Please be patient as the page loads
|
RCOR3_HUMAN
|
||||||
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RCOR3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 161 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
RCOR3_MOUSE
|
||||||
| θ value | 1.97579e-164 (rank : 2) | NC score | 0.973363 (rank : 2) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q6PGA0 | Gene names | Rcor3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
RCOR1_MOUSE
|
||||||
| θ value | 2.4321e-138 (rank : 3) | NC score | 0.967115 (rank : 3) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CFE3, Q3V092, Q8BK28, Q8CHI2 | Gene names | Rcor1, D12Wsu95e, Kiaa0071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
RCOR1_HUMAN
|
||||||
| θ value | 1.33454e-136 (rank : 4) | NC score | 0.966469 (rank : 4) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UKL0, Q15044, Q6P2I9, Q86VG5 | Gene names | RCOR1, KIAA0071, RCOR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
RCOR2_MOUSE
|
||||||
| θ value | 6.41512e-131 (rank : 5) | NC score | 0.962831 (rank : 6) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8C796, Q9JMK4 | Gene names | Rcor2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2 (M-CoREST). | |||||
|
RCOR2_HUMAN
|
||||||
| θ value | 1.86652e-130 (rank : 6) | NC score | 0.965384 (rank : 5) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8IZ40, Q96FP3 | Gene names | RCOR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2. | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 3.75424e-22 (rank : 7) | NC score | 0.504995 (rank : 11) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 1.42661e-21 (rank : 8) | NC score | 0.516738 (rank : 8) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 5.07402e-19 (rank : 9) | NC score | 0.510851 (rank : 10) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 5.07402e-19 (rank : 10) | NC score | 0.516623 (rank : 9) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
MTA3_MOUSE
|
||||||
| θ value | 1.133e-10 (rank : 11) | NC score | 0.477731 (rank : 12) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q924K8, Q8VC18 | Gene names | Mta3 | |||
|
Domain Architecture |
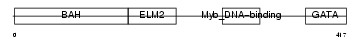
|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
TREF1_MOUSE
|
||||||
| θ value | 1.9326e-10 (rank : 12) | NC score | 0.388543 (rank : 21) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
MTA2_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 13) | NC score | 0.470551 (rank : 13) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94776, Q9UQB5 | Gene names | MTA2, MTA1L1, PID | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1) (MTA1-L1 protein) (p53 target protein in deacetylase complex). | |||||
|
TREF1_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 14) | NC score | 0.389603 (rank : 20) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
ZN541_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 15) | NC score | 0.529220 (rank : 7) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
MTA2_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 16) | NC score | 0.467154 (rank : 15) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9R190, Q3TVT8 | Gene names | Mta2, Mta1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1). | |||||
|
MTA3_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 17) | NC score | 0.468355 (rank : 14) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BTC8, Q9NSP2, Q9ULF4 | Gene names | MTA3, KIAA1266 | |||
|
Domain Architecture |
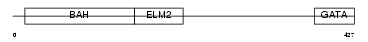
|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
MTA1_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 18) | NC score | 0.439946 (rank : 18) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13330, Q8NFI8, Q96GI8 | Gene names | MTA1 | |||
|
Domain Architecture |
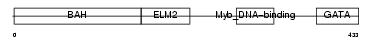
|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
|
MTA1_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 19) | NC score | 0.435568 (rank : 19) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8K4B0, Q80UI1, Q8K4D4, Q924K9 | Gene names | Mta1 | |||
|
Domain Architecture |
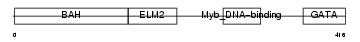
|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
|
MIER1_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 20) | NC score | 0.454024 (rank : 16) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5UAK0, Q5UAK1, Q5UAK2, Q5UAK3, Q6ZPL6, Q9D402 | Gene names | Mier1, Kiaa1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1). | |||||
|
MIER1_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 21) | NC score | 0.449597 (rank : 17) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
|
MYSM1_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 22) | NC score | 0.263840 (rank : 24) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q69Z66, Q3TPV7, Q8BRP5, Q8C4N1 | Gene names | Mysm1, Kiaa1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 23) | NC score | 0.337832 (rank : 22) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 24) | NC score | 0.334796 (rank : 23) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
MYSM1_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 25) | NC score | 0.263080 (rank : 25) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5VVJ2, Q7Z3G8, Q96PX3 | Gene names | MYSM1, KIAA1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
DCP1A_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 26) | NC score | 0.092803 (rank : 26) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NPI6 | Gene names | DCP1A, SMIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (Smad4-interacting transcriptional co-activator). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 27) | NC score | 0.072997 (rank : 35) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 28) | NC score | 0.068137 (rank : 38) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 29) | NC score | 0.057042 (rank : 42) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
RED_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 30) | NC score | 0.091278 (rank : 27) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z1M8 | Gene names | Ik, Red, Rer | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Red (Protein RER) (IK factor) (Cytokine IK). | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 31) | NC score | 0.055446 (rank : 43) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
RED_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 32) | NC score | 0.087927 (rank : 29) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13123 | Gene names | IK, RED, RER | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Red (Protein RER) (IK factor) (Cytokine IK). | |||||
|
SOX30_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 33) | NC score | 0.047757 (rank : 63) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 34) | NC score | 0.084441 (rank : 30) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
FOG1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 35) | NC score | 0.017517 (rank : 121) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
GA2L2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 36) | NC score | 0.079080 (rank : 33) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 37) | NC score | 0.048199 (rank : 62) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 0.125558 (rank : 38) | NC score | 0.053749 (rank : 47) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 39) | NC score | 0.032024 (rank : 85) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
HSF1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 40) | NC score | 0.042264 (rank : 69) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
NUMBL_HUMAN
|
||||||
| θ value | 0.163984 (rank : 41) | NC score | 0.048734 (rank : 60) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 42) | NC score | 0.066561 (rank : 40) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
CCD55_MOUSE
|
||||||
| θ value | 0.365318 (rank : 43) | NC score | 0.044578 (rank : 65) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
CJ108_HUMAN
|
||||||
| θ value | 0.365318 (rank : 44) | NC score | 0.084129 (rank : 31) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N8Z3 | Gene names | C10orf108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative uncharacterized protein C10orf108. | |||||
|
DCC_MOUSE
|
||||||
| θ value | 0.365318 (rank : 45) | NC score | 0.015705 (rank : 129) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
HNRPU_HUMAN
|
||||||
| θ value | 0.365318 (rank : 46) | NC score | 0.027710 (rank : 99) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q00839, O75507, Q96HY9 | Gene names | HNRPU, SAFA, U21.1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U (hnRNP U) (Scaffold attachment factor A) (SAF-A) (p120) (pp120). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.365318 (rank : 47) | NC score | 0.054768 (rank : 45) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
STAR8_HUMAN
|
||||||
| θ value | 0.365318 (rank : 48) | NC score | 0.024922 (rank : 107) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
DDX10_HUMAN
|
||||||
| θ value | 0.47712 (rank : 49) | NC score | 0.010003 (rank : 151) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 50) | NC score | 0.080008 (rank : 32) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
MD1L1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 51) | NC score | 0.026041 (rank : 102) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
NCOA1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 52) | NC score | 0.035108 (rank : 78) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 53) | NC score | 0.066586 (rank : 39) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
TRI56_HUMAN
|
||||||
| θ value | 0.62314 (rank : 54) | NC score | 0.016268 (rank : 126) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BRZ2, Q86VT6, Q8N2H8, Q8NAC0, Q9H031 | Gene names | TRIM56, RNF109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 56 (RING finger protein 109). | |||||
|
GAK6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.032863 (rank : 81) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK11_HUMAN
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.035425 (rank : 76) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.035151 (rank : 77) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
MAD3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.039831 (rank : 72) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
MYB_MOUSE
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.041609 (rank : 70) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P06876, Q61929 | Gene names | Myb | |||
|
Domain Architecture |

|
|||||
| Description | Myb proto-oncogene protein (C-myb). | |||||
|
SC24A_MOUSE
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.049305 (rank : 59) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
TM131_MOUSE
|
||||||
| θ value | 1.06291 (rank : 61) | NC score | 0.038111 (rank : 73) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O70472 | Gene names | Tmem131, D1Bwg0491e, Rw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 131 (Protein RW1). | |||||
|
BIN1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.028889 (rank : 95) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00499, O00297, O00545, O43867, O60552, O60553, O60554, O60555, O75514, O75515, O75516, O75517, O75518, Q92944, Q99688 | Gene names | BIN1, AMPHL | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc- interacting protein 1). | |||||
|
GAK2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.032886 (rank : 80) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
MAD3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.037369 (rank : 74) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.076769 (rank : 34) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NCOA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.031439 (rank : 88) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.043225 (rank : 67) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
ATX2L_HUMAN
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.052599 (rank : 50) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.054757 (rank : 46) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.070503 (rank : 37) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MEF2B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.039996 (rank : 71) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02080 | Gene names | MEF2B, XMEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B (Serum response factor-like protein 2) (XMEF2) (RSRFR2). | |||||
|
SFR11_HUMAN
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.025341 (rank : 105) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
EZH2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.030285 (rank : 91) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15910, Q15755, Q92857 | Gene names | EZH2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
EZH2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.032682 (rank : 82) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61188, Q9R090 | Gene names | Ezh2, Enx1h | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
FGD1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.014841 (rank : 132) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
KAISO_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.003218 (rank : 167) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BN78, Q8C226, Q9WTZ7 | Gene names | Zbtb33, Kaiso | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional regulator Kaiso (Zinc finger and BTB domain-containing protein 33). | |||||
|
MAST2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.000350 (rank : 171) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MTG8R_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.023274 (rank : 109) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O70374 | Gene names | Cbfa2t2, Cbfa2t2h, Mtgr1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1). | |||||
|
PDLI7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.030251 (rank : 92) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
SOX13_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.023030 (rank : 110) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
US6NL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.016040 (rank : 127) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92738, Q15400, Q7L0K9 | Gene names | USP6NL, KIAA0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein (Related to the N-terminus of tre) (RN- tre). | |||||
|
4ET_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.025652 (rank : 103) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9EST3, Q9CSS3 | Gene names | Eif4enif1, Clast4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
BAG3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.045261 (rank : 64) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
BCAR1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.048330 (rank : 61) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.052711 (rank : 48) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
CCD96_MOUSE
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.021750 (rank : 113) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.006441 (rank : 160) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
MYB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.036237 (rank : 75) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P10242, P78391, P78392, P78525, P78526, Q14023, Q14024, Q9UE83 | Gene names | MYB | |||
|
Domain Architecture |
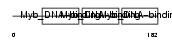
|
|||||
| Description | Myb proto-oncogene protein (C-myb). | |||||
|
P73_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.021677 (rank : 114) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15350, O15351, Q5TBV5, Q5TBV6, Q8NHW9, Q8TDY5, Q8TDY6, Q9NTK8 | Gene names | TP73, P73 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein p73 (p53-like transcription factor) (p53-related protein). | |||||
|
ZN473_MOUSE
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | -0.001890 (rank : 172) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 824 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BI67, Q8BI98, Q8BIB7 | Gene names | Znf473, Zfp100, Zfp473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 473 homolog (Zinc finger protein 100) (Zfp-100). | |||||
|
CB013_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.016867 (rank : 123) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IW19, Q53P47, Q53PB9, Q53QU0 | Gene names | C2orf13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C2orf13. | |||||
|
CNOT2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.031904 (rank : 87) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZN8, Q9H3E0, Q9NSX5, Q9NWR6, Q9P028 | Gene names | CNOT2, CDC36, NOT2 | |||
|
Domain Architecture |
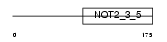
|
|||||
| Description | CCR4-NOT transcription complex subunit 2 (CCR4-associated factor 2). | |||||
|
CNOT2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.032153 (rank : 83) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C5L3, Q3UE39, Q80YA5, Q9D0P1 | Gene names | Cnot2 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 2 (CCR4-associated factor 2). | |||||
|
HSF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.030632 (rank : 90) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q00613 | Gene names | HSF1, HSTF1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
INCE_MOUSE
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.018924 (rank : 119) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.011580 (rank : 142) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
KI21A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.011497 (rank : 143) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
MTG8R_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.019915 (rank : 117) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43439, Q5TGE4, Q5TGE7, Q8IWF3, Q96B06, Q96L00, Q9H436, Q9UJP8, Q9UJP9, Q9UP24 | Gene names | CBFA2T2, EHT, MTGR1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1) (Myeloid translocation-related protein 1) (ETO homologous on chromosome 20) (p85). | |||||
|
MYLK_MOUSE
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.001707 (rank : 170) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
NTBP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.016351 (rank : 125) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BRM2, Q8CAA8 | Gene names | Scyl1bp1, Ntklbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like-binding protein 1 (NTKL-binding protein 1) (NTKL-BP1) (mNTKL-BP1) (SCY1-like 1-binding protein 1) (SCYL1-binding protein 1) (SCYL1-BP1). | |||||
|
P66A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.014855 (rank : 131) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CHY6, Q8BTQ2, Q8VEC9 | Gene names | Gatad2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 alpha (GATA zinc finger domain- containing protein 2A). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.042752 (rank : 68) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
PP16A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.007963 (rank : 156) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96I34 | Gene names | PPP1R16A, MYPT3 | |||
|
Domain Architecture |
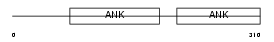
|
|||||
| Description | Protein phosphatase 1 regulatory subunit 16A (Myosin phosphatase- targeting subunit 3). | |||||
|
RHG09_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.014244 (rank : 134) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
UBQL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.009501 (rank : 153) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UMX0, Q8IXS9, Q8N2Q3, Q9H0T8, Q9H3R4, Q9HAZ5 | Gene names | UBQLN1, DA41, PLIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-1 (Protein linking IAP with cytoskeleton 1) (PLIC-1) (hPLIC- 1). | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.014995 (rank : 130) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
CSPG5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.025424 (rank : 104) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95196, Q71M39, Q71M40 | Gene names | CSPG5, CALEB, NGC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.032150 (rank : 84) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
DDX23_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.011272 (rank : 145) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
ETV5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.013960 (rank : 135) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
FA76B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.010152 (rank : 150) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5HYJ3, Q6PIU3, Q8TC53 | Gene names | FAM76B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM76B. | |||||
|
KAISO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.002305 (rank : 169) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86T24, O00319, Q7Z361, Q8IVP6, Q8N3P0 | Gene names | ZBTB33, KAISO, ZNF348 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional regulator Kaiso (Zinc finger and BTB domain-containing protein 33). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.031926 (rank : 86) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
MYT1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.010840 (rank : 148) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
NIN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.022849 (rank : 111) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.012722 (rank : 140) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.019545 (rank : 118) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
NUMBL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.028349 (rank : 96) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O08919, Q6NVG8 | Gene names | Numbl, Nbl | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein. | |||||
|
PBX4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.006019 (rank : 161) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99NE9, Q8CFL2, Q99NE8 | Gene names | Pbx4 | |||
|
Domain Architecture |
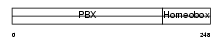
|
|||||
| Description | Pre-B-cell leukemia transcription factor 4 (Homeobox protein PBX4). | |||||
|
QSCN6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.013071 (rank : 137) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00391, Q59G29, Q5T2X0, Q8TDL6, Q8WVP4 | Gene names | QSCN6, QSOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfhydryl oxidase 1 precursor (EC 1.8.3.2) (Quiescin Q6) (hQSOX). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.026763 (rank : 100) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
RBM16_HUMAN
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.025311 (rank : 106) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.043822 (rank : 66) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
ANKR6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.003845 (rank : 166) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
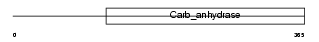
CAH9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.004375 (rank : 165) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VHB5, Q8K1G1, Q8VDE4 | Gene names | Ca9, Car9 | |||
|
Domain Architecture |
|
|||||
| Description | Carbonic anhydrase 9 precursor (EC 4.2.1.1) (Carbonic anhydrase IX) (Carbonate dehydratase IX) (CA-IX) (CAIX) (Membrane antigen MN homolog). | |||||
|
CJ086_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.029021 (rank : 93) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NXX6, Q5SQQ5, Q6P673, Q8WY66, Q9BS90 | Gene names | C10orf86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf86. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.020623 (rank : 115) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
IF4B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.015884 (rank : 128) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
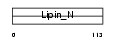
LPIN3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.012848 (rank : 139) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99PI4, Q3TQ75, Q8C7R9 | Gene names | Lpin3 | |||
|
Domain Architecture |
|
|||||
| Description | Lipin-3. | |||||
|
MAGI1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.009305 (rank : 154) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.033269 (rank : 79) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MD1L1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.013726 (rank : 136) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.072987 (rank : 36) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
PC11X_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.002415 (rank : 168) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BZA7, Q2TJH0, Q2TJH1, Q2TJH3, Q5JVZ0, Q70LR8, Q70LS7, Q70LS8, Q70LS9, Q70LT7, Q70LT8, Q70LT9, Q70LU0, Q70LU1, Q96RV4, Q96RW0, Q9BZA6, Q9H4E0, Q9P2M0, Q9P2X5 | Gene names | PCDH11X, PCDH11, PCDHX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 X-linked precursor (Protocadherin-11) (Protocadherin on the X chromosome) (PCDH-X) (Protocadherin-S). | |||||
|
PEX2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.011372 (rank : 144) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55098, O35467 | Gene names | Pxmp3, Paf1, Pex2, Pmp35 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 1 (PAF-1) (Peroxin-2) (Peroxisomal membrane protein 3). | |||||
|
SMAD4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.011016 (rank : 147) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
SMRD2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.006726 (rank : 158) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99JR8 | Gene names | Smarcd2, Baf60b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 2 (60 kDa BRG-1/Brm-associated factor subunit B) (BRG1-associated factor 60B). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.030734 (rank : 89) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.016552 (rank : 124) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CCD21_MOUSE
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.014602 (rank : 133) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.021809 (rank : 112) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
FA47A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.052120 (rank : 53) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
FNBP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.005362 (rank : 162) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
GAK12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.028001 (rank : 98) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
INCE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.020606 (rank : 116) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
IRX2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.006502 (rank : 159) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P81066, O55121, Q9ERN1 | Gene names | Irx2, Irxa2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-2 (Iroquois homeobox protein 2) (Homeodomain protein IRXA2). | |||||
|
K0174_MOUSE
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.011124 (rank : 146) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CX00, Q80U68 | Gene names | Kiaa0174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0174. | |||||
|
K0256_MOUSE
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.007628 (rank : 157) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6A098 | Gene names | Kiaa0256 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0256. | |||||
|
LARP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.028269 (rank : 97) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
NDF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.004573 (rank : 164) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13562, O00343, Q13340, Q5U095, Q96TH0, Q99455, Q9UEC8 | Gene names | NEUROD1, NEUROD | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1) (NeuroD). | |||||
|
NOC2L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.016870 (rank : 122) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y3T9, Q5SVA3, Q9BTN6 | Gene names | NOC2L | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar complex protein 2 homolog (Protein NOC2 homolog) (NOC2- like). | |||||
|
ONEC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.009153 (rank : 155) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6XBJ3 | Gene names | Onecut2, Oc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | One cut domain family member 2 (Transcription factor ONECUT-2) (OC-2). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.026662 (rank : 101) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PF21A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.013021 (rank : 138) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.024136 (rank : 108) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
SIX2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.004583 (rank : 163) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62232, P70179 | Gene names | Six2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX2 (Sine oculis homeobox homolog 2). | |||||
|
SMAD4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.010729 (rank : 149) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13485 | Gene names | SMAD4, DPC4, MADH4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4) (hSMAD4). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.029012 (rank : 94) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.012192 (rank : 141) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.009964 (rank : 152) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
ZCHC8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.018907 (rank : 120) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.087941 (rank : 28) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CCD95_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.052665 (rank : 49) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NBZ0 | Gene names | CCDC95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 95. | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.051053 (rank : 57) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.054802 (rank : 44) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.052428 (rank : 52) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.051787 (rank : 56) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.052099 (rank : 54) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.052559 (rank : 51) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.051916 (rank : 55) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.057145 (rank : 41) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.050136 (rank : 58) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
RCOR3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 161 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
RCOR3_MOUSE
|
||||||
| NC score | 0.973363 (rank : 2) | θ value | 1.97579e-164 (rank : 2) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q6PGA0 | Gene names | Rcor3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
RCOR1_MOUSE
|
||||||
| NC score | 0.967115 (rank : 3) | θ value | 2.4321e-138 (rank : 3) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CFE3, Q3V092, Q8BK28, Q8CHI2 | Gene names | Rcor1, D12Wsu95e, Kiaa0071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
RCOR1_HUMAN
|
||||||
| NC score | 0.966469 (rank : 4) | θ value | 1.33454e-136 (rank : 4) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UKL0, Q15044, Q6P2I9, Q86VG5 | Gene names | RCOR1, KIAA0071, RCOR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 1 (Protein CoREST). | |||||
|
RCOR2_HUMAN
|
||||||
| NC score | 0.965384 (rank : 5) | θ value | 1.86652e-130 (rank : 6) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8IZ40, Q96FP3 | Gene names | RCOR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2. | |||||
|
RCOR2_MOUSE
|
||||||
| NC score | 0.962831 (rank : 6) | θ value | 6.41512e-131 (rank : 5) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8C796, Q9JMK4 | Gene names | Rcor2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 2 (M-CoREST). | |||||
|
ZN541_HUMAN
|
||||||
| NC score | 0.529220 (rank : 7) | θ value | 3.29651e-10 (rank : 15) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H0D2, Q8NDK8 | Gene names | ZNF541 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 541. | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.516738 (rank : 8) | θ value | 1.42661e-21 (rank : 8) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.516623 (rank : 9) | θ value | 5.07402e-19 (rank : 10) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.510851 (rank : 10) | θ value | 5.07402e-19 (rank : 9) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.504995 (rank : 11) | θ value | 3.75424e-22 (rank : 7) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
MTA3_MOUSE
|
||||||
| NC score | 0.477731 (rank : 12) | θ value | 1.133e-10 (rank : 11) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q924K8, Q8VC18 | Gene names | Mta3 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
MTA2_HUMAN
|
||||||
| NC score | 0.470551 (rank : 13) | θ value | 2.52405e-10 (rank : 13) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94776, Q9UQB5 | Gene names | MTA2, MTA1L1, PID | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1) (MTA1-L1 protein) (p53 target protein in deacetylase complex). | |||||
|
MTA3_HUMAN
|
||||||
| NC score | 0.468355 (rank : 14) | θ value | 9.59137e-10 (rank : 17) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BTC8, Q9NSP2, Q9ULF4 | Gene names | MTA3, KIAA1266 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA3. | |||||
|
MTA2_MOUSE
|
||||||
| NC score | 0.467154 (rank : 15) | θ value | 5.62301e-10 (rank : 16) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9R190, Q3TVT8 | Gene names | Mta2, Mta1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA2 (Metastasis-associated 1-like 1). | |||||
|
MIER1_MOUSE
|
||||||
| NC score | 0.454024 (rank : 16) | θ value | 2.88788e-06 (rank : 20) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5UAK0, Q5UAK1, Q5UAK2, Q5UAK3, Q6ZPL6, Q9D402 | Gene names | Mier1, Kiaa1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1). | |||||
|
MIER1_HUMAN
|
||||||
| NC score | 0.449597 (rank : 17) | θ value | 4.92598e-06 (rank : 21) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8N108, Q5T104, Q6AHY8, Q86TB4, Q8N156, Q8N161, Q8NC37, Q8NES4, Q8NES5, Q8NES6, Q8WWG2, Q9HCG2 | Gene names | MIER1, KIAA1610 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mesoderm induction early response protein 1 (Mi-er1) (hmi-er1) (Early response 1) (Er1). | |||||
|
MTA1_HUMAN
|
||||||
| NC score | 0.439946 (rank : 18) | θ value | 4.0297e-08 (rank : 18) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13330, Q8NFI8, Q96GI8 | Gene names | MTA1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
|
MTA1_MOUSE
|
||||||
| NC score | 0.435568 (rank : 19) | θ value | 5.26297e-08 (rank : 19) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8K4B0, Q80UI1, Q8K4D4, Q924K9 | Gene names | Mta1 | |||
|
Domain Architecture |

|
|||||
| Description | Metastasis-associated protein MTA1. | |||||
|
TREF1_HUMAN
|
||||||
| NC score | 0.389603 (rank : 20) | θ value | 3.29651e-10 (rank : 14) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96PN7, Q7Z6T2, Q7Z6T3, Q9NQ72, Q9NQ73, Q9NUN9 | Gene names | TRERF1, RAPA, TREP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132) (Zinc finger protein rapa). | |||||
|
TREF1_MOUSE
|
||||||
| NC score | 0.388543 (rank : 21) | θ value | 1.9326e-10 (rank : 12) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.337832 (rank : 22) | θ value | 3.19293e-05 (rank : 23) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.334796 (rank : 23) | θ value | 5.44631e-05 (rank : 24) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
MYSM1_MOUSE
|
||||||
| NC score | 0.263840 (rank : 24) | θ value | 1.43324e-05 (rank : 22) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q69Z66, Q3TPV7, Q8BRP5, Q8C4N1 | Gene names | Mysm1, Kiaa1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
MYSM1_HUMAN
|
||||||
| NC score | 0.263080 (rank : 25) | θ value | 0.000461057 (rank : 25) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5VVJ2, Q7Z3G8, Q96PX3 | Gene names | MYSM1, KIAA1915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MYSM1 (Myb-like, SWIRM and MPN domain-containing protein 1). | |||||
|
DCP1A_HUMAN
|
||||||
| NC score | 0.092803 (rank : 26) | θ value | 0.00298849 (rank : 26) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NPI6 | Gene names | DCP1A, SMIF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (Smad4-interacting transcriptional co-activator). | |||||
|
RED_MOUSE
|
||||||
| NC score | 0.091278 (rank : 27) | θ value | 0.0252991 (rank : 30) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z1M8 | Gene names | Ik, Red, Rer | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Red (Protein RER) (IK factor) (Cytokine IK). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.087941 (rank : 28) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
RED_HUMAN
|
||||||
| NC score | 0.087927 (rank : 29) | θ value | 0.0330416 (rank : 32) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13123 | Gene names | IK, RED, RER | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Red (Protein RER) (IK factor) (Cytokine IK). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.084441 (rank : 30) | θ value | 0.0736092 (rank : 34) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CJ108_HUMAN
|
||||||
| NC score | 0.084129 (rank : 31) | θ value | 0.365318 (rank : 44) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N8Z3 | Gene names | C10orf108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative uncharacterized protein C10orf108. | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.080008 (rank : 32) | θ value | 0.47712 (rank : 50) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
GA2L2_MOUSE
|
||||||
| NC score | 0.079080 (rank : 33) | θ value | 0.0961366 (rank : 36) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.076769 (rank : 34) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.072997 (rank : 35) | θ value | 0.00869519 (rank : 27) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.072987 (rank : 36) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.070503 (rank : 37) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.068137 (rank : 38) | θ value | 0.00869519 (rank : 28) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.066586 (rank : 39) | θ value | 0.47712 (rank : 53) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.066561 (rank : 40) | θ value | 0.279714 (rank : 42) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.057145 (rank : 41) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.057042 (rank : 42) | θ value | 0.0252991 (rank : 29) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.055446 (rank : 43) | θ value | 0.0330416 (rank : 31) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.054802 (rank : 44) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.054768 (rank : 45) | θ value | 0.365318 (rank : 47) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.054757 (rank : 46) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.053749 (rank : 47) | θ value | 0.125558 (rank : 38) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.052711 (rank : 48) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
CCD95_HUMAN
|
||||||
| NC score | 0.052665 (rank : 49) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NBZ0 | Gene names | CCDC95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 95. | |||||
|
ATX2L_HUMAN
|
||||||
| NC score | 0.052599 (rank : 50) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.052559 (rank : 51) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.052428 (rank : 52) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
FA47A_HUMAN
|
||||||
| NC score | 0.052120 (rank : 53) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.052099 (rank : 54) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.051916 (rank : 55) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.051787 (rank : 56) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.051053 (rank : 57) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.050136 (rank : 58) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
SC24A_MOUSE
|
||||||
| NC score | 0.049305 (rank : 59) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
NUMBL_HUMAN
|
||||||
| NC score | 0.048734 (rank : 60) | θ value | 0.163984 (rank : 41) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
BCAR1_HUMAN
|
||||||
| NC score | 0.048330 (rank : 61) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.048199 (rank : 62) | θ value | 0.0961366 (rank : 37) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
SOX30_MOUSE
|
||||||
| NC score | 0.047757 (rank : 63) | θ value | 0.0563607 (rank : 33) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
BAG3_MOUSE
|
||||||
| NC score | 0.045261 (rank : 64) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
CCD55_MOUSE
|
||||||
| NC score | 0.044578 (rank : 65) | θ value | 0.365318 (rank : 43) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.043822 (rank : 66) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.043225 (rank : 67) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.042752 (rank : 68) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
HSF1_MOUSE
|
||||||
| NC score | 0.042264 (rank : 69) | θ value | 0.163984 (rank : 40) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
MYB_MOUSE
|
||||||
| NC score | 0.041609 (rank : 70) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P06876, Q61929 | Gene names | Myb | |||
|
Domain Architecture |

|
|||||
| Description | Myb proto-oncogene protein (C-myb). | |||||
|
MEF2B_HUMAN
|
||||||
| NC score | 0.039996 (rank : 71) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02080 | Gene names | MEF2B, XMEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B (Serum response factor-like protein 2) (XMEF2) (RSRFR2). | |||||
|
MAD3_HUMAN
|
||||||
| NC score | 0.039831 (rank : 72) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BW11, Q53HK1, Q7Z4Y0, Q8NDJ7, Q96ME3 | Gene names | MXD3, MAD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3) (Myx). | |||||
|
TM131_MOUSE
|
||||||
| NC score | 0.038111 (rank : 73) | θ value | 1.06291 (rank : 61) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O70472 | Gene names | Tmem131, D1Bwg0491e, Rw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 131 (Protein RW1). | |||||
|
MAD3_MOUSE
|
||||||
| NC score | 0.037369 (rank : 74) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80US8, Q60947 | Gene names | Mxd3, Mad3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Max-interacting transcriptional repressor MAD3 (Max-associated protein 3) (MAX dimerization protein 3). | |||||
|
MYB_HUMAN
|
||||||
| NC score | 0.036237 (rank : 75) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P10242, P78391, P78392, P78525, P78526, Q14023, Q14024, Q9UE83 | Gene names | MYB | |||
|
Domain Architecture |
|
|||||
| Description | Myb proto-oncogene protein (C-myb). | |||||
|
GAK11_HUMAN
|
||||||
| NC score | 0.035425 (rank : 76) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| NC score | 0.035151 (rank : 77) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
NCOA1_MOUSE
|
||||||
| NC score | 0.035108 (rank : 78) | θ value | 0.47712 (rank : 52) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70365, P70366, Q61202, Q8CBI9 | Gene names | Ncoa1, Src1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (Nuclear receptor coactivator protein 1) (mNRC-1). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.033269 (rank : 79) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
GAK2_HUMAN
|
||||||
| NC score | 0.032886 (rank : 80) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK6_HUMAN
|
||||||
| NC score | 0.032863 (rank : 81) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
EZH2_MOUSE
|
||||||
| NC score | 0.032682 (rank : 82) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61188, Q9R090 | Gene names | Ezh2, Enx1h | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
CNOT2_MOUSE
|
||||||
| NC score | 0.032153 (rank : 83) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C5L3, Q3UE39, Q80YA5, Q9D0P1 | Gene names | Cnot2 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 2 (CCR4-associated factor 2). | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.032150 (rank : 84) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.032024 (rank : 85) | θ value | 0.125558 (rank : 39) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.031926 (rank : 86) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
CNOT2_HUMAN
|
||||||
| NC score | 0.031904 (rank : 87) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZN8, Q9H3E0, Q9NSX5, Q9NWR6, Q9P028 | Gene names | CNOT2, CDC36, NOT2 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 2 (CCR4-associated factor 2). | |||||
|
NCOA1_HUMAN
|
||||||
| NC score | 0.031439 (rank : 88) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15788, O00150, O43792, O43793, Q13071, Q13420, Q6GVI5, Q7KYV3 | Gene names | NCOA1, SRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 1 (EC 2.3.1.48) (NCoA-1) (Steroid receptor coactivator 1) (SRC-1) (RIP160) (Protein Hin-2) (NY-REN-52 antigen). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.030734 (rank : 89) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
HSF1_HUMAN
|
||||||
| NC score | 0.030632 (rank : 90) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q00613 | Gene names | HSF1, HSTF1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
EZH2_HUMAN
|
||||||
| NC score | 0.030285 (rank : 91) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15910, Q15755, Q92857 | Gene names | EZH2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
PDLI7_HUMAN
|
||||||
| NC score | 0.030251 (rank : 92) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
CJ086_HUMAN
|
||||||
| NC score | 0.029021 (rank : 93) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NXX6, Q5SQQ5, Q6P673, Q8WY66, Q9BS90 | Gene names | C10orf86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf86. | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.029012 (rank : 94) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
BIN1_HUMAN
|
||||||
| NC score | 0.028889 (rank : 95) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00499, O00297, O00545, O43867, O60552, O60553, O60554, O60555, O75514, O75515, O75516, O75517, O75518, Q92944, Q99688 | Gene names | BIN1, AMPHL | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc- interacting protein 1). | |||||
|
NUMBL_MOUSE
|
||||||
| NC score | 0.028349 (rank : 96) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O08919, Q6NVG8 | Gene names | Numbl, Nbl | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein. | |||||
|
LARP1_HUMAN
|
||||||
| NC score | 0.028269 (rank : 97) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6PKG0, O94836, Q8N4M2, Q8NB73, Q9UFD7 | Gene names | LARP1, KIAA0731, LARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 1 (La ribonucleoprotein domain family member 1). | |||||
|
GAK12_HUMAN
|
||||||
| NC score | 0.028001 (rank : 98) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
HNRPU_HUMAN
|
||||||
| NC score | 0.027710 (rank : 99) | θ value | 0.365318 (rank : 46) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q00839, O75507, Q96HY9 | Gene names | HNRPU, SAFA, U21.1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U (hnRNP U) (Scaffold attachment factor A) (SAF-A) (p120) (pp120). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.026763 (rank : 100) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.026662 (rank : 101) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
MD1L1_HUMAN
|
||||||
| NC score | 0.026041 (rank : 102) | θ value | 0.47712 (rank : 51) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
4ET_MOUSE
|
||||||
| NC score | 0.025652 (rank : 103) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9EST3, Q9CSS3 | Gene names | Eif4enif1, Clast4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
CSPG5_HUMAN
|
||||||
| NC score | 0.025424 (rank : 104) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95196, Q71M39, Q71M40 | Gene names | CSPG5, CALEB, NGC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
SFR11_HUMAN
|
||||||
| NC score | 0.025341 (rank : 105) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
RBM16_HUMAN
|
||||||
| NC score | 0.025311 (rank : 106) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
STAR8_HUMAN
|
||||||
| NC score | 0.024922 (rank : 107) | θ value | 0.365318 (rank : 48) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.024136 (rank : 108) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
MTG8R_MOUSE
|
||||||
| NC score | 0.023274 (rank : 109) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O70374 | Gene names | Cbfa2t2, Cbfa2t2h, Mtgr1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1). | |||||
|
SOX13_HUMAN
|
||||||
| NC score | 0.023030 (rank : 110) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.022849 (rank : 111) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.021809 (rank : 112) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
CCD96_MOUSE
|
||||||
| NC score | 0.021750 (rank : 113) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
P73_HUMAN
|
||||||
| NC score | 0.021677 (rank : 114) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15350, O15351, Q5TBV5, Q5TBV6, Q8NHW9, Q8TDY5, Q8TDY6, Q9NTK8 | Gene names | TP73, P73 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein p73 (p53-like transcription factor) (p53-related protein). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.020623 (rank : 115) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.020606 (rank : 116) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
MTG8R_HUMAN
|
||||||
| NC score | 0.019915 (rank : 117) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43439, Q5TGE4, Q5TGE7, Q8IWF3, Q96B06, Q96L00, Q9H436, Q9UJP8, Q9UJP9, Q9UP24 | Gene names | CBFA2T2, EHT, MTGR1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein CBFA2T2 (MTG8-like protein) (MTG8-related protein 1) (Myeloid translocation-related protein 1) (ETO homologous on chromosome 20) (p85). | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.019545 (rank : 118) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.018924 (rank : 119) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
ZCHC8_MOUSE
|
||||||
| NC score | 0.018907 (rank : 120) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CYA6, Q3TK71, Q8CCB7, Q91WQ1 | Gene names | Zcchc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 8. | |||||
|
FOG1_MOUSE
|
||||||
| NC score | 0.017517 (rank : 121) | θ value | 0.0961366 (rank : 35) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
NOC2L_HUMAN
|
||||||
| NC score | 0.016870 (rank : 122) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y3T9, Q5SVA3, Q9BTN6 | Gene names | NOC2L | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar complex protein 2 homolog (Protein NOC2 homolog) (NOC2- like). | |||||
|
CB013_HUMAN
|
||||||
| NC score | 0.016867 (rank : 123) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IW19, Q53P47, Q53PB9, Q53QU0 | Gene names | C2orf13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C2orf13. | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.016552 (rank : 124) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
NTBP1_MOUSE
|
||||||
| NC score | 0.016351 (rank : 125) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BRM2, Q8CAA8 | Gene names | Scyl1bp1, Ntklbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-terminal kinase-like-binding protein 1 (NTKL-binding protein 1) (NTKL-BP1) (mNTKL-BP1) (SCY1-like 1-binding protein 1) (SCYL1-binding protein 1) (SCYL1-BP1). | |||||
|
TRI56_HUMAN
|
||||||
| NC score | 0.016268 (rank : 126) | θ value | 0.62314 (rank : 54) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BRZ2, Q86VT6, Q8N2H8, Q8NAC0, Q9H031 | Gene names | TRIM56, RNF109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 56 (RING finger protein 109). | |||||
|
US6NL_HUMAN
|
||||||
| NC score | 0.016040 (rank : 127) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92738, Q15400, Q7L0K9 | Gene names | USP6NL, KIAA0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein (Related to the N-terminus of tre) (RN- tre). | |||||
|
IF4B_MOUSE
|
||||||
| NC score | 0.015884 (rank : 128) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
DCC_MOUSE
|
||||||
| NC score | 0.015705 (rank : 129) | θ value | 0.365318 (rank : 45) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.014995 (rank : 130) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
P66A_MOUSE
|
||||||
| NC score | 0.014855 (rank : 131) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CHY6, Q8BTQ2, Q8VEC9 | Gene names | Gatad2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional repressor p66 alpha (GATA zinc finger domain- containing protein 2A). | |||||
|
FGD1_MOUSE
|
||||||
| NC score | 0.014841 (rank : 132) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
CCD21_MOUSE
|
||||||
| NC score | 0.014602 (rank : 133) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
RHG09_HUMAN
|
||||||
| NC score | 0.014244 (rank : 134) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
ETV5_MOUSE
|
||||||
| NC score | 0.013960 (rank : 135) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
MD1L1_MOUSE
|
||||||
| NC score | 0.013726 (rank : 136) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
QSCN6_HUMAN
|
||||||
| NC score | 0.013071 (rank : 137) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00391, Q59G29, Q5T2X0, Q8TDL6, Q8WVP4 | Gene names | QSCN6, QSOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfhydryl oxidase 1 precursor (EC 1.8.3.2) (Quiescin Q6) (hQSOX). | |||||
|
PF21A_MOUSE
|
||||||
| NC score | 0.013021 (rank : 138) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
LPIN3_MOUSE
|
||||||
| NC score | 0.012848 (rank : 139) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99PI4, Q3TQ75, Q8C7R9 | Gene names | Lpin3 | |||
|
Domain Architecture |

|
|||||
| Description | Lipin-3. | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.012722 (rank : 140) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.012192 (rank : 141) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.011580 (rank : 142) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
KI21A_HUMAN
|
||||||
| NC score | 0.011497 (rank : 143) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
PEX2_MOUSE
|
||||||
| NC score | 0.011372 (rank : 144) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55098, O35467 | Gene names | Pxmp3, Paf1, Pex2, Pmp35 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 1 (PAF-1) (Peroxin-2) (Peroxisomal membrane protein 3). | |||||
|
DDX23_HUMAN
|
||||||
| NC score | 0.011272 (rank : 145) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
K0174_MOUSE
|
||||||
| NC score | 0.011124 (rank : 146) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CX00, Q80U68 | Gene names | Kiaa0174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0174. | |||||
|
SMAD4_MOUSE
|
||||||
| NC score | 0.011016 (rank : 147) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
MYT1_MOUSE
|
||||||
| NC score | 0.010840 (rank : 148) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
SMAD4_HUMAN
|
||||||
| NC score | 0.010729 (rank : 149) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13485 | Gene names | SMAD4, DPC4, MADH4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4) (hSMAD4). | |||||
|
FA76B_HUMAN
|
||||||
| NC score | 0.010152 (rank : 150) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5HYJ3, Q6PIU3, Q8TC53 | Gene names | FAM76B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM76B. | |||||
|
DDX10_HUMAN
|
||||||
| NC score | 0.010003 (rank : 151) | θ value | 0.47712 (rank : 49) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13206, Q5BJD8 | Gene names | DDX10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.009964 (rank : 152) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
UBQL1_HUMAN
|
||||||
| NC score | 0.009501 (rank : 153) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UMX0, Q8IXS9, Q8N2Q3, Q9H0T8, Q9H3R4, Q9HAZ5 | Gene names | UBQLN1, DA41, PLIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-1 (Protein linking IAP with cytoskeleton 1) (PLIC-1) (hPLIC- 1). | |||||
|
MAGI1_HUMAN
|
||||||
| NC score | 0.009305 (rank : 154) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
ONEC2_MOUSE
|
||||||
| NC score | 0.009153 (rank : 155) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6XBJ3 | Gene names | Onecut2, Oc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | One cut domain family member 2 (Transcription factor ONECUT-2) (OC-2). | |||||
|
PP16A_HUMAN
|
||||||
| NC score | 0.007963 (rank : 156) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96I34 | Gene names | PPP1R16A, MYPT3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 16A (Myosin phosphatase- targeting subunit 3). | |||||
|
K0256_MOUSE
|
||||||
| NC score | 0.007628 (rank : 157) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6A098 | Gene names | Kiaa0256 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0256. | |||||
|
SMRD2_MOUSE
|
||||||
| NC score | 0.006726 (rank : 158) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99JR8 | Gene names | Smarcd2, Baf60b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 2 (60 kDa BRG-1/Brm-associated factor subunit B) (BRG1-associated factor 60B). | |||||
|
IRX2_MOUSE
|
||||||
| NC score | 0.006502 (rank : 159) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P81066, O55121, Q9ERN1 | Gene names | Irx2, Irxa2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-2 (Iroquois homeobox protein 2) (Homeodomain protein IRXA2). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.006441 (rank : 160) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
PBX4_MOUSE
|
||||||
| NC score | 0.006019 (rank : 161) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99NE9, Q8CFL2, Q99NE8 | Gene names | Pbx4 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-B-cell leukemia transcription factor 4 (Homeobox protein PBX4). | |||||
|
FNBP2_HUMAN
|
||||||
| NC score | 0.005362 (rank : 162) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
SIX2_MOUSE
|
||||||
| NC score | 0.004583 (rank : 163) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62232, P70179 | Gene names | Six2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX2 (Sine oculis homeobox homolog 2). | |||||
|
NDF1_HUMAN
|
||||||
| NC score | 0.004573 (rank : 164) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13562, O00343, Q13340, Q5U095, Q96TH0, Q99455, Q9UEC8 | Gene names | NEUROD1, NEUROD | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1) (NeuroD). | |||||
|
CAH9_MOUSE
|
||||||
| NC score | 0.004375 (rank : 165) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VHB5, Q8K1G1, Q8VDE4 | Gene names | Ca9, Car9 | |||
|
Domain Architecture |

|
|||||
| Description | Carbonic anhydrase 9 precursor (EC 4.2.1.1) (Carbonic anhydrase IX) (Carbonate dehydratase IX) (CA-IX) (CAIX) (Membrane antigen MN homolog). | |||||
|
ANKR6_HUMAN
|
||||||
| NC score | 0.003845 (rank : 166) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
KAISO_MOUSE
|
||||||
| NC score | 0.003218 (rank : 167) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BN78, Q8C226, Q9WTZ7 | Gene names | Zbtb33, Kaiso | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional regulator Kaiso (Zinc finger and BTB domain-containing protein 33). | |||||
|
PC11X_HUMAN
|
||||||
| NC score | 0.002415 (rank : 168) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BZA7, Q2TJH0, Q2TJH1, Q2TJH3, Q5JVZ0, Q70LR8, Q70LS7, Q70LS8, Q70LS9, Q70LT7, Q70LT8, Q70LT9, Q70LU0, Q70LU1, Q96RV4, Q96RW0, Q9BZA6, Q9H4E0, Q9P2M0, Q9P2X5 | Gene names | PCDH11X, PCDH11, PCDHX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 X-linked precursor (Protocadherin-11) (Protocadherin on the X chromosome) (PCDH-X) (Protocadherin-S). | |||||
|
KAISO_HUMAN
|
||||||
| NC score | 0.002305 (rank : 169) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86T24, O00319, Q7Z361, Q8IVP6, Q8N3P0 | Gene names | ZBTB33, KAISO, ZNF348 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional regulator Kaiso (Zinc finger and BTB domain-containing protein 33). | |||||
|
MYLK_MOUSE
|
||||||
| NC score | 0.001707 (rank : 170) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
MAST2_HUMAN
|
||||||
| NC score | 0.000350 (rank : 171) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
ZN473_MOUSE
|
||||||
| NC score | -0.001890 (rank : 172) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 824 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BI67, Q8BI98, Q8BIB7 | Gene names | Znf473, Zfp100, Zfp473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 473 homolog (Zinc finger protein 100) (Zfp-100). | |||||