Please be patient as the page loads
|
MYSM1_MOUSE
|
||||||
| SwissProt Accessions | Q69Z66, Q3TPV7, Q8BRP5, Q8C4N1 | Gene names | Mysm1, Kiaa1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MYSM1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.964448 (rank : 2) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5VVJ2, Q7Z3G8, Q96PX3 | Gene names | MYSM1, KIAA1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
MYSM1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 134 | |
| SwissProt Accessions | Q69Z66, Q3TPV7, Q8BRP5, Q8C4N1 | Gene names | Mysm1, Kiaa1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
SMRC2_MOUSE
|
||||||
| θ value | 9.26847e-13 (rank : 3) | NC score | 0.391063 (rank : 4) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 1.2105e-12 (rank : 4) | NC score | 0.390624 (rank : 5) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SMRC1_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 5) | NC score | 0.393637 (rank : 3) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
SMRC1_MOUSE
|
||||||
| θ value | 1.9326e-10 (rank : 6) | NC score | 0.389622 (rank : 6) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
PSDE_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 7) | NC score | 0.303773 (rank : 7) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00487, O00176 | Gene names | PSMD14, POH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 14 (26S proteasome regulatory subunit rpn11) (26S proteasome-associated PAD1 homolog 1). | |||||
|
PSDE_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 8) | NC score | 0.303773 (rank : 8) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35593, Q3UB50, Q9CZY6 | Gene names | Psmd14, Pad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 14 (26S proteasome regulatory subunit rpn11) (MAD1). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 9) | NC score | 0.262459 (rank : 11) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
RCOR3_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 10) | NC score | 0.263840 (rank : 10) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 11) | NC score | 0.267688 (rank : 9) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 12) | NC score | 0.260955 (rank : 12) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 13) | NC score | 0.254071 (rank : 13) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
LSD1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 14) | NC score | 0.154527 (rank : 30) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
LSD1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 15) | NC score | 0.162106 (rank : 29) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZQ88, Q6PB53, Q8VEA1 | Gene names | Aof2, Kiaa0601, Lsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
MYBA_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 16) | NC score | 0.197777 (rank : 23) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10243 | Gene names | MYBL1, AMYB | |||
|
Domain Architecture |
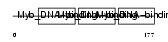
|
|||||
| Description | Myb-related protein A (A-Myb). | |||||
|
MYBA_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 17) | NC score | 0.197564 (rank : 24) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51960 | Gene names | Mybl1, Amyb | |||
|
Domain Architecture |
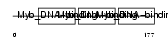
|
|||||
| Description | Myb-related protein A (A-Myb). | |||||
|
RCOR1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 18) | NC score | 0.244799 (rank : 14) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UKL0, Q15044, Q6P2I9, Q86VG5 | Gene names | RCOR1, KIAA0071, RCOR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
MYB_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 19) | NC score | 0.193991 (rank : 25) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P06876, Q61929 | Gene names | Myb | |||
|
Domain Architecture |

|
|||||
| Description | Myb proto-oncogene protein (C-myb). | |||||
|
RCOR2_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 20) | NC score | 0.235229 (rank : 16) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IZ40, Q96FP3 | Gene names | RCOR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2. | |||||
|
RCOR2_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 21) | NC score | 0.234365 (rank : 17) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8C796, Q9JMK4 | Gene names | Rcor2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2 (M-CoREST). | |||||
|
BRCC3_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 22) | NC score | 0.229493 (rank : 18) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P46736, Q16107, Q9BTZ6 | Gene names | BRCC3, C6.1A, CXorf53 | |||
|
Domain Architecture |
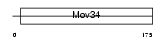
|
|||||
| Description | BRCA1/BRCA2-containing complex subunit 3. | |||||
|
BRCC3_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 23) | NC score | 0.228173 (rank : 19) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P46737, Q9D025 | Gene names | Brcc3, C6.1A | |||
|
Domain Architecture |
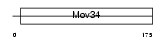
|
|||||
| Description | BRCA1/BRCA2-containing complex subunit 3. | |||||
|
CSN5_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 24) | NC score | 0.226418 (rank : 20) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92905, O15386, Q6AW95, Q86WQ4, Q9BQ17 | Gene names | COPS5, CSN5, JAB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | COP9 signalosome complex subunit 5 (EC 3.4.-.-) (Signalosome subunit 5) (SGN5) (Jun activation domain-binding protein 1). | |||||
|
CSN5_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 25) | NC score | 0.226418 (rank : 21) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35864, Q3UA70, Q8C1S1 | Gene names | Cops5, Csn5, Jab1, Kic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | COP9 signalosome complex subunit 5 (EC 3.4.-.-) (Signalosome subunit 5) (SGN5) (Jun activation domain-binding protein 1) (Kip1 C-terminus- interacting protein 2). | |||||
|
MYB_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 26) | NC score | 0.187541 (rank : 26) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P10242, P78391, P78392, P78525, P78526, Q14023, Q14024, Q9UE83 | Gene names | MYB | |||
|
Domain Architecture |
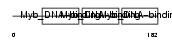
|
|||||
| Description | Myb proto-oncogene protein (C-myb). | |||||
|
RCOR1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 27) | NC score | 0.239399 (rank : 15) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CFE3, Q3V092, Q8BK28, Q8CHI2 | Gene names | Rcor1, D12Wsu95e, Kiaa0071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 28) | NC score | 0.050145 (rank : 75) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RCOR3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 29) | NC score | 0.216448 (rank : 22) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6PGA0 | Gene names | Rcor3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 30) | NC score | 0.047943 (rank : 80) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
DDX23_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 31) | NC score | 0.024853 (rank : 134) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
NIN_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 32) | NC score | 0.051285 (rank : 70) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 33) | NC score | 0.041638 (rank : 93) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
MYBB_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 34) | NC score | 0.164506 (rank : 28) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P10244 | Gene names | MYBL2, BMYB | |||
|
Domain Architecture |

|
|||||
| Description | Myb-related protein B (B-Myb). | |||||
|
MYBB_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 35) | NC score | 0.166724 (rank : 27) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P48972 | Gene names | Mybl2, Bmyb | |||
|
Domain Architecture |

|
|||||
| Description | Myb-related protein B (B-Myb). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 36) | NC score | 0.048140 (rank : 79) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
TEX14_MOUSE
|
||||||
| θ value | 0.125558 (rank : 37) | NC score | 0.028136 (rank : 124) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1333 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7M6U3, Q3UT36, Q5NC10, Q5NC11, Q8CGK1, Q8CGK2, Q99MV8 | Gene names | Tex14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
INCE_MOUSE
|
||||||
| θ value | 0.163984 (rank : 38) | NC score | 0.046727 (rank : 83) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
INVO_HUMAN
|
||||||
| θ value | 0.21417 (rank : 39) | NC score | 0.043121 (rank : 88) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 40) | NC score | 0.017746 (rank : 148) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
KI13A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 41) | NC score | 0.029688 (rank : 118) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H1H9, Q9H1H8 | Gene names | KIF13A, RBKIN | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13A (Kinesin-like protein RBKIN). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 42) | NC score | 0.093581 (rank : 34) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
NCKX1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 43) | NC score | 0.043378 (rank : 87) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.279714 (rank : 44) | NC score | 0.038719 (rank : 98) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PININ_HUMAN
|
||||||
| θ value | 0.279714 (rank : 45) | NC score | 0.054882 (rank : 58) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
AP3B1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 46) | NC score | 0.031027 (rank : 117) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
NP1L3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 47) | NC score | 0.049851 (rank : 76) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
PDXL2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 48) | NC score | 0.046789 (rank : 82) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NZ53, Q6UVY4, Q8WUV6 | Gene names | PODXL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
CCAR1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 49) | NC score | 0.052599 (rank : 64) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8CH18, Q6AXC9, Q6PAR2, Q80XE4, Q8BJY0, Q8BVN2, Q8CGG1, Q9CSR5 | Gene names | Ccar1, Carp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1). | |||||
|
G3BP2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 50) | NC score | 0.027926 (rank : 125) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UN86, O60606, O75149, Q9UPA1 | Gene names | G3BP2, KIAA0660 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 2 (GAP SH3-domain- binding protein 2) (G3BP-2). | |||||
|
INVO_MOUSE
|
||||||
| θ value | 0.47712 (rank : 51) | NC score | 0.054676 (rank : 60) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P48997 | Gene names | Ivl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
MYT1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 52) | NC score | 0.058190 (rank : 55) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
SMC3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 53) | NC score | 0.050419 (rank : 74) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
SMC3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 54) | NC score | 0.050451 (rank : 73) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
TREF1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 55) | NC score | 0.101874 (rank : 31) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
TREF1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 56) | NC score | 0.099760 (rank : 32) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
FBXW7_HUMAN
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.013276 (rank : 152) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q969H0, Q96A16, Q96LE0, Q96RI2, Q9NUX6 | Gene names | FBXW7, FBW7, FBX30, SEL10 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 7 (F-box and WD-40 domain protein 7) (F-box protein FBX30) (hCdc4) (Archipelago homolog) (hAgo) (SEL-10). | |||||
|
GLU2B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.038593 (rank : 99) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08795, Q921X2 | Gene names | Prkcsh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
RFC1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 59) | NC score | 0.044063 (rank : 85) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
VCX1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 60) | NC score | 0.066762 (rank : 46) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
VCX3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 61) | NC score | 0.076085 (rank : 40) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 1.06291 (rank : 62) | NC score | 0.048354 (rank : 78) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
ARS2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 63) | NC score | 0.042387 (rank : 90) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99MR6, Q99MR4, Q99MR5, Q99MR7 | Gene names | Ars2, Asr2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.028872 (rank : 121) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CCD41_HUMAN
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.041726 (rank : 92) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
HDAC5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.026593 (rank : 129) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z2V6, Q9JL73 | Gene names | Hdac5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Histone deacetylase mHDA1). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.033312 (rank : 112) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 1.06291 (rank : 68) | NC score | 0.034019 (rank : 109) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 69) | NC score | 0.052223 (rank : 65) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
SCG1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 70) | NC score | 0.037332 (rank : 105) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P16014 | Gene names | Chgb, Scg-1, Scg1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB). | |||||
|
TXLNB_MOUSE
|
||||||
| θ value | 1.06291 (rank : 71) | NC score | 0.025446 (rank : 132) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VBT1, Q3UVB8, Q8BUK2 | Gene names | Txlnb, Mdp77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.054018 (rank : 62) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MFAP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.029269 (rank : 120) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CQU1, Q3TU29, Q8CCL1, Q9CSJ5 | Gene names | Mfap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microfibrillar-associated protein 1. | |||||
|
NEUM_MOUSE
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.043993 (rank : 86) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
NP1L3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.041934 (rank : 91) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99457, O60788 | Gene names | NAP1L3, BNAP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome assembly protein 1-like 3. | |||||
|
PELP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 76) | NC score | 0.035843 (rank : 106) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
RRMJ3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 77) | NC score | 0.032340 (rank : 115) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IY81, Q8N3A3, Q8WXX1, Q9BWM4, Q9NXT6 | Gene names | FTSJ3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative rRNA methyltransferase 3 (EC 2.1.1.-) (rRNA (uridine-2'-O-)- methyltransferase 3). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 1.38821 (rank : 78) | NC score | 0.032672 (rank : 114) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 1.38821 (rank : 79) | NC score | 0.040107 (rank : 97) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
TUB_MOUSE
|
||||||
| θ value | 1.38821 (rank : 80) | NC score | 0.023016 (rank : 140) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50586 | Gene names | Tub, Rd5 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby protein. | |||||
|
ABTAP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 81) | NC score | 0.042959 (rank : 89) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
BCL7A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 82) | NC score | 0.034118 (rank : 107) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q4VC05, Q13843 | Gene names | BCL7A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member A. | |||||
|
CD2L1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 83) | NC score | 0.017939 (rank : 147) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
HDAC5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 84) | NC score | 0.026345 (rank : 130) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UQL6, O60340, O60528, Q96DY4 | Gene names | HDAC5, KIAA0600 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Antigen NY-CO-9). | |||||
|
SEPP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | 0.033803 (rank : 111) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70274, Q9Z2T7 | Gene names | Sepp1, Selp | |||
|
Domain Architecture |

|
|||||
| Description | Selenoprotein P precursor (SeP) (Plasma selenoprotein P). | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | 0.015190 (rank : 151) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
CENPB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.037533 (rank : 102) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
MYT1L_HUMAN
|
||||||
| θ value | 2.36792 (rank : 88) | NC score | 0.049699 (rank : 77) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
MYT1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 89) | NC score | 0.047884 (rank : 81) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 2.36792 (rank : 90) | NC score | 0.033087 (rank : 113) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
ZN291_HUMAN
|
||||||
| θ value | 2.36792 (rank : 91) | NC score | 0.027323 (rank : 127) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 647 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BY12 | Gene names | ZNF291 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 291. | |||||
|
ZRF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 92) | NC score | 0.057018 (rank : 56) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.009443 (rank : 155) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CCD27_MOUSE
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.024875 (rank : 133) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3V036 | Gene names | Ccdc27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
ELOA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.040509 (rank : 96) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.037342 (rank : 104) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
MYO10_HUMAN
|
||||||
| θ value | 3.0926 (rank : 97) | NC score | 0.016696 (rank : 149) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
SMC2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 98) | NC score | 0.041464 (rank : 94) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
VCXC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 99) | NC score | 0.051500 (rank : 69) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
CCHCR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.027634 (rank : 126) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
CP053_MOUSE
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.026878 (rank : 128) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99L02 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf53 homolog. | |||||
|
INCE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.040706 (rank : 95) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
LMO7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.022711 (rank : 141) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
OPTN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.038423 (rank : 100) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
OSBL7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.011030 (rank : 154) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZF2 | Gene names | OSBPL7, ORP7 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 7 (OSBP-related protein 7) (ORP-7). | |||||
|
ZFP27_MOUSE
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | -0.002677 (rank : 160) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10077, Q6PCZ6, Q8CED4 | Gene names | Zfp27, Mkr4, Zfp-27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 27 (Zfp-27) (Protein mKR4). | |||||
|
AF9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.025778 (rank : 131) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
AT1B4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.012612 (rank : 153) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99ME6, Q99ME5 | Gene names | Atp1b4 | |||
|
Domain Architecture |
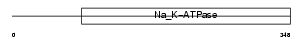
|
|||||
| Description | X/potassium-transporting ATPase subunit beta-m (X,K-ATPase beta-m subunit). | |||||
|
CENPB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.034091 (rank : 108) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P07199 | Gene names | CENPB | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
DDIT3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.031449 (rank : 116) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35638 | Gene names | DDIT3, CHOP, GADD153 | |||
|
Domain Architecture |

|
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
GP137_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.020476 (rank : 144) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14444, Q15074 | Gene names | M11S1, GPIP137 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI-anchored protein p137 (p137GPI) (Membrane component chromosome 11 surface marker 1). | |||||
|
NIN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.037627 (rank : 101) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
RM19_MOUSE
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.021608 (rank : 143) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D338, Q543I0, Q8R1R0 | Gene names | Mrpl19 | |||
|
Domain Architecture |
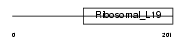
|
|||||
| Description | 39S ribosomal protein L19, mitochondrial precursor (L19mt) (MRP-L19). | |||||
|
TERF2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.051543 (rank : 68) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O35144 | Gene names | Terf2, Trf2 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
TNNT3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.023550 (rank : 138) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QZ47, O35575, O35576, O35577, O35578, O35579, O35580, O35581, O35582, O35583, O35584, O35585, P97456, Q99L89, Q9QZ46 | Gene names | Tnnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, fast skeletal muscle (TnTf) (Fast skeletal muscle troponin T) (fTnT). | |||||
|
USH1C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.008666 (rank : 157) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y6N9, Q96B29, Q9UM04, Q9UM17, Q9UPC3 | Gene names | USH1C, AIE75 | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein) (Autoimmune enteropathy- related antigen AIE-75) (Antigen NY-CO-38/NY-CO-37) (PDZ-73 protein) (NY-REN-3 antigen). | |||||
|
WDR81_HUMAN
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.015462 (rank : 150) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q562E7, Q24JP6, Q8N277, Q8N3F3 | Gene names | WDR81 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 81. | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.052018 (rank : 66) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.022658 (rank : 142) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
CCAR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.037407 (rank : 103) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IX12, Q32NE3, Q5VUP6, Q6PIZ0, Q6X935, Q9H8N4, Q9NVA7, Q9NVQ0, Q9NWM6 | Gene names | CCAR1, CARP1, DIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain). | |||||
|
DMD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.023662 (rank : 137) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.020003 (rank : 146) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.020107 (rank : 145) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
NUCKS_MOUSE
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.029590 (rank : 119) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
PRP4B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.005912 (rank : 159) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61136, O88378, Q8BND8, Q8R4Y5, Q9CTL9, Q9CTT0 | Gene names | Prpf4b, Cbp143, Prp4h, Prp4k, Prp4m, Prpk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (Pre-mRNA protein kinase). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.024005 (rank : 135) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
BCL7A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.023391 (rank : 139) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CXE2, Q8C361, Q8C8M8, Q8VD15 | Gene names | Bcl7a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member A. | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.008992 (rank : 156) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
CMGA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.033933 (rank : 110) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P10645, Q96E84, Q96GL7, Q9BQB5 | Gene names | CHGA | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) (Pituitary secretory protein I) (SP-I) [Contains: Vasostatin-1 (Vasostatin I); Vasostatin-2 (Vasostatin II); EA-92; ES-43; Pancreastatin; SS-18; WA-8; WE-14; LF-19; AL-11; GV-19; GR-44; ER-37]. | |||||
|
MACOI_MOUSE
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.023860 (rank : 136) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 620 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7TQE6 | Gene names | Tmem57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57) (Brain-specific adaptor protein C61). | |||||
|
MYT1L_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.045128 (rank : 84) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
PRC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.028409 (rank : 123) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99K43, Q8CE25 | Gene names | Prc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein regulator of cytokinesis 1. | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.028419 (rank : 122) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.007166 (rank : 158) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.050653 (rank : 71) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.074268 (rank : 44) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CCD21_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.050523 (rank : 72) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CDC5L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.064369 (rank : 51) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99459, Q76N46, Q99974 | Gene names | CDC5L, KIAA0432, PCDC5RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 5-like protein (Cdc5-like protein) (Pombe cdc5- related protein). | |||||
|
CDC5L_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.064704 (rank : 49) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6A068, Q8K1J9 | Gene names | Cdc5l, Kiaa0432 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 5-related protein (Cdc5-like protein). | |||||
|
CF010_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.065607 (rank : 47) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.054962 (rank : 57) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.076986 (rank : 38) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.080151 (rank : 36) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.074979 (rank : 41) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.071384 (rank : 45) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.063128 (rank : 53) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MIER1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.079218 (rank : 37) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
|
MIER1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.082798 (rank : 35) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5UAK0, Q5UAK1, Q5UAK2, Q5UAK3, Q6ZPL6, Q9D402 | Gene names | Mier1, Kiaa1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1). | |||||
|
NASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.074848 (rank : 43) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.063187 (rank : 52) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.051812 (rank : 67) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.064459 (rank : 50) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.054878 (rank : 59) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.053519 (rank : 63) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.074952 (rank : 42) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
SNPC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.065469 (rank : 48) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BP86, Q6PGG7, Q80UG9, Q810L1 | Gene names | Snapc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.076962 (rank : 39) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.054434 (rank : 61) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.061632 (rank : 54) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
ZN541_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.096733 (rank : 33) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
MYSM1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 134 | |
| SwissProt Accessions | Q69Z66, Q3TPV7, Q8BRP5, Q8C4N1 | Gene names | Mysm1, Kiaa1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
MYSM1_HUMAN
|
||||||
| NC score | 0.964448 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5VVJ2, Q7Z3G8, Q96PX3 | Gene names | MYSM1, KIAA1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
SMRC1_HUMAN
|
||||||
| NC score | 0.393637 (rank : 3) | θ value | 1.9326e-10 (rank : 5) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
SMRC2_MOUSE
|
||||||
| NC score | 0.391063 (rank : 4) | θ value | 9.26847e-13 (rank : 3) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.390624 (rank : 5) | θ value | 1.2105e-12 (rank : 4) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
SMRC1_MOUSE
|
||||||
| NC score | 0.389622 (rank : 6) | θ value | 1.9326e-10 (rank : 6) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
PSDE_HUMAN
|
||||||
| NC score | 0.303773 (rank : 7) | θ value | 2.21117e-06 (rank : 7) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00487, O00176 | Gene names | PSMD14, POH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 14 (26S proteasome regulatory subunit rpn11) (26S proteasome-associated PAD1 homolog 1). | |||||
|
PSDE_MOUSE
|
||||||
| NC score | 0.303773 (rank : 8) | θ value | 2.21117e-06 (rank : 8) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35593, Q3UB50, Q9CZY6 | Gene names | Psmd14, Pad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 14 (26S proteasome regulatory subunit rpn11) (MAD1). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.267688 (rank : 9) | θ value | 2.44474e-05 (rank : 11) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
RCOR3_HUMAN
|
||||||
| NC score | 0.263840 (rank : 10) | θ value | 1.43324e-05 (rank : 10) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.262459 (rank : 11) | θ value | 8.40245e-06 (rank : 9) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.260955 (rank : 12) | θ value | 9.29e-05 (rank : 12) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.254071 (rank : 13) | θ value | 0.000461057 (rank : 13) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
RCOR1_HUMAN
|
||||||
| NC score | 0.244799 (rank : 14) | θ value | 0.00228821 (rank : 18) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UKL0, Q15044, Q6P2I9, Q86VG5 | Gene names | RCOR1, KIAA0071, RCOR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
RCOR1_MOUSE
|
||||||
| NC score | 0.239399 (rank : 15) | θ value | 0.00869519 (rank : 27) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CFE3, Q3V092, Q8BK28, Q8CHI2 | Gene names | Rcor1, D12Wsu95e, Kiaa0071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
RCOR2_HUMAN
|
||||||
| NC score | 0.235229 (rank : 16) | θ value | 0.00390308 (rank : 20) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IZ40, Q96FP3 | Gene names | RCOR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2. | |||||
|
RCOR2_MOUSE
|
||||||
| NC score | 0.234365 (rank : 17) | θ value | 0.00390308 (rank : 21) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8C796, Q9JMK4 | Gene names | Rcor2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2 (M-CoREST). | |||||
|
BRCC3_HUMAN
|
||||||
| NC score | 0.229493 (rank : 18) | θ value | 0.00509761 (rank : 22) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P46736, Q16107, Q9BTZ6 | Gene names | BRCC3, C6.1A, CXorf53 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1/BRCA2-containing complex subunit 3. | |||||
|
BRCC3_MOUSE
|
||||||
| NC score | 0.228173 (rank : 19) | θ value | 0.00509761 (rank : 23) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P46737, Q9D025 | Gene names | Brcc3, C6.1A | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1/BRCA2-containing complex subunit 3. | |||||
|
CSN5_HUMAN
|
||||||
| NC score | 0.226418 (rank : 20) | θ value | 0.00509761 (rank : 24) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92905, O15386, Q6AW95, Q86WQ4, Q9BQ17 | Gene names | COPS5, CSN5, JAB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | COP9 signalosome complex subunit 5 (EC 3.4.-.-) (Signalosome subunit 5) (SGN5) (Jun activation domain-binding protein 1). | |||||
|
CSN5_MOUSE
|
||||||
| NC score | 0.226418 (rank : 21) | θ value | 0.00509761 (rank : 25) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35864, Q3UA70, Q8C1S1 | Gene names | Cops5, Csn5, Jab1, Kic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | COP9 signalosome complex subunit 5 (EC 3.4.-.-) (Signalosome subunit 5) (SGN5) (Jun activation domain-binding protein 1) (Kip1 C-terminus- interacting protein 2). | |||||
|
RCOR3_MOUSE
|
||||||
| NC score | 0.216448 (rank : 22) | θ value | 0.0252991 (rank : 29) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6PGA0 | Gene names | Rcor3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
MYBA_HUMAN
|
||||||
| NC score | 0.197777 (rank : 23) | θ value | 0.00175202 (rank : 16) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10243 | Gene names | MYBL1, AMYB | |||
|
Domain Architecture |

|
|||||
| Description | Myb-related protein A (A-Myb). | |||||
|
MYBA_MOUSE
|
||||||
| NC score | 0.197564 (rank : 24) | θ value | 0.00175202 (rank : 17) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51960 | Gene names | Mybl1, Amyb | |||
|
Domain Architecture |

|
|||||
| Description | Myb-related protein A (A-Myb). | |||||
|
MYB_MOUSE
|
||||||
| NC score | 0.193991 (rank : 25) | θ value | 0.00390308 (rank : 19) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P06876, Q61929 | Gene names | Myb | |||
|
Domain Architecture |

|
|||||
| Description | Myb proto-oncogene protein (C-myb). | |||||
|
MYB_HUMAN
|
||||||
| NC score | 0.187541 (rank : 26) | θ value | 0.00869519 (rank : 26) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P10242, P78391, P78392, P78525, P78526, Q14023, Q14024, Q9UE83 | Gene names | MYB | |||
|
Domain Architecture |
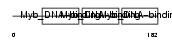
|
|||||
| Description | Myb proto-oncogene protein (C-myb). | |||||
|
MYBB_MOUSE
|
||||||
| NC score | 0.166724 (rank : 27) | θ value | 0.0961366 (rank : 35) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P48972 | Gene names | Mybl2, Bmyb | |||
|
Domain Architecture |

|
|||||
| Description | Myb-related protein B (B-Myb). | |||||
|
MYBB_HUMAN
|
||||||
| NC score | 0.164506 (rank : 28) | θ value | 0.0961366 (rank : 34) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P10244 | Gene names | MYBL2, BMYB | |||
|
Domain Architecture |

|
|||||
| Description | Myb-related protein B (B-Myb). | |||||
|
LSD1_MOUSE
|
||||||
| NC score | 0.162106 (rank : 29) | θ value | 0.00134147 (rank : 15) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZQ88, Q6PB53, Q8VEA1 | Gene names | Aof2, Kiaa0601, Lsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
LSD1_HUMAN
|
||||||
| NC score | 0.154527 (rank : 30) | θ value | 0.00134147 (rank : 14) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
TREF1_HUMAN
|
||||||
| NC score | 0.101874 (rank : 31) | θ value | 0.62314 (rank : 55) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
TREF1_MOUSE
|
||||||
| NC score | 0.099760 (rank : 32) | θ value | 0.62314 (rank : 56) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
ZN541_HUMAN
|
||||||
| NC score | 0.096733 (rank : 33) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.093581 (rank : 34) | θ value | 0.279714 (rank : 42) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
MIER1_MOUSE
|
||||||
| NC score | 0.082798 (rank : 35) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5UAK0, Q5UAK1, Q5UAK2, Q5UAK3, Q6ZPL6, Q9D402 | Gene names | Mier1, Kiaa1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.080151 (rank : 36) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MIER1_HUMAN
|
||||||
| NC score | 0.079218 (rank : 37) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.076986 (rank : 38) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.076962 (rank : 39) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
VCX3_HUMAN
|
||||||
| NC score | 0.076085 (rank : 40) | θ value | 0.813845 (rank : 61) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.074979 (rank : 41) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.074952 (rank : 42) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.074848 (rank : 43) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.074268 (rank : 44) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.071384 (rank : 45) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
VCX1_HUMAN
|
||||||
| NC score | 0.066762 (rank : 46) | θ value | 0.813845 (rank : 60) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
CF010_HUMAN
|
||||||
| NC score | 0.065607 (rank : 47) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
SNPC4_MOUSE
|
||||||
| NC score | 0.065469 (rank : 48) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BP86, Q6PGG7, Q80UG9, Q810L1 | Gene names | Snapc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit). | |||||
|
CDC5L_MOUSE
|
||||||
| NC score | 0.064704 (rank : 49) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6A068, Q8K1J9 | Gene names | Cdc5l, Kiaa0432 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 5-related protein (Cdc5-like protein). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.064459 (rank : 50) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
CDC5L_HUMAN
|
||||||
| NC score | 0.064369 (rank : 51) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99459, Q76N46, Q99974 | Gene names | CDC5L, KIAA0432, PCDC5RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 5-like protein (Cdc5-like protein) (Pombe cdc5- related protein). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.063187 (rank : 52) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.063128 (rank : 53) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.061632 (rank : 54) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
MYT1_MOUSE
|
||||||
| NC score | 0.058190 (rank : 55) | θ value | 0.47712 (rank : 52) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
ZRF1_HUMAN
|
||||||
| NC score | 0.057018 (rank : 56) | θ value | 2.36792 (rank : 92) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.054962 (rank : 57) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.054882 (rank : 58) | θ value | 0.279714 (rank : 45) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.054878 (rank : 59) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
INVO_MOUSE
|
||||||
| NC score | 0.054676 (rank : 60) | θ value | 0.47712 (rank : 51) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P48997 | Gene names | Ivl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.054434 (rank : 61) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.054018 (rank : 62) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.053519 (rank : 63) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
CCAR1_MOUSE
|
||||||
| NC score | 0.052599 (rank : 64) | θ value | 0.47712 (rank : 49) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8CH18, Q6AXC9, Q6PAR2, Q80XE4, Q8BJY0, Q8BVN2, Q8CGG1, Q9CSR5 | Gene names | Ccar1, Carp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.052223 (rank : 65) | θ value | 1.06291 (rank : 69) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.052018 (rank : 66) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.051812 (rank : 67) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
TERF2_MOUSE
|
||||||
| NC score | 0.051543 (rank : 68) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O35144 | Gene names | Terf2, Trf2 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 2 (TTAGGG repeat-binding factor 2) (Telomeric DNA-binding protein). | |||||
|
VCXC_HUMAN
|
||||||
| NC score | 0.051500 (rank : 69) | θ value | 3.0926 (rank : 99) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.051285 (rank : 70) | θ value | 0.0736092 (rank : 32) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.050653 (rank : 71) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.050523 (rank : 72) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
SMC3_MOUSE
|
||||||
| NC score | 0.050451 (rank : 73) | θ value | 0.47712 (rank : 54) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
SMC3_HUMAN
|
||||||
| NC score | 0.050419 (rank : 74) | θ value | 0.47712 (rank : 53) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.050145 (rank : 75) | θ value | 0.0148317 (rank : 28) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NP1L3_MOUSE
|
||||||
| NC score | 0.049851 (rank : 76) | θ value | 0.365318 (rank : 47) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
MYT1L_HUMAN
|
||||||
| NC score | 0.049699 (rank : 77) | θ value | 2.36792 (rank : 88) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.048354 (rank : 78) | θ value | 1.06291 (rank : 62) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.048140 (rank : 79) | θ value | 0.125558 (rank : 36) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.047943 (rank : 80) | θ value | 0.0563607 (rank : 30) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
MYT1_HUMAN
|
||||||
| NC score | 0.047884 (rank : 81) | θ value | 2.36792 (rank : 89) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
PDXL2_HUMAN
|
||||||
| NC score | 0.046789 (rank : 82) | θ value | 0.365318 (rank : 48) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NZ53, Q6UVY4, Q8WUV6 | Gene names | PODXL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Podocalyxin-like protein 2 precursor (Endoglycan). | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.046727 (rank : 83) | θ value | 0.163984 (rank : 38) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
MYT1L_MOUSE
|
||||||
| NC score | 0.045128 (rank : 84) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
RFC1_MOUSE
|
||||||
| NC score | 0.044063 (rank : 85) | θ value | 0.813845 (rank : 59) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.043993 (rank : 86) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
NCKX1_HUMAN
|
||||||
| NC score | 0.043378 (rank : 87) | θ value | 0.279714 (rank : 43) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
INVO_HUMAN
|
||||||
| NC score | 0.043121 (rank : 88) | θ value | 0.21417 (rank : 39) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
ABTAP_MOUSE
|
||||||
| NC score | 0.042959 (rank : 89) | θ value | 1.81305 (rank : 81) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
ARS2_MOUSE
|
||||||
| NC score | 0.042387 (rank : 90) | θ value | 1.06291 (rank : 63) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99MR6, Q99MR4, Q99MR5, Q99MR7 | Gene names | Ars2, Asr2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
NP1L3_HUMAN
|
||||||
| NC score | 0.041934 (rank : 91) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99457, O60788 | Gene names | NAP1L3, BNAP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome assembly protein 1-like 3. | |||||
|
CCD41_HUMAN
|
||||||
| NC score | 0.041726 (rank : 92) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.041638 (rank : 93) | θ value | 0.0961366 (rank : 33) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
SMC2_MOUSE
|
||||||
| NC score | 0.041464 (rank : 94) | θ value | 3.0926 (rank : 98) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.040706 (rank : 95) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
ELOA1_HUMAN
|
||||||
| NC score | 0.040509 (rank : 96) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.040107 (rank : 97) | θ value | 1.38821 (rank : 79) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.038719 (rank : 98) | θ value | 0.279714 (rank : 44) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
GLU2B_MOUSE
|
||||||
| NC score | 0.038593 (rank : 99) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08795, Q921X2 | Gene names | Prkcsh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glucosidase 2 subunit beta precursor (Glucosidase II subunit beta) (Protein kinase C substrate, 60.1 kDa protein, heavy chain) (PKCSH) (80K-H protein). | |||||
|
OPTN_MOUSE
|
||||||
| NC score | 0.038423 (rank : 100) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.037627 (rank : 101) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
CENPB_MOUSE
|
||||||
| NC score | 0.037533 (rank : 102) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
CCAR1_HUMAN
|
||||||
| NC score | 0.037407 (rank : 103) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IX12, Q32NE3, Q5VUP6, Q6PIZ0, Q6X935, Q9H8N4, Q9NVA7, Q9NVQ0, Q9NWM6 | Gene names | CCAR1, CARP1, DIS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1) (Death inducer with SAP domain). | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.037342 (rank : 104) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
SCG1_MOUSE
|
||||||
| NC score | 0.037332 (rank : 105) | θ value | 1.06291 (rank : 70) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P16014 | Gene names | Chgb, Scg-1, Scg1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB). | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.035843 (rank : 106) | θ value | 1.38821 (rank : 76) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
BCL7A_HUMAN
|
||||||
| NC score | 0.034118 (rank : 107) | θ value | 1.81305 (rank : 82) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q4VC05, Q13843 | Gene names | BCL7A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member A. | |||||
|
CENPB_HUMAN
|
||||||
| NC score | 0.034091 (rank : 108) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P07199 | Gene names | CENPB | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.034019 (rank : 109) | θ value | 1.06291 (rank : 68) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
CMGA_HUMAN
|
||||||
| NC score | 0.033933 (rank : 110) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P10645, Q96E84, Q96GL7, Q9BQB5 | Gene names | CHGA | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) (Pituitary secretory protein I) (SP-I) [Contains: Vasostatin-1 (Vasostatin I); Vasostatin-2 (Vasostatin II); EA-92; ES-43; Pancreastatin; SS-18; WA-8; WE-14; LF-19; AL-11; GV-19; GR-44; ER-37]. | |||||
|
SEPP1_MOUSE
|
||||||
| NC score | 0.033803 (rank : 111) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70274, Q9Z2T7 | Gene names | Sepp1, Selp | |||
|
Domain Architecture |

|
|||||
| Description | Selenoprotein P precursor (SeP) (Plasma selenoprotein P). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.033312 (rank : 112) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.033087 (rank : 113) | θ value | 2.36792 (rank : 90) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.032672 (rank : 114) | θ value | 1.38821 (rank : 78) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
RRMJ3_HUMAN
|
||||||
| NC score | 0.032340 (rank : 115) | θ value | 1.38821 (rank : 77) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IY81, Q8N3A3, Q8WXX1, Q9BWM4, Q9NXT6 | Gene names | FTSJ3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative rRNA methyltransferase 3 (EC 2.1.1.-) (rRNA (uridine-2'-O-)- methyltransferase 3). | |||||
|
DDIT3_HUMAN
|
||||||
| NC score | 0.031449 (rank : 116) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35638 | Gene names | DDIT3, CHOP, GADD153 | |||
|
Domain Architecture |

|
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
AP3B1_HUMAN
|
||||||
| NC score | 0.031027 (rank : 117) | θ value | 0.365318 (rank : 46) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
KI13A_HUMAN
|
||||||
| NC score | 0.029688 (rank : 118) | θ value | 0.279714 (rank : 41) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H1H9, Q9H1H8 | Gene names | KIF13A, RBKIN | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13A (Kinesin-like protein RBKIN). | |||||
|
NUCKS_MOUSE
|
||||||
| NC score | 0.029590 (rank : 119) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
MFAP1_MOUSE
|
||||||
| NC score | 0.029269 (rank : 120) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CQU1, Q3TU29, Q8CCL1, Q9CSJ5 | Gene names | Mfap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microfibrillar-associated protein 1. | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.028872 (rank : 121) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.028419 (rank : 122) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
PRC1_MOUSE
|
||||||
| NC score | 0.028409 (rank : 123) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99K43, Q8CE25 | Gene names | Prc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein regulator of cytokinesis 1. | |||||
|
TEX14_MOUSE
|
||||||
| NC score | 0.028136 (rank : 124) | θ value | 0.125558 (rank : 37) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1333 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7M6U3, Q3UT36, Q5NC10, Q5NC11, Q8CGK1, Q8CGK2, Q99MV8 | Gene names | Tex14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
G3BP2_HUMAN
|
||||||
| NC score | 0.027926 (rank : 125) | θ value | 0.47712 (rank : 50) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UN86, O60606, O75149, Q9UPA1 | Gene names | G3BP2, KIAA0660 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 2 (GAP SH3-domain- binding protein 2) (G3BP-2). | |||||
|
CCHCR_HUMAN
|
||||||
| NC score | 0.027634 (rank : 126) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
ZN291_HUMAN
|
||||||
| NC score | 0.027323 (rank : 127) | θ value | 2.36792 (rank : 91) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 647 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BY12 | Gene names | ZNF291 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 291. | |||||
|
CP053_MOUSE
|
||||||
| NC score | 0.026878 (rank : 128) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99L02 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf53 homolog. | |||||
|
HDAC5_MOUSE
|
||||||
| NC score | 0.026593 (rank : 129) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z2V6, Q9JL73 | Gene names | Hdac5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Histone deacetylase mHDA1). | |||||
|
HDAC5_HUMAN
|
||||||
| NC score | 0.026345 (rank : 130) | θ value | 1.81305 (rank : 84) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UQL6, O60340, O60528, Q96DY4 | Gene names | HDAC5, KIAA0600 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Antigen NY-CO-9). | |||||
|
AF9_HUMAN
|
||||||
| NC score | 0.025778 (rank : 131) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
TXLNB_MOUSE
|
||||||
| NC score | 0.025446 (rank : 132) | θ value | 1.06291 (rank : 71) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VBT1, Q3UVB8, Q8BUK2 | Gene names | Txlnb, Mdp77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77). | |||||
|
CCD27_MOUSE
|
||||||
| NC score | 0.024875 (rank : 133) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3V036 | Gene names | Ccdc27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
DDX23_HUMAN
|
||||||
| NC score | 0.024853 (rank : 134) | θ value | 0.0563607 (rank : 31) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.024005 (rank : 135) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
MACOI_MOUSE
|
||||||
| NC score | 0.023860 (rank : 136) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 620 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q7TQE6 | Gene names | Tmem57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57) (Brain-specific adaptor protein C61). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.023662 (rank : 137) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
TNNT3_MOUSE
|
||||||
| NC score | 0.023550 (rank : 138) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QZ47, O35575, O35576, O35577, O35578, O35579, O35580, O35581, O35582, O35583, O35584, O35585, P97456, Q99L89, Q9QZ46 | Gene names | Tnnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin T, fast skeletal muscle (TnTf) (Fast skeletal muscle troponin T) (fTnT). | |||||
|
BCL7A_MOUSE
|
||||||
| NC score | 0.023391 (rank : 139) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CXE2, Q8C361, Q8C8M8, Q8VD15 | Gene names | Bcl7a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 7 protein family member A. | |||||
|
TUB_MOUSE
|
||||||
| NC score | 0.023016 (rank : 140) | θ value | 1.38821 (rank : 80) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50586 | Gene names | Tub, Rd5 | |||
|
Domain Architecture |

|
|||||
| Description | Tubby protein. | |||||
|
LMO7_HUMAN
|
||||||
| NC score | 0.022711 (rank : 141) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.022658 (rank : 142) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
RM19_MOUSE
|
||||||
| NC score | 0.021608 (rank : 143) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D338, Q543I0, Q8R1R0 | Gene names | Mrpl19 | |||
|
Domain Architecture |

|
|||||
| Description | 39S ribosomal protein L19, mitochondrial precursor (L19mt) (MRP-L19). | |||||
|
GP137_HUMAN
|
||||||
| NC score | 0.020476 (rank : 144) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14444, Q15074 | Gene names | M11S1, GPIP137 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI-anchored protein p137 (p137GPI) (Membrane component chromosome 11 surface marker 1). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.020107 (rank : 145) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.020003 (rank : 146) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
CD2L1_MOUSE
|
||||||
| NC score | 0.017939 (rank : 147) | θ value | 1.81305 (rank : 83) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | 0.017746 (rank : 148) | θ value | 0.279714 (rank : 40) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.016696 (rank : 149) | θ value | 3.0926 (rank : 97) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
WDR81_HUMAN
|
||||||
| NC score | 0.015462 (rank : 150) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q562E7, Q24JP6, Q8N277, Q8N3F3 | Gene names | WDR81 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 81. | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.015190 (rank : 151) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
FBXW7_HUMAN
|
||||||
| NC score | 0.013276 (rank : 152) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q969H0, Q96A16, Q96LE0, Q96RI2, Q9NUX6 | Gene names | FBXW7, FBW7, FBX30, SEL10 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 7 (F-box and WD-40 domain protein 7) (F-box protein FBX30) (hCdc4) (Archipelago homolog) (hAgo) (SEL-10). | |||||
|
AT1B4_MOUSE
|
||||||
| NC score | 0.012612 (rank : 153) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99ME6, Q99ME5 | Gene names | Atp1b4 | |||
|
Domain Architecture |

|
|||||
| Description | X/potassium-transporting ATPase subunit beta-m (X,K-ATPase beta-m subunit). | |||||
|
OSBL7_HUMAN
|
||||||
| NC score | 0.011030 (rank : 154) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZF2 | Gene names | OSBPL7, ORP7 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 7 (OSBP-related protein 7) (ORP-7). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.009443 (rank : 155) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.008992 (rank : 156) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
USH1C_HUMAN
|
||||||
| NC score | 0.008666 (rank : 157) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y6N9, Q96B29, Q9UM04, Q9UM17, Q9UPC3 | Gene names | USH1C, AIE75 | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein) (Autoimmune enteropathy- related antigen AIE-75) (Antigen NY-CO-38/NY-CO-37) (PDZ-73 protein) (NY-REN-3 antigen). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.007166 (rank : 158) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
PRP4B_MOUSE
|
||||||
| NC score | 0.005912 (rank : 159) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61136, O88378, Q8BND8, Q8R4Y5, Q9CTL9, Q9CTT0 | Gene names | Prpf4b, Cbp143, Prp4h, Prp4k, Prp4m, Prpk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (Pre-mRNA protein kinase). | |||||
|
ZFP27_MOUSE
|
||||||
| NC score | -0.002677 (rank : 160) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 134 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10077, Q6PCZ6, Q8CED4 | Gene names | Zfp27, Mkr4, Zfp-27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 27 (Zfp-27) (Protein mKR4). | |||||