Please be patient as the page loads
|
BRCC3_MOUSE
|
||||||
| SwissProt Accessions | P46737, Q9D025 | Gene names | Brcc3, C6.1A | |||
|
Domain Architecture |
|
|||||
| Description | BRCA1/BRCA2-containing complex subunit 3. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
BRCC3_MOUSE
|
||||||
| θ value | 2.68277e-145 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P46737, Q9D025 | Gene names | Brcc3, C6.1A | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1/BRCA2-containing complex subunit 3. | |||||
|
BRCC3_HUMAN
|
||||||
| θ value | 5.41814e-138 (rank : 2) | NC score | 0.998535 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P46736, Q16107, Q9BTZ6 | Gene names | BRCC3, C6.1A, CXorf53 | |||
|
Domain Architecture |
|
|||||
| Description | BRCA1/BRCA2-containing complex subunit 3. | |||||
|
PSDE_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 3) | NC score | 0.515457 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00487, O00176 | Gene names | PSMD14, POH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 14 (26S proteasome regulatory subunit rpn11) (26S proteasome-associated PAD1 homolog 1). | |||||
|
PSDE_MOUSE
|
||||||
| θ value | 1.63604e-09 (rank : 4) | NC score | 0.515457 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35593, Q3UB50, Q9CZY6 | Gene names | Psmd14, Pad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 14 (26S proteasome regulatory subunit rpn11) (MAD1). | |||||
|
CSN5_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 5) | NC score | 0.424768 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92905, O15386, Q6AW95, Q86WQ4, Q9BQ17 | Gene names | COPS5, CSN5, JAB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | COP9 signalosome complex subunit 5 (EC 3.4.-.-) (Signalosome subunit 5) (SGN5) (Jun activation domain-binding protein 1). | |||||
|
CSN5_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 6) | NC score | 0.424768 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35864, Q3UA70, Q8C1S1 | Gene names | Cops5, Csn5, Jab1, Kic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | COP9 signalosome complex subunit 5 (EC 3.4.-.-) (Signalosome subunit 5) (SGN5) (Jun activation domain-binding protein 1) (Kip1 C-terminus- interacting protein 2). | |||||
|
MYSM1_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 7) | NC score | 0.235090 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5VVJ2, Q7Z3G8, Q96PX3 | Gene names | MYSM1, KIAA1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
MYSM1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 8) | NC score | 0.228173 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q69Z66, Q3TPV7, Q8BRP5, Q8C4N1 | Gene names | Mysm1, Kiaa1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
STABP_HUMAN
|
||||||
| θ value | 0.125558 (rank : 9) | NC score | 0.180383 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95630, Q3MJE7 | Gene names | STAMBP, AMSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STAM-binding protein (EC 3.1.2.15) (Associated molecule with the SH3 domain of STAM). | |||||
|
STABP_MOUSE
|
||||||
| θ value | 0.163984 (rank : 10) | NC score | 0.171682 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9CQ26, Q3UTI9 | Gene names | Stambp, Amsh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STAM-binding protein (EC 3.1.2.15) (Associated molecule with the SH3 domain of STAM). | |||||
|
STALP_HUMAN
|
||||||
| θ value | 0.21417 (rank : 11) | NC score | 0.189064 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96FJ0, Q5T9N4, Q5T9N9, Q7Z420, Q9P2H4 | Gene names | STAMBPL1, AMSHLP, KIAA1373 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AMSH-like protease (EC 3.1.2.15) (AMSH-LP) (STAM-binding protein-like 1). | |||||
|
STALP_MOUSE
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.186160 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q76N33, Q76LY0, Q8VEK5 | Gene names | Stambpl1, Amshlp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AMSH-like protease (EC 3.1.2.15) (AMSH-LP) (STAM-binding protein-like 1) (AMSH family protein) (AMSH-FP). | |||||
|
K0555_HUMAN
|
||||||
| θ value | 1.06291 (rank : 13) | NC score | 0.020227 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.020653 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
AT10A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 15) | NC score | 0.012820 (rank : 23) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54827, Q8R3B8 | Gene names | Atp10a, Atpc5, Pfatp | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (P-locus fat-associated ATPase). | |||||
|
CP135_HUMAN
|
||||||
| θ value | 5.27518 (rank : 16) | NC score | 0.008793 (rank : 24) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
ZN699_HUMAN
|
||||||
| θ value | 5.27518 (rank : 17) | NC score | -0.000053 (rank : 26) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q32M78, Q8N9A1 | Gene names | ZNF699 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 699 (Hangover homolog). | |||||
|
DMD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | 0.014549 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
NCAM2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.004360 (rank : 25) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35136, O35962 | Gene names | Ncam2, Ocam, Rncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 2 precursor (N-CAM 2) (RB-8 neural cell adhesion molecule) (R4B12). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 20) | NC score | 0.020921 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.020571 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
DMD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.014788 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
IF33_HUMAN
|
||||||
| θ value | θ > 10 (rank : 23) | NC score | 0.068613 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15372 | Gene names | EIF3S3 | |||
|
Domain Architecture |
|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 3 (eIF-3 gamma) (eIF3 p40 subunit) (eIF3h). | |||||
|
IF33_MOUSE
|
||||||
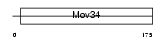
| θ value | θ > 10 (rank : 24) | NC score | 0.068917 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91WK2 | Gene names | Eif3s3 | |||
|
Domain Architecture |
|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 3 (eIF-3 gamma) (eIF3 p40 subunit) (eIF3h). | |||||
|
PSD7_HUMAN
|
||||||
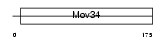
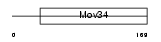
| θ value | θ > 10 (rank : 25) | NC score | 0.054010 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51665, Q6PKI2, Q96E97 | Gene names | PSMD7, MOV34L | |||
|
Domain Architecture |
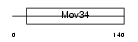
|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 7 (26S proteasome regulatory subunit rpn8) (26S proteasome regulatory subunit S12) (Proteasome subunit p40) (Mov34 protein homolog). | |||||
|
PSD7_MOUSE
|
||||||
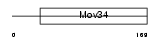
| θ value | θ > 10 (rank : 26) | NC score | 0.054094 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26516, Q3TW61, Q8C0I8 | Gene names | Psmd7, Mov-34, Mov34 | |||
|
Domain Architecture |
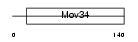
|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 7 (26S proteasome regulatory subunit rpn8) (26S proteasome regulatory subunit S12) (Proteasome subunit p40) (Mov34 protein). | |||||
|
BRCC3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.68277e-145 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P46737, Q9D025 | Gene names | Brcc3, C6.1A | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1/BRCA2-containing complex subunit 3. | |||||
|
BRCC3_HUMAN
|
||||||
| NC score | 0.998535 (rank : 2) | θ value | 5.41814e-138 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P46736, Q16107, Q9BTZ6 | Gene names | BRCC3, C6.1A, CXorf53 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1/BRCA2-containing complex subunit 3. | |||||
|
PSDE_HUMAN
|
||||||
| NC score | 0.515457 (rank : 3) | θ value | 1.63604e-09 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00487, O00176 | Gene names | PSMD14, POH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 14 (26S proteasome regulatory subunit rpn11) (26S proteasome-associated PAD1 homolog 1). | |||||
|
PSDE_MOUSE
|
||||||
| NC score | 0.515457 (rank : 4) | θ value | 1.63604e-09 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35593, Q3UB50, Q9CZY6 | Gene names | Psmd14, Pad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 14 (26S proteasome regulatory subunit rpn11) (MAD1). | |||||
|
CSN5_HUMAN
|
||||||
| NC score | 0.424768 (rank : 5) | θ value | 4.1701e-05 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92905, O15386, Q6AW95, Q86WQ4, Q9BQ17 | Gene names | COPS5, CSN5, JAB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | COP9 signalosome complex subunit 5 (EC 3.4.-.-) (Signalosome subunit 5) (SGN5) (Jun activation domain-binding protein 1). | |||||
|
CSN5_MOUSE
|
||||||
| NC score | 0.424768 (rank : 6) | θ value | 4.1701e-05 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35864, Q3UA70, Q8C1S1 | Gene names | Cops5, Csn5, Jab1, Kic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | COP9 signalosome complex subunit 5 (EC 3.4.-.-) (Signalosome subunit 5) (SGN5) (Jun activation domain-binding protein 1) (Kip1 C-terminus- interacting protein 2). | |||||
|
MYSM1_HUMAN
|
||||||
| NC score | 0.235090 (rank : 7) | θ value | 0.00509761 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5VVJ2, Q7Z3G8, Q96PX3 | Gene names | MYSM1, KIAA1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
MYSM1_MOUSE
|
||||||
| NC score | 0.228173 (rank : 8) | θ value | 0.00509761 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q69Z66, Q3TPV7, Q8BRP5, Q8C4N1 | Gene names | Mysm1, Kiaa1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
STALP_HUMAN
|
||||||
| NC score | 0.189064 (rank : 9) | θ value | 0.21417 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96FJ0, Q5T9N4, Q5T9N9, Q7Z420, Q9P2H4 | Gene names | STAMBPL1, AMSHLP, KIAA1373 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AMSH-like protease (EC 3.1.2.15) (AMSH-LP) (STAM-binding protein-like 1). | |||||
|
STALP_MOUSE
|
||||||
| NC score | 0.186160 (rank : 10) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q76N33, Q76LY0, Q8VEK5 | Gene names | Stambpl1, Amshlp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AMSH-like protease (EC 3.1.2.15) (AMSH-LP) (STAM-binding protein-like 1) (AMSH family protein) (AMSH-FP). | |||||
|
STABP_HUMAN
|
||||||
| NC score | 0.180383 (rank : 11) | θ value | 0.125558 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95630, Q3MJE7 | Gene names | STAMBP, AMSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STAM-binding protein (EC 3.1.2.15) (Associated molecule with the SH3 domain of STAM). | |||||
|
STABP_MOUSE
|
||||||
| NC score | 0.171682 (rank : 12) | θ value | 0.163984 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9CQ26, Q3UTI9 | Gene names | Stambp, Amsh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STAM-binding protein (EC 3.1.2.15) (Associated molecule with the SH3 domain of STAM). | |||||
|
IF33_MOUSE
|
||||||
| NC score | 0.068917 (rank : 13) | θ value | θ > 10 (rank : 24) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91WK2 | Gene names | Eif3s3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 3 (eIF-3 gamma) (eIF3 p40 subunit) (eIF3h). | |||||
|
IF33_HUMAN
|
||||||
| NC score | 0.068613 (rank : 14) | θ value | θ > 10 (rank : 23) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15372 | Gene names | EIF3S3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 3 (eIF-3 gamma) (eIF3 p40 subunit) (eIF3h). | |||||
|
PSD7_MOUSE
|
||||||
| NC score | 0.054094 (rank : 15) | θ value | θ > 10 (rank : 26) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26516, Q3TW61, Q8C0I8 | Gene names | Psmd7, Mov-34, Mov34 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 7 (26S proteasome regulatory subunit rpn8) (26S proteasome regulatory subunit S12) (Proteasome subunit p40) (Mov34 protein). | |||||
|
PSD7_HUMAN
|
||||||
| NC score | 0.054010 (rank : 16) | θ value | θ > 10 (rank : 25) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51665, Q6PKI2, Q96E97 | Gene names | PSMD7, MOV34L | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 7 (26S proteasome regulatory subunit rpn8) (26S proteasome regulatory subunit S12) (Proteasome subunit p40) (Mov34 protein homolog). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.020921 (rank : 17) | θ value | 6.88961 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.020653 (rank : 18) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.020571 (rank : 19) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
K0555_HUMAN
|
||||||
| NC score | 0.020227 (rank : 20) | θ value | 1.06291 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.014788 (rank : 21) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.014549 (rank : 22) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
AT10A_MOUSE
|
||||||
| NC score | 0.012820 (rank : 23) | θ value | 5.27518 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54827, Q8R3B8 | Gene names | Atp10a, Atpc5, Pfatp | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (P-locus fat-associated ATPase). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.008793 (rank : 24) | θ value | 5.27518 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
NCAM2_MOUSE
|
||||||
| NC score | 0.004360 (rank : 25) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35136, O35962 | Gene names | Ncam2, Ocam, Rncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 2 precursor (N-CAM 2) (RB-8 neural cell adhesion molecule) (R4B12). | |||||
|
ZN699_HUMAN
|
||||||
| NC score | -0.000053 (rank : 26) | θ value | 5.27518 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 773 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q32M78, Q8N9A1 | Gene names | ZNF699 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 699 (Hangover homolog). | |||||