Please be patient as the page loads
|
NOC2L_HUMAN
|
||||||
| SwissProt Accessions | Q9Y3T9, Q5SVA3, Q9BTN6 | Gene names | NOC2L | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar complex protein 2 homolog (Protein NOC2 homolog) (NOC2- like). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NOC2L_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y3T9, Q5SVA3, Q9BTN6 | Gene names | NOC2L | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar complex protein 2 homolog (Protein NOC2 homolog) (NOC2- like). | |||||
|
NOC2L_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.975972 (rank : 2) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WV70, Q8VE16 | Gene names | Noc2l | |||
|
Domain Architecture |
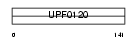
|
|||||
| Description | Nucleolar complex protein 2 homolog (Protein NOC2 homolog) (NOC2- like). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 3) | NC score | 0.047542 (rank : 7) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 4) | NC score | 0.055005 (rank : 4) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 5) | NC score | 0.057065 (rank : 3) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
HOOK1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 6) | NC score | 0.026599 (rank : 14) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
WDR60_MOUSE
|
||||||
| θ value | 0.365318 (rank : 7) | NC score | 0.049918 (rank : 5) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C761, Q3UNM4 | Gene names | Wdr60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
HOOK1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 8) | NC score | 0.025775 (rank : 19) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BIL5, Q8BIZ2, Q8K0P1, Q8K454, Q9CTN6 | Gene names | Hook1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1. | |||||
|
RFIP4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 9) | NC score | 0.027359 (rank : 13) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86YS3, Q8N829, Q8NDT7, Q969D8 | Gene names | RAB11FIP4, KIAA1821 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 4 (Rab11-FIP4). | |||||
|
LMD1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 10) | NC score | 0.030635 (rank : 10) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29536 | Gene names | LMOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Leiomodin-1 (Leiomodin, muscle form) (64 kDa autoantigen D1) (64 kDa autoantigen 1D) (64 kDa autoantigen 1D3) (Thyroid-associated ophthalmopathy autoantigen) (Smooth muscle leiomodin) (SM-Lmod). | |||||
|
CA065_HUMAN
|
||||||
| θ value | 2.36792 (rank : 11) | NC score | 0.026312 (rank : 15) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N715, Q8N746, Q8NA93 | Gene names | C1orf65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf65. | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 12) | NC score | 0.042432 (rank : 8) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 13) | NC score | 0.040693 (rank : 9) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
CENPK_HUMAN
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.025791 (rank : 18) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BS16, Q9H4L0 | Gene names | CENPK, ICEN37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein K (CENP-K) (Interphase centromere complex protein 37) (Protein AF-5alpha) (p33). | |||||
|
FBX38_HUMAN
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.028374 (rank : 12) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PIJ6, Q6PK72, Q7Z2U0, Q86VN3, Q9BXY6, Q9H837, Q9HC40 | Gene names | FBXO38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 38 (Modulator of KLF7 activity homolog) (MoKA). | |||||
|
HS90B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.026067 (rank : 16) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11499, O89078 | Gene names | Hsp90ab1, Hsp84, Hsp84-1, Hspcb | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (Tumor-specific transplantation 84 kDa antigen) (TSTA). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.047702 (rank : 6) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
TLK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.015211 (rank : 35) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UKI8, Q14150, Q8N591, Q9NYH2, Q9Y4F6 | Gene names | TLK1, KIAA0137 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 1 (EC 2.7.11.1) (Tousled- like kinase 1) (PKU-beta). | |||||
|
TLK1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.015059 (rank : 37) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C0V0 | Gene names | Tlk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 1 (EC 2.7.11.1) (Tousled- like kinase 1). | |||||
|
ARHGG_MOUSE
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.013511 (rank : 39) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3U5C8, Q501M8, Q8VCE8 | Gene names | Arhgef16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
CCAR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.025432 (rank : 21) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CH18, Q6AXC9, Q6PAR2, Q80XE4, Q8BJY0, Q8BVN2, Q8CGG1, Q9CSR5 | Gene names | Ccar1, Carp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1). | |||||
|
DHX37_HUMAN
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.014816 (rank : 38) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IY37, Q9BUI7, Q9P211 | Gene names | DHX37, DDX37, KIAA1517 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX37 (EC 3.6.1.-) (DEAH box protein 37). | |||||
|
ELL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.020212 (rank : 27) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55199 | Gene names | ELL, C19orf17 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL (Eleven-nineteen lysine-rich leukemia protein). | |||||
|
FYV1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.025699 (rank : 20) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
FYV1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.025948 (rank : 17) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z1T6 | Gene names | Pip5k3, Pikfyve | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (PIP5K) (PtdIns(4)P-5- kinase) (PIKfyve) (p235). | |||||
|
HS90B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.025225 (rank : 22) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P08238, Q5T9W7, Q9NQW0 | Gene names | HSP90AB1, HSP90B, HSPC2, HSPCB | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (HSP 90). | |||||
|
K0555_HUMAN
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.017204 (rank : 32) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
LATS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.007698 (rank : 44) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
TLK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.015243 (rank : 34) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86UE8, Q9UKI7, Q9Y4F7 | Gene names | TLK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 2 (EC 2.7.11.1) (Tousled- like kinase 2) (PKU-alpha). | |||||
|
TLK2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.015070 (rank : 36) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O55047, Q9D5Y5 | Gene names | Tlk2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 2 (EC 2.7.11.1) (Tousled- like kinase 2). | |||||
|
CALL3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.007793 (rank : 43) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
LRRF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.024576 (rank : 23) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.019747 (rank : 29) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
RCOR3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.019871 (rank : 28) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PGA0 | Gene names | Rcor3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
RFIP4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.023877 (rank : 24) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BQP8, Q80T87 | Gene names | Rab11fip4, Kiaa1821 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 4 (Rab11-FIP4). | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.007595 (rank : 46) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.007674 (rank : 45) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
ERCC5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.018602 (rank : 30) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P28715, Q7Z2V3, Q8IZL6, Q8N1B7, Q9HD59 | Gene names | ERCC5, XPG, XPGC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells (Xeroderma pigmentosum group G-complementing protein) (DNA excision repair protein ERCC-5). | |||||
|
FMNL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.018160 (rank : 31) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JL26, Q6KAN4, Q9Z2V7 | Gene names | Fmnl1, Frl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-like protein 1 (Formin-related protein). | |||||
|
LATS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.006975 (rank : 47) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95835, Q6PKD0 | Gene names | LATS1, WARTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase) (h-warts). | |||||
|
SRBS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.011554 (rank : 40) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BX66, Q7LBE5, Q8IVK0, Q8IVQ4, Q96KF3, Q96KF4, Q9BX64, Q9BX65, Q9P2Q0, Q9UFT2, Q9UHN7, Q9Y338 | Gene names | SORBS1, KIAA1296, SH3D5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
UBF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.023084 (rank : 25) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
CCD55_MOUSE
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.028656 (rank : 11) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.008918 (rank : 42) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
IF39_MOUSE
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.020628 (rank : 26) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
RAC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.004401 (rank : 48) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P60763, O14658, Q5U0M8 | Gene names | RAC3 | |||
|
Domain Architecture |
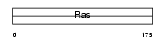
|
|||||
| Description | Ras-related C3 botulinum toxin substrate 3 (p21-Rac3). | |||||
|
RAC3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.004401 (rank : 49) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P60764, O14658 | Gene names | Rac3 | |||
|
Domain Architecture |
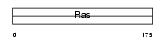
|
|||||
| Description | Ras-related C3 botulinum toxin substrate 3 (p21-Rac3). | |||||
|
RCOR3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.016870 (rank : 33) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
RIMB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.009794 (rank : 41) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15034, Q96ID2 | Gene names | RIMBP2, KIAA0318, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | RIM-binding protein 2 (RIM-BP2). | |||||
|
NOC2L_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y3T9, Q5SVA3, Q9BTN6 | Gene names | NOC2L | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar complex protein 2 homolog (Protein NOC2 homolog) (NOC2- like). | |||||
|
NOC2L_MOUSE
|
||||||
| NC score | 0.975972 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WV70, Q8VE16 | Gene names | Noc2l | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar complex protein 2 homolog (Protein NOC2 homolog) (NOC2- like). | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.057065 (rank : 3) | θ value | 0.279714 (rank : 5) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.055005 (rank : 4) | θ value | 0.163984 (rank : 4) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
WDR60_MOUSE
|
||||||
| NC score | 0.049918 (rank : 5) | θ value | 0.365318 (rank : 7) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C761, Q3UNM4 | Gene names | Wdr60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.047702 (rank : 6) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.047542 (rank : 7) | θ value | 0.163984 (rank : 3) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.042432 (rank : 8) | θ value | 2.36792 (rank : 12) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.040693 (rank : 9) | θ value | 2.36792 (rank : 13) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
LMD1_HUMAN
|
||||||
| NC score | 0.030635 (rank : 10) | θ value | 1.81305 (rank : 10) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29536 | Gene names | LMOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Leiomodin-1 (Leiomodin, muscle form) (64 kDa autoantigen D1) (64 kDa autoantigen 1D) (64 kDa autoantigen 1D3) (Thyroid-associated ophthalmopathy autoantigen) (Smooth muscle leiomodin) (SM-Lmod). | |||||
|
CCD55_MOUSE
|
||||||
| NC score | 0.028656 (rank : 11) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
FBX38_HUMAN
|
||||||
| NC score | 0.028374 (rank : 12) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PIJ6, Q6PK72, Q7Z2U0, Q86VN3, Q9BXY6, Q9H837, Q9HC40 | Gene names | FBXO38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 38 (Modulator of KLF7 activity homolog) (MoKA). | |||||
|
RFIP4_HUMAN
|
||||||
| NC score | 0.027359 (rank : 13) | θ value | 1.38821 (rank : 9) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86YS3, Q8N829, Q8NDT7, Q969D8 | Gene names | RAB11FIP4, KIAA1821 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 4 (Rab11-FIP4). | |||||
|
HOOK1_HUMAN
|
||||||
| NC score | 0.026599 (rank : 14) | θ value | 0.365318 (rank : 6) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
CA065_HUMAN
|
||||||
| NC score | 0.026312 (rank : 15) | θ value | 2.36792 (rank : 11) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N715, Q8N746, Q8NA93 | Gene names | C1orf65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf65. | |||||
|
HS90B_MOUSE
|
||||||
| NC score | 0.026067 (rank : 16) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11499, O89078 | Gene names | Hsp90ab1, Hsp84, Hsp84-1, Hspcb | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (Tumor-specific transplantation 84 kDa antigen) (TSTA). | |||||
|
FYV1_MOUSE
|
||||||
| NC score | 0.025948 (rank : 17) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z1T6 | Gene names | Pip5k3, Pikfyve | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (PIP5K) (PtdIns(4)P-5- kinase) (PIKfyve) (p235). | |||||
|
CENPK_HUMAN
|
||||||
| NC score | 0.025791 (rank : 18) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BS16, Q9H4L0 | Gene names | CENPK, ICEN37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein K (CENP-K) (Interphase centromere complex protein 37) (Protein AF-5alpha) (p33). | |||||
|
HOOK1_MOUSE
|
||||||
| NC score | 0.025775 (rank : 19) | θ value | 0.62314 (rank : 8) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BIL5, Q8BIZ2, Q8K0P1, Q8K454, Q9CTN6 | Gene names | Hook1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1. | |||||
|
FYV1_HUMAN
|
||||||
| NC score | 0.025699 (rank : 20) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2I7, Q8NB67 | Gene names | PIP5K3, KIAA0981, PIKFYVE | |||
|
Domain Architecture |

|
|||||
| Description | FYVE finger-containing phosphoinositide kinase (EC 2.7.1.68) (1- phosphatidylinositol-4-phosphate 5-kinase) (Phosphatidylinositol-3- phosphate 5-kinase type III) (PIP5K) (PtdIns(4)P-5-kinase) (PIKfyve) (p235). | |||||
|
CCAR1_MOUSE
|
||||||
| NC score | 0.025432 (rank : 21) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CH18, Q6AXC9, Q6PAR2, Q80XE4, Q8BJY0, Q8BVN2, Q8CGG1, Q9CSR5 | Gene names | Ccar1, Carp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle and apoptosis regulator protein 1 (Cell cycle and apoptosis regulatory protein 1) (CARP-1). | |||||
|
HS90B_HUMAN
|
||||||
| NC score | 0.025225 (rank : 22) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P08238, Q5T9W7, Q9NQW0 | Gene names | HSP90AB1, HSP90B, HSPC2, HSPCB | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (HSP 90). | |||||
|
LRRF1_MOUSE
|
||||||
| NC score | 0.024576 (rank : 23) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
RFIP4_MOUSE
|
||||||
| NC score | 0.023877 (rank : 24) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BQP8, Q80T87 | Gene names | Rab11fip4, Kiaa1821 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 4 (Rab11-FIP4). | |||||
|
UBF1_HUMAN
|
||||||
| NC score | 0.023084 (rank : 25) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P17480 | Gene names | UBTF, UBF, UBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar transcription factor 1 (Upstream-binding factor 1) (UBF-1) (Autoantigen NOR-90). | |||||
|
IF39_MOUSE
|
||||||
| NC score | 0.020628 (rank : 26) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8JZQ9, Q922K2 | Gene names | Eif3s9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116). | |||||
|
ELL_HUMAN
|
||||||
| NC score | 0.020212 (rank : 27) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55199 | Gene names | ELL, C19orf17 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL (Eleven-nineteen lysine-rich leukemia protein). | |||||
|
RCOR3_MOUSE
|
||||||
| NC score | 0.019871 (rank : 28) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PGA0 | Gene names | Rcor3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.019747 (rank : 29) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
ERCC5_HUMAN
|
||||||
| NC score | 0.018602 (rank : 30) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P28715, Q7Z2V3, Q8IZL6, Q8N1B7, Q9HD59 | Gene names | ERCC5, XPG, XPGC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells (Xeroderma pigmentosum group G-complementing protein) (DNA excision repair protein ERCC-5). | |||||
|
FMNL_MOUSE
|
||||||
| NC score | 0.018160 (rank : 31) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JL26, Q6KAN4, Q9Z2V7 | Gene names | Fmnl1, Frl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-like protein 1 (Formin-related protein). | |||||
|
K0555_HUMAN
|
||||||
| NC score | 0.017204 (rank : 32) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
RCOR3_HUMAN
|
||||||
| NC score | 0.016870 (rank : 33) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
TLK2_HUMAN
|
||||||
| NC score | 0.015243 (rank : 34) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86UE8, Q9UKI7, Q9Y4F7 | Gene names | TLK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 2 (EC 2.7.11.1) (Tousled- like kinase 2) (PKU-alpha). | |||||
|
TLK1_HUMAN
|
||||||
| NC score | 0.015211 (rank : 35) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 967 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UKI8, Q14150, Q8N591, Q9NYH2, Q9Y4F6 | Gene names | TLK1, KIAA0137 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 1 (EC 2.7.11.1) (Tousled- like kinase 1) (PKU-beta). | |||||
|
TLK2_MOUSE
|
||||||
| NC score | 0.015070 (rank : 36) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O55047, Q9D5Y5 | Gene names | Tlk2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 2 (EC 2.7.11.1) (Tousled- like kinase 2). | |||||
|
TLK1_MOUSE
|
||||||
| NC score | 0.015059 (rank : 37) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C0V0 | Gene names | Tlk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase tousled-like 1 (EC 2.7.11.1) (Tousled- like kinase 1). | |||||
|
DHX37_HUMAN
|
||||||
| NC score | 0.014816 (rank : 38) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IY37, Q9BUI7, Q9P211 | Gene names | DHX37, DDX37, KIAA1517 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DHX37 (EC 3.6.1.-) (DEAH box protein 37). | |||||
|
ARHGG_MOUSE
|
||||||
| NC score | 0.013511 (rank : 39) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3U5C8, Q501M8, Q8VCE8 | Gene names | Arhgef16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
SRBS1_HUMAN
|
||||||
| NC score | 0.011554 (rank : 40) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BX66, Q7LBE5, Q8IVK0, Q8IVQ4, Q96KF3, Q96KF4, Q9BX64, Q9BX65, Q9P2Q0, Q9UFT2, Q9UHN7, Q9Y338 | Gene names | SORBS1, KIAA1296, SH3D5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
RIMB2_HUMAN
|
||||||
| NC score | 0.009794 (rank : 41) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15034, Q96ID2 | Gene names | RIMBP2, KIAA0318, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | RIM-binding protein 2 (RIM-BP2). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.008918 (rank : 42) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
CALL3_HUMAN
|
||||||
| NC score | 0.007793 (rank : 43) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
LATS1_MOUSE
|
||||||
| NC score | 0.007698 (rank : 44) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | 0.007674 (rank : 45) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.007595 (rank : 46) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
LATS1_HUMAN
|
||||||
| NC score | 0.006975 (rank : 47) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95835, Q6PKD0 | Gene names | LATS1, WARTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase) (h-warts). | |||||
|
RAC3_HUMAN
|
||||||
| NC score | 0.004401 (rank : 48) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P60763, O14658, Q5U0M8 | Gene names | RAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-related C3 botulinum toxin substrate 3 (p21-Rac3). | |||||
|
RAC3_MOUSE
|
||||||
| NC score | 0.004401 (rank : 49) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P60764, O14658 | Gene names | Rac3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-related C3 botulinum toxin substrate 3 (p21-Rac3). | |||||