Please be patient as the page loads
|
PSD7_MOUSE
|
||||||
| SwissProt Accessions | P26516, Q3TW61, Q8C0I8 | Gene names | Psmd7, Mov-34, Mov34 | |||
|
Domain Architecture |
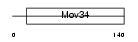
|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 7 (26S proteasome regulatory subunit rpn8) (26S proteasome regulatory subunit S12) (Proteasome subunit p40) (Mov34 protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PSD7_HUMAN
|
||||||
| θ value | 3.16907e-146 (rank : 1) | NC score | 0.997111 (rank : 2) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P51665, Q6PKI2, Q96E97 | Gene names | PSMD7, MOV34L | |||
|
Domain Architecture |
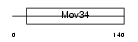
|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 7 (26S proteasome regulatory subunit rpn8) (26S proteasome regulatory subunit S12) (Proteasome subunit p40) (Mov34 protein homolog). | |||||
|
PSD7_MOUSE
|
||||||
| θ value | 7.05993e-146 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P26516, Q3TW61, Q8C0I8 | Gene names | Psmd7, Mov-34, Mov34 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 7 (26S proteasome regulatory subunit rpn8) (26S proteasome regulatory subunit S12) (Proteasome subunit p40) (Mov34 protein). | |||||
|
IF35_HUMAN
|
||||||
| θ value | 5.59698e-26 (rank : 3) | NC score | 0.749525 (rank : 3) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00303, Q8N978 | Gene names | EIF3S5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
IF35_MOUSE
|
||||||
| θ value | 2.12685e-25 (rank : 4) | NC score | 0.709708 (rank : 6) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
CSN6_MOUSE
|
||||||
| θ value | 2.5182e-18 (rank : 5) | NC score | 0.722123 (rank : 4) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88545 | Gene names | Cops6, Csn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | COP9 signalosome complex subunit 6 (Signalosome subunit 6) (SGN6) (JAB1-containing signalosome subunit 6). | |||||
|
CSN6_HUMAN
|
||||||
| θ value | 3.28887e-18 (rank : 6) | NC score | 0.719749 (rank : 5) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7L5N1, O15387 | Gene names | COPS6, CSN6, HVIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | COP9 signalosome complex subunit 6 (Signalosome subunit 6) (SGN6) (JAB1-containing signalosome subunit 6) (Vpr-interacting protein) (hVIP) (MOV34 homolog). | |||||
|
PSDE_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 7) | NC score | 0.302163 (rank : 7) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O00487, O00176 | Gene names | PSMD14, POH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 14 (26S proteasome regulatory subunit rpn11) (26S proteasome-associated PAD1 homolog 1). | |||||
|
PSDE_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 8) | NC score | 0.302163 (rank : 8) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35593, Q3UB50, Q9CZY6 | Gene names | Psmd14, Pad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 14 (26S proteasome regulatory subunit rpn11) (MAD1). | |||||
|
CSN5_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 9) | NC score | 0.299443 (rank : 9) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92905, O15386, Q6AW95, Q86WQ4, Q9BQ17 | Gene names | COPS5, CSN5, JAB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | COP9 signalosome complex subunit 5 (EC 3.4.-.-) (Signalosome subunit 5) (SGN5) (Jun activation domain-binding protein 1). | |||||
|
CSN5_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 10) | NC score | 0.299443 (rank : 10) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35864, Q3UA70, Q8C1S1 | Gene names | Cops5, Csn5, Jab1, Kic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | COP9 signalosome complex subunit 5 (EC 3.4.-.-) (Signalosome subunit 5) (SGN5) (Jun activation domain-binding protein 1) (Kip1 C-terminus- interacting protein 2). | |||||
|
RGS9_HUMAN
|
||||||
| θ value | 0.163984 (rank : 11) | NC score | 0.051662 (rank : 15) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75916, O75573, Q696R2, Q8TD64, Q8TD65, Q9HC32, Q9HC33 | Gene names | RGS9 | |||
|
Domain Architecture |
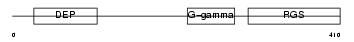
|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
RGS9_MOUSE
|
||||||
| θ value | 0.47712 (rank : 12) | NC score | 0.047117 (rank : 16) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O54828, Q9Z0S0 | Gene names | Rgs9 | |||
|
Domain Architecture |
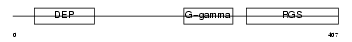
|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
IF33_MOUSE
|
||||||
| θ value | 0.813845 (rank : 13) | NC score | 0.225714 (rank : 11) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91WK2 | Gene names | Eif3s3 | |||
|
Domain Architecture |
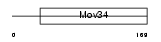
|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 3 (eIF-3 gamma) (eIF3 p40 subunit) (eIF3h). | |||||
|
IF33_HUMAN
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.219885 (rank : 12) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15372 | Gene names | EIF3S3 | |||
|
Domain Architecture |
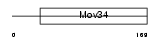
|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 3 (eIF-3 gamma) (eIF3 p40 subunit) (eIF3h). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 1.81305 (rank : 15) | NC score | 0.009535 (rank : 22) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
SMC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.017330 (rank : 19) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
ACSL5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 17) | NC score | 0.021663 (rank : 18) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULC5, Q6UX44, Q9UIU4 | Gene names | ACSL5, ACS5, FACL5 | |||
|
Domain Architecture |

|
|||||
| Description | Long-chain-fatty-acid--CoA ligase 5 (EC 6.2.1.3) (Long-chain acyl-CoA synthetase 5) (LACS 5). | |||||
|
ACSL5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.021832 (rank : 17) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8JZR0 | Gene names | Acsl5, Facl5 | |||
|
Domain Architecture |

|
|||||
| Description | Long-chain-fatty-acid--CoA ligase 5 (EC 6.2.1.3) (Long-chain acyl-CoA synthetase 5) (LACS 5). | |||||
|
LIPA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.006079 (rank : 25) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.011135 (rank : 21) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
UBP48_MOUSE
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.008263 (rank : 23) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
ANC5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.013714 (rank : 20) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BTZ4, Q3TGA7, Q80UJ6, Q80W43, Q8BWW7, Q8C0F7, Q9CRX0, Q9CSL1, Q9JJB8 | Gene names | Anapc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 5 (APC5) (Cyclosome subunit 5). | |||||
|
K1H2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 23) | NC score | 0.004328 (rank : 26) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62168 | Gene names | Krtha2, Hka2, Krt1-2 | |||
|
Domain Architecture |
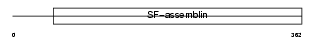
|
|||||
| Description | Keratin, type I cuticular Ha2 (Hair keratin, type I Ha2). | |||||
|
PK3CG_MOUSE
|
||||||
| θ value | 8.99809 (rank : 24) | NC score | 0.006501 (rank : 24) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHG7 | Gene names | Pik3cg, Pi3kg1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
BRCC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 25) | NC score | 0.056640 (rank : 13) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P46736, Q16107, Q9BTZ6 | Gene names | BRCC3, C6.1A, CXorf53 | |||
|
Domain Architecture |
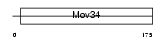
|
|||||
| Description | BRCA1/BRCA2-containing complex subunit 3. | |||||
|
BRCC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 26) | NC score | 0.054094 (rank : 14) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P46737, Q9D025 | Gene names | Brcc3, C6.1A | |||
|
Domain Architecture |
|
|||||
| Description | BRCA1/BRCA2-containing complex subunit 3. | |||||
|
PSD7_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 7.05993e-146 (rank : 2) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P26516, Q3TW61, Q8C0I8 | Gene names | Psmd7, Mov-34, Mov34 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 7 (26S proteasome regulatory subunit rpn8) (26S proteasome regulatory subunit S12) (Proteasome subunit p40) (Mov34 protein). | |||||
|
PSD7_HUMAN
|
||||||
| NC score | 0.997111 (rank : 2) | θ value | 3.16907e-146 (rank : 1) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P51665, Q6PKI2, Q96E97 | Gene names | PSMD7, MOV34L | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 7 (26S proteasome regulatory subunit rpn8) (26S proteasome regulatory subunit S12) (Proteasome subunit p40) (Mov34 protein homolog). | |||||
|
IF35_HUMAN
|
||||||
| NC score | 0.749525 (rank : 3) | θ value | 5.59698e-26 (rank : 3) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00303, Q8N978 | Gene names | EIF3S5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
CSN6_MOUSE
|
||||||
| NC score | 0.722123 (rank : 4) | θ value | 2.5182e-18 (rank : 5) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88545 | Gene names | Cops6, Csn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | COP9 signalosome complex subunit 6 (Signalosome subunit 6) (SGN6) (JAB1-containing signalosome subunit 6). | |||||
|
CSN6_HUMAN
|
||||||
| NC score | 0.719749 (rank : 5) | θ value | 3.28887e-18 (rank : 6) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7L5N1, O15387 | Gene names | COPS6, CSN6, HVIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | COP9 signalosome complex subunit 6 (Signalosome subunit 6) (SGN6) (JAB1-containing signalosome subunit 6) (Vpr-interacting protein) (hVIP) (MOV34 homolog). | |||||
|
IF35_MOUSE
|
||||||
| NC score | 0.709708 (rank : 6) | θ value | 2.12685e-25 (rank : 4) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
PSDE_HUMAN
|
||||||
| NC score | 0.302163 (rank : 7) | θ value | 0.00020696 (rank : 7) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O00487, O00176 | Gene names | PSMD14, POH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 14 (26S proteasome regulatory subunit rpn11) (26S proteasome-associated PAD1 homolog 1). | |||||
|
PSDE_MOUSE
|
||||||
| NC score | 0.302163 (rank : 8) | θ value | 0.00020696 (rank : 8) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35593, Q3UB50, Q9CZY6 | Gene names | Psmd14, Pad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 14 (26S proteasome regulatory subunit rpn11) (MAD1). | |||||
|
CSN5_HUMAN
|
||||||
| NC score | 0.299443 (rank : 9) | θ value | 0.00175202 (rank : 9) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92905, O15386, Q6AW95, Q86WQ4, Q9BQ17 | Gene names | COPS5, CSN5, JAB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | COP9 signalosome complex subunit 5 (EC 3.4.-.-) (Signalosome subunit 5) (SGN5) (Jun activation domain-binding protein 1). | |||||
|
CSN5_MOUSE
|
||||||
| NC score | 0.299443 (rank : 10) | θ value | 0.00175202 (rank : 10) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35864, Q3UA70, Q8C1S1 | Gene names | Cops5, Csn5, Jab1, Kic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | COP9 signalosome complex subunit 5 (EC 3.4.-.-) (Signalosome subunit 5) (SGN5) (Jun activation domain-binding protein 1) (Kip1 C-terminus- interacting protein 2). | |||||
|
IF33_MOUSE
|
||||||
| NC score | 0.225714 (rank : 11) | θ value | 0.813845 (rank : 13) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91WK2 | Gene names | Eif3s3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 3 (eIF-3 gamma) (eIF3 p40 subunit) (eIF3h). | |||||
|
IF33_HUMAN
|
||||||
| NC score | 0.219885 (rank : 12) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15372 | Gene names | EIF3S3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 3 (eIF-3 gamma) (eIF3 p40 subunit) (eIF3h). | |||||
|
BRCC3_HUMAN
|
||||||
| NC score | 0.056640 (rank : 13) | θ value | θ > 10 (rank : 25) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P46736, Q16107, Q9BTZ6 | Gene names | BRCC3, C6.1A, CXorf53 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1/BRCA2-containing complex subunit 3. | |||||
|
BRCC3_MOUSE
|
||||||
| NC score | 0.054094 (rank : 14) | θ value | θ > 10 (rank : 26) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P46737, Q9D025 | Gene names | Brcc3, C6.1A | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1/BRCA2-containing complex subunit 3. | |||||
|
RGS9_HUMAN
|
||||||
| NC score | 0.051662 (rank : 15) | θ value | 0.163984 (rank : 11) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75916, O75573, Q696R2, Q8TD64, Q8TD65, Q9HC32, Q9HC33 | Gene names | RGS9 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
RGS9_MOUSE
|
||||||
| NC score | 0.047117 (rank : 16) | θ value | 0.47712 (rank : 12) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O54828, Q9Z0S0 | Gene names | Rgs9 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 9 (RGS9). | |||||
|
ACSL5_MOUSE
|
||||||
| NC score | 0.021832 (rank : 17) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8JZR0 | Gene names | Acsl5, Facl5 | |||
|
Domain Architecture |

|
|||||
| Description | Long-chain-fatty-acid--CoA ligase 5 (EC 6.2.1.3) (Long-chain acyl-CoA synthetase 5) (LACS 5). | |||||
|
ACSL5_HUMAN
|
||||||
| NC score | 0.021663 (rank : 18) | θ value | 5.27518 (rank : 17) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ULC5, Q6UX44, Q9UIU4 | Gene names | ACSL5, ACS5, FACL5 | |||
|
Domain Architecture |

|
|||||
| Description | Long-chain-fatty-acid--CoA ligase 5 (EC 6.2.1.3) (Long-chain acyl-CoA synthetase 5) (LACS 5). | |||||
|
SMC2_MOUSE
|
||||||
| NC score | 0.017330 (rank : 19) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
ANC5_MOUSE
|
||||||
| NC score | 0.013714 (rank : 20) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BTZ4, Q3TGA7, Q80UJ6, Q80W43, Q8BWW7, Q8C0F7, Q9CRX0, Q9CSL1, Q9JJB8 | Gene names | Anapc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 5 (APC5) (Cyclosome subunit 5). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.011135 (rank : 21) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.009535 (rank : 22) | θ value | 1.81305 (rank : 15) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
UBP48_MOUSE
|
||||||
| NC score | 0.008263 (rank : 23) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3V0C5, Q3TM39, Q571J9, Q8VDJ1 | Gene names | Usp48, Kiaa4202 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 48 (EC 3.1.2.15) (Ubiquitin thioesterase 48) (Ubiquitin-specific-processing protease 48) (Deubiquitinating enzyme 48). | |||||
|
PK3CG_MOUSE
|
||||||
| NC score | 0.006501 (rank : 24) | θ value | 8.99809 (rank : 24) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHG7 | Gene names | Pik3cg, Pi3kg1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
LIPA1_HUMAN
|
||||||
| NC score | 0.006079 (rank : 25) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
K1H2_MOUSE
|
||||||
| NC score | 0.004328 (rank : 26) | θ value | 8.99809 (rank : 23) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62168 | Gene names | Krtha2, Hka2, Krt1-2 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha2 (Hair keratin, type I Ha2). | |||||