|
||||||
| θ value | 4.74913e-17 (rank : 26) | NC score | 0.865943 (rank : 10) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9JL25, Q3TBD1, Q3TC18, Q3TD56 | Gene names | Rgs1 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1). | |||||
|
RGS8_HUMAN
|
||||||
| θ value | 4.74913e-17 (rank : 27) | NC score | 0.862253 (rank : 18) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P57771, Q3SYD2 | Gene names | RGS8 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 8 (RGS8). | |||||
|
RGS17_MOUSE
|
||||||
| θ value | 6.20254e-17 (rank : 28) | NC score | 0.864865 (rank : 12) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QZB0, Q543T9 | Gene names | Rgs17, Rgsz2 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 17 (RGS17) (Regulator of Gz-selective protein signaling 2). | |||||
|
RGS2_HUMAN
|
||||||
| θ value | 6.20254e-17 (rank : 29) | NC score | 0.854140 (rank : 24) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P41220 | Gene names | RGS2, G0S8 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 2 (RGS2) (G0/G1 switch regulatory protein 8). | |||||
|
RGS8_MOUSE
|
||||||
| θ value | 8.10077e-17 (rank : 30) | NC score | 0.861405 (rank : 21) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BXT1 | Gene names | Rgs8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 8 (RGS8). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 31) | NC score | 0.611447 (rank : 37) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 1.38178e-16 (rank : 32) | NC score | 0.545917 (rank : 40) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
RGS1_HUMAN
|
||||||
| θ value | 6.85773e-16 (rank : 33) | NC score | 0.864532 (rank : 14) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q08116, Q07918, Q9H1W2 | Gene names | RGS1, 1R20, BL34 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1) (Early response protein 1R20) (B-cell activation protein BL34). | |||||
|
RGS6_HUMAN
|
||||||
| θ value | 1.16975e-15 (rank : 34) | NC score | 0.770881 (rank : 34) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49758, O75576, O75577, Q8TE13, Q8TE14, Q8TE15, Q8TE16, Q8TE17, Q8TE18, Q8TE19, Q8TE20, Q8TE21, Q8TE22, Q9Y245, Q9Y647 | Gene names | RGS6 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 6 (RGS6) (S914). | |||||
|
RGS13_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 35) | NC score | 0.851355 (rank : 27) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14921, Q6PGR2, Q8TD63, Q9BX45 | Gene names | RGS13 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 13 (RGS13). | |||||
|
RGS13_MOUSE
|
||||||
| θ value | 7.09661e-13 (rank : 36) | NC score | 0.851805 (rank : 26) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8K443 | Gene names | Rgs13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 13 (RGS13). | |||||
|
AXN1_MOUSE
|
||||||
| θ value | 1.133e-10 (rank : 37) | NC score | 0.640013 (rank : 35) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O35625 | Gene names | Axin1, Axin, Fu | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (Fused protein). | |||||
|
AXN1_HUMAN
|
||||||
| θ value | 7.34386e-10 (rank : 38) | NC score | 0.621802 (rank : 36) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O15169, Q4TT26, Q4TT27, Q86YA7, Q8WVW6, Q96S28 | Gene names | AXIN1, AXIN | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (hAxin). | |||||
|
AXN2_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 39) | NC score | 0.608491 (rank : 38) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Y2T1, Q9H3M6, Q9UH84 | Gene names | AXIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
AXN2_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 40) | NC score | 0.596286 (rank : 39) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O88566, Q9QXJ6 | Gene names | Axin2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
AKA10_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 41) | NC score | 0.340573 (rank : 42) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43572, Q96AJ7 | Gene names | AKAP10 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
AKA10_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 42) | NC score | 0.340628 (rank : 41) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O88845, Q5SUB5, Q5SUB7, Q7TPE7, Q8BQL6 | Gene names | Akap10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
CCD22_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 43) | NC score | 0.042290 (rank : 48) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60826 | Gene names | CCDC22, CXorf37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 22. | |||||
|
SNX13_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 44) | NC score | 0.278545 (rank : 43) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6PHS6 | Gene names | Snx13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-13. | |||||
|
CTDP1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 45) | NC score | 0.043365 (rank : 47) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
SNX13_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 46) | NC score | 0.272355 (rank : 44) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y5W8, O94821, Q8WVZ2, Q8WXH8 | Gene names | SNX13, KIAA0713 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-13 (RGS domain- and PHOX domain-containing protein) (RGS-PX1). | |||||
|
CABL2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 47) | NC score | 0.029118 (rank : 51) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BTV7, Q5JWL0, Q9BYK0 | Gene names | CABLES2, C20orf150 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
LYSM3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 48) | NC score | 0.033087 (rank : 50) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99LE3 | Gene names | Lysmd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LysM and putative peptidoglycan-binding domain-containing protein 3. | |||||
|
PTN23_HUMAN
|
||||||
| θ value | 0.62314 (rank : 49) | NC score | 0.016044 (rank : 57) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
ZNT3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.020795 (rank : 54) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99726 | Gene names | SLC30A3, ZNT3 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc transporter 3 (ZnT-3) (Solute carrier family 30 member 3). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.016304 (rank : 56) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
MAST3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.000544 (rank : 76) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1128 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60307, Q7LDZ8, Q9UPI0 | Gene names | MAST3, KIAA0561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 3 (EC 2.7.11.1). | |||||
|
SPTA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.008121 (rank : 65) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
KCNH2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.013127 (rank : 61) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
RPP25_HUMAN
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.025801 (rank : 52) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BUL9, Q9NX88 | Gene names | RPP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease P protein subunit p25 (EC 3.1.26.5) (RNase P protein subunit p25). | |||||
|
CS029_MOUSE
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.015923 (rank : 58) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CS00 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf29 homolog. | |||||
|
KCNH2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.014110 (rank : 60) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.015385 (rank : 59) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
PCNT_MOUSE
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.006336 (rank : 71) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
CRCM_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.009188 (rank : 64) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23508 | Gene names | MCC | |||
|
Domain Architecture |
|
|||||
| Description | Colorectal mutant cancer protein (Protein MCC). | |||||
|
PELP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.023804 (rank : 53) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
KLF4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | -0.003312 (rank : 79) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43474, P78338, Q9UNP3 | Gene names | KLF4, EZF, GKLF | |||
|
Domain Architecture |

|
|||||
| Description | Krueppel-like factor 4 (Epithelial zinc-finger protein EZF) (Gut- enriched krueppel-like factor). | |||||
|
SPTA1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.007901 (rank : 66) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P08032, P97502 | Gene names | Spta1, Spna1, Spta | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
ITA2B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.004457 (rank : 74) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QUM0, Q64229, Q9Z2M0 | Gene names | Itga2b | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-IIb precursor (Platelet membrane glycoprotein IIb) (GPalpha IIb) (GPIIb) (CD41 antigen) [Contains: Integrin alpha-IIb heavy chain; Integrin alpha-IIb light chain]. | |||||
|
JIP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.019944 (rank : 55) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13387, Q96G62, Q99771, Q9NZ59, Q9UKQ4 | Gene names | MAPK8IP2, IB2, JIP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
RGNEF_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.006962 (rank : 68) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97433 | Gene names | Rgnef, Rhoip2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-guanine nucleotide exchange factor (Rho-interacting protein 2) (RhoGEF) (RIP2). | |||||
|
ARHGB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.034327 (rank : 49) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
KI21A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | -0.000478 (rank : 78) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
KI21A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.000417 (rank : 77) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
MELPH_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.011092 (rank : 63) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91V27, Q99N53 | Gene names | Mlph, Ln, Slac2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Leaden protein) (Synaptotagmin-like protein 2a) (Slac2-a) (Slp homolog lacking C2 domains a). | |||||
|
MTMR5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.005013 (rank : 73) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
ZFY28_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.006446 (rank : 70) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HCC9 | Gene names | ZFYVE28, KIAA1643 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 28. | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.007448 (rank : 67) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
KIF1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.002761 (rank : 75) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60333, Q96Q94, Q9BV80, Q9P280 | Gene names | KIF1B, KIAA0591, KIAA1448 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1B (Klp). | |||||
|
LY10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.005285 (rank : 72) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
PLVAP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.006740 (rank : 69) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91VC4, Q99JB1 | Gene names | Plvap, Pv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plasmalemma vesicle-associated protein (Plasmalemma vesicle protein 1) (PV-1) (MECA-32 antigen). | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.011412 (rank : 62) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SNX14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.052200 (rank : 46) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BHY8 | Gene names | Snx14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-14. | |||||
|
SNX25_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.124365 (rank : 45) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H3E2 | Gene names | SNX25 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-25. | |||||
|
RGS14_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O43566, O43565, Q8TD62 | Gene names | RGS14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14). | |||||
|
RGS14_MOUSE
|
||||||
| NC score | 0.994594 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P97492, Q9DCD1 | Gene names | Rgs14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14) (RAP1/RAP2-interacting protein). | |||||
|
RGS10_HUMAN
|
||||||
| NC score | 0.923426 (rank : 3) | θ value | 4.42448e-31 (rank : 4) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O43665 | Gene names | RGS10 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 10 (RGS10). | |||||
|
RGS10_MOUSE
|
||||||
| NC score | 0.921864 (rank : 4) | θ value | 5.77852e-31 (rank : 5) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9CQE5, Q9D3L2 | Gene names | Rgs10 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 10 (RGS10). | |||||
|
RGS5_MOUSE
|
||||||
| NC score | 0.872379 (rank : 5) | θ value | 5.99374e-20 (rank : 7) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08850, Q543B1, Q9D0Z2 | Gene names | Rgs5 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 5 (RGS5). | |||||
|
RGS19_MOUSE
|
||||||
| NC score | 0.868310 (rank : 6) | θ value | 1.02238e-19 (rank : 8) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9CX84, Q99L50 | Gene names | Rgs19 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 19 (RGS19). | |||||
|
RGS4_HUMAN
|
||||||
| NC score | 0.867696 (rank : 7) | θ value | 8.65492e-19 (rank : 14) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P49798 | Gene names | RGS4 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 4 (RGS4) (RGP4). | |||||
|
RGS20_MOUSE
|
||||||
| NC score | 0.867559 (rank : 8) | θ value | 1.02238e-19 (rank : 10) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QZB1, Q3TY63, Q9CUV8, Q9QZB2 | Gene names | Rgs20, Rgsz1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 20 (RGS20) (Regulator of G-protein signaling Z1). | |||||
|
RGS4_MOUSE
|
||||||
| NC score | 0.866938 (rank : 9) | θ value | 1.92812e-18 (rank : 18) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08899, Q99L30 | Gene names | Rgs4 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 4 (RGS4). | |||||
|
RGS1_MOUSE
|
||||||
| NC score | 0.865943 (rank : 10) | θ value | 4.74913e-17 (rank : 26) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9JL25, Q3TBD1, Q3TC18, Q3TD56 | Gene names | Rgs1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1). | |||||
|
RGS17_HUMAN
|
||||||
| NC score | 0.864983 (rank : 11) | θ value | 2.7842e-17 (rank : 24) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UGC6, Q5TF49, Q8TD61, Q9UJS8 | Gene names | RGS17 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 17 (RGS17). | |||||
|
RGS17_MOUSE
|
||||||
| NC score | 0.864865 (rank : 12) | θ value | 6.20254e-17 (rank : 28) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9QZB0, Q543T9 | Gene names | Rgs17, Rgsz2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 17 (RGS17) (Regulator of Gz-selective protein signaling 2). | |||||
|
RGS19_HUMAN
|
||||||
| NC score | 0.864745 (rank : 13) | θ value | 3.88503e-19 (rank : 12) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P49795, Q53XN0, Q8TD60 | Gene names | RGS19, GAIP, GNAI3IP | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 19 (RGS19) (G-alpha-interacting protein) (GAIP protein). | |||||
|
RGS1_HUMAN
|
||||||
| NC score | 0.864532 (rank : 14) | θ value | 6.85773e-16 (rank : 33) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q08116, Q07918, Q9H1W2 | Gene names | RGS1, 1R20, BL34 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 1 (RGS1) (Early response protein 1R20) (B-cell activation protein BL34). | |||||
|
RGS5_HUMAN
|
||||||
| NC score | 0.863928 (rank : 15) | θ value | 3.28887e-18 (rank : 20) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15539 | Gene names | RGS5 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 5 (RGS5). | |||||
|
RGS20_HUMAN
|
||||||
| NC score | 0.863381 (rank : 16) | θ value | 1.02238e-19 (rank : 9) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O76081, Q96BG9 | Gene names | RGS20, RGSZ1, ZGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 20 (RGS20) (Regulator of Gz-selective protein signaling 1) (Gz-selective GTPase-activating protein) (G(z)GAP). | |||||
|
RGS18_HUMAN
|
||||||
| NC score | 0.862274 (rank : 17) | θ value | 1.92812e-18 (rank : 17) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NS28 | Gene names | RGS18, RGS13 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 18 (RGS18). | |||||
|
RGS8_HUMAN
|
||||||
| NC score | 0.862253 (rank : 18) | θ value | 4.74913e-17 (rank : 27) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P57771, Q3SYD2 | Gene names | RGS8 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 8 (RGS8). | |||||
|
RGS16_MOUSE
|
||||||
| NC score | 0.861573 (rank : 19) | θ value | 9.56915e-18 (rank : 21) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P97428, O09091, P97420, Q80V16 | Gene names | Rgs16, Rgsr | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 16 (RGS16) (Retinally abundant regulator of G-protein signaling) (RGS-R) (A28-RGS14P). | |||||
|
RGS16_HUMAN
|
||||||
| NC score | 0.861507 (rank : 20) | θ value | 1.24977e-17 (rank : 23) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O15492, Q99701 | Gene names | RGS16, RGSR | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 16 (RGS16) (Retinally abundant regulator of G-protein signaling) (RGS-R) (A28-RGS14P). | |||||
|
RGS8_MOUSE
|
||||||
| NC score | 0.861405 (rank : 21) | θ value | 8.10077e-17 (rank : 30) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BXT1 | Gene names | Rgs8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 8 (RGS8). | |||||
|
RGS18_MOUSE
|
||||||
| NC score | 0.861383 (rank : 22) | θ value | 1.13037e-18 (rank : 15) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99PG4 | Gene names | Rgs18 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 18 (RGS18). | |||||
|
RGS12_HUMAN
|
||||||
| NC score | 0.859969 (rank : 23) | θ value | 6.27148e-94 (rank : 3) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
RGS2_HUMAN
|
||||||
| NC score | 0.854140 (rank : 24) | θ value | 6.20254e-17 (rank : 29) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P41220 | Gene names | RGS2, G0S8 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 2 (RGS2) (G0/G1 switch regulatory protein 8). | |||||
|
RGS2_MOUSE
|
||||||
| NC score | 0.852976 (rank : 25) | θ value | 3.63628e-17 (rank : 25) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O08849, Q544S7, Q91WX1, Q9JL24 | Gene names | Rgs2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 2 (RGS2). | |||||
|
RGS13_MOUSE
|
||||||
| NC score | 0.851805 (rank : 26) | θ value | 7.09661e-13 (rank : 36) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8K443 | Gene names | Rgs13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 13 (RGS13). | |||||
|
RGS13_HUMAN
|
||||||
| NC score | 0.851355 (rank : 27) | θ value | 7.09661e-13 (rank : 35) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14921, Q6PGR2, Q8TD63, Q9BX45 | Gene names | RGS13 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 13 (RGS13). | |||||
|
RGS11_HUMAN
|
||||||
| NC score | 0.817982 (rank : 28) | θ value | 5.99374e-20 (rank : 6) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O94810, O75883 | Gene names | RGS11 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 11 (RGS11). | |||||
|
RGS9_MOUSE
|
||||||
| NC score | 0.805591 (rank : 29) | θ value | 2.97466e-19 (rank : 11) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O54828, Q9Z0S0 | Gene names | Rgs9 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
RGS9_HUMAN
|
||||||
| NC score | 0.804666 (rank : 30) | θ value | 5.07402e-19 (rank : 13) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O75916, O75573, Q696R2, Q8TD64, Q8TD65, Q9HC32, Q9HC33 | Gene names | RGS9 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
RGS7_MOUSE
|
||||||
| NC score | 0.796584 (rank : 31) | θ value | 1.47631e-18 (rank : 16) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O54829 | Gene names | Rgs7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RGS7_HUMAN
|
||||||
| NC score | 0.796460 (rank : 32) | θ value | 9.56915e-18 (rank : 22) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49802, Q8TD66, Q8TD67, Q9UNU7, Q9Y6B9 | Gene names | RGS7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RGS6_MOUSE
|
||||||
| NC score | 0.794371 (rank : 33) | θ value | 1.92812e-18 (rank : 19) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Z2H2, Q8BGI8 | Gene names | Rgs6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of G-protein signaling 6 (RGS6). | |||||
|
RGS6_HUMAN
|
||||||
| NC score | 0.770881 (rank : 34) | θ value | 1.16975e-15 (rank : 34) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P49758, O75576, O75577, Q8TE13, Q8TE14, Q8TE15, Q8TE16, Q8TE17, Q8TE18, Q8TE19, Q8TE20, Q8TE21, Q8TE22, Q9Y245, Q9Y647 | Gene names | RGS6 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 6 (RGS6) (S914). | |||||
|
AXN1_MOUSE
|
||||||
| NC score | 0.640013 (rank : 35) | θ value | 1.133e-10 (rank : 37) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O35625 | Gene names | Axin1, Axin, Fu | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (Fused protein). | |||||
|
AXN1_HUMAN
|
||||||
| NC score | 0.621802 (rank : 36) | θ value | 7.34386e-10 (rank : 38) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O15169, Q4TT26, Q4TT27, Q86YA7, Q8WVW6, Q96S28 | Gene names | AXIN1, AXIN | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (hAxin). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.611447 (rank : 37) | θ value | 1.38178e-16 (rank : 31) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
AXN2_HUMAN
|
||||||
| NC score | 0.608491 (rank : 38) | θ value | 1.38499e-08 (rank : 39) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Y2T1, Q9H3M6, Q9UH84 | Gene names | AXIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
AXN2_MOUSE
|
||||||
| NC score | 0.596286 (rank : 39) | θ value | 1.53129e-07 (rank : 40) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O88566, Q9QXJ6 | Gene names | Axin2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.545917 (rank : 40) | θ value | 1.38178e-16 (rank : 32) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
AKA10_MOUSE
|
||||||
| NC score | 0.340628 (rank : 41) | θ value | 7.1131e-05 (rank : 42) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O88845, Q5SUB5, Q5SUB7, Q7TPE7, Q8BQL6 | Gene names | Akap10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
AKA10_HUMAN
|
||||||
| NC score | 0.340573 (rank : 42) | θ value | 7.1131e-05 (rank : 41) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43572, Q96AJ7 | Gene names | AKAP10 | |||
|
Domain Architecture |

|
|||||
| Description | A kinase anchor protein 10, mitochondrial precursor (Protein kinase A- anchoring protein 10) (PRKA10) (Dual specificity A kinase-anchoring protein 2) (D-AKAP-2). | |||||
|
SNX13_MOUSE
|
||||||
| NC score | 0.278545 (rank : 43) | θ value | 0.0736092 (rank : 44) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6PHS6 | Gene names | Snx13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-13. | |||||
|
SNX13_HUMAN
|
||||||
| NC score | 0.272355 (rank : 44) | θ value | 0.0961366 (rank : 46) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y5W8, O94821, Q8WVZ2, Q8WXH8 | Gene names | SNX13, KIAA0713 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-13 (RGS domain- and PHOX domain-containing protein) (RGS-PX1). | |||||
|
SNX25_HUMAN
|
||||||
| NC score | 0.124365 (rank : 45) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H3E2 | Gene names | SNX25 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-25. | |||||
|
SNX14_MOUSE
|
||||||
| NC score | 0.052200 (rank : 46) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BHY8 | Gene names | Snx14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-14. | |||||
|
CTDP1_HUMAN
|
||||||
| NC score | 0.043365 (rank : 47) | θ value | 0.0961366 (rank : 45) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
CCD22_HUMAN
|
||||||
| NC score | 0.042290 (rank : 48) | θ value | 0.0431538 (rank : 43) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60826 | Gene names | CCDC22, CXorf37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 22. | |||||
|
ARHGB_HUMAN
|
||||||
| NC score | 0.034327 (rank : 49) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15085 | Gene names | ARHGEF11, KIAA0380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 11 (PDZ-RhoGEF). | |||||
|
LYSM3_MOUSE
|
||||||
| NC score | 0.033087 (rank : 50) | θ value | 0.279714 (rank : 48) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99LE3 | Gene names | Lysmd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LysM and putative peptidoglycan-binding domain-containing protein 3. | |||||
|
CABL2_HUMAN
|
||||||
| NC score | 0.029118 (rank : 51) | θ value | 0.21417 (rank : 47) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BTV7, Q5JWL0, Q9BYK0 | Gene names | CABLES2, C20orf150 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
RPP25_HUMAN
|
||||||
| NC score | 0.025801 (rank : 52) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BUL9, Q9NX88 | Gene names | RPP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease P protein subunit p25 (EC 3.1.26.5) (RNase P protein subunit p25). | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.023804 (rank : 53) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
ZNT3_HUMAN
|
||||||
| NC score | 0.020795 (rank : 54) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99726 | Gene names | SLC30A3, ZNT3 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc transporter 3 (ZnT-3) (Solute carrier family 30 member 3). | |||||
|
JIP2_HUMAN
|
||||||
| NC score | 0.019944 (rank : 55) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13387, Q96G62, Q99771, Q9NZ59, Q9UKQ4 | Gene names | MAPK8IP2, IB2, JIP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.016304 (rank : 56) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.016044 (rank : 57) | θ value | 0.62314 (rank : 49) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
CS029_MOUSE
|
||||||
| NC score | 0.015923 (rank : 58) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CS00 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf29 homolog. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.015385 (rank : 59) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
KCNH2_HUMAN
|
||||||
| NC score | 0.014110 (rank : 60) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
KCNH2_MOUSE
|
||||||
| NC score | 0.013127 (rank : 61) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.011412 (rank : 62) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
MELPH_MOUSE
|
||||||
| NC score | 0.011092 (rank : 63) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91V27, Q99N53 | Gene names | Mlph, Ln, Slac2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Leaden protein) (Synaptotagmin-like protein 2a) (Slac2-a) (Slp homolog lacking C2 domains a). | |||||
|
CRCM_HUMAN
|
||||||
| NC score | 0.009188 (rank : 64) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23508 | Gene names | MCC | |||
|
Domain Architecture |

|
|||||
| Description | Colorectal mutant cancer protein (Protein MCC). | |||||
|
SPTA1_HUMAN
|
||||||
| NC score | 0.008121 (rank : 65) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
SPTA1_MOUSE
|
||||||
| NC score | 0.007901 (rank : 66) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P08032, P97502 | Gene names | Spta1, Spna1, Spta | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.007448 (rank : 67) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
RGNEF_MOUSE
|
||||||
| NC score | 0.006962 (rank : 68) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97433 | Gene names | Rgnef, Rhoip2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-guanine nucleotide exchange factor (Rho-interacting protein 2) (RhoGEF) (RIP2). | |||||
|
PLVAP_MOUSE
|
||||||
| NC score | 0.006740 (rank : 69) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91VC4, Q99JB1 | Gene names | Plvap, Pv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plasmalemma vesicle-associated protein (Plasmalemma vesicle protein 1) (PV-1) (MECA-32 antigen). | |||||
|
ZFY28_HUMAN
|
||||||
| NC score | 0.006446 (rank : 70) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HCC9 | Gene names | ZFYVE28, KIAA1643 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 28. | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.006336 (rank : 71) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
LY10_HUMAN
|
||||||
| NC score | 0.005285 (rank : 72) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
MTMR5_HUMAN
|
||||||
| NC score | 0.005013 (rank : 73) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
ITA2B_MOUSE
|
||||||
| NC score | 0.004457 (rank : 74) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QUM0, Q64229, Q9Z2M0 | Gene names | Itga2b | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-IIb precursor (Platelet membrane glycoprotein IIb) (GPalpha IIb) (GPIIb) (CD41 antigen) [Contains: Integrin alpha-IIb heavy chain; Integrin alpha-IIb light chain]. | |||||
|
KIF1B_HUMAN
|
||||||
| NC score | 0.002761 (rank : 75) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60333, Q96Q94, Q9BV80, Q9P280 | Gene names | KIF1B, KIAA0591, KIAA1448 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF1B (Klp). | |||||
|
MAST3_HUMAN
|
||||||
| NC score | 0.000544 (rank : 76) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1128 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60307, Q7LDZ8, Q9UPI0 | Gene names | MAST3, KIAA0561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 3 (EC 2.7.11.1). | |||||
|
KI21A_MOUSE
|
||||||
| NC score | 0.000417 (rank : 77) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
KI21A_HUMAN
|
||||||
| NC score | -0.000478 (rank : 78) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
KLF4_HUMAN
|
||||||
| NC score | -0.003312 (rank : 79) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43474, P78338, Q9UNP3 | Gene names | KLF4, EZF, GKLF | |||
|
Domain Architecture |

|
|||||
| Description | Krueppel-like factor 4 (Epithelial zinc-finger protein EZF) (Gut- enriched krueppel-like factor). | |||||