Please be patient as the page loads
|
HS90B_MOUSE
|
||||||
| SwissProt Accessions | P11499, O89078 | Gene names | Hsp90ab1, Hsp84, Hsp84-1, Hspcb | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (Tumor-specific transplantation 84 kDa antigen) (TSTA). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HS90A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.977869 (rank : 4) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P07900, Q2PP14, Q9BVQ5 | Gene names | HSP90AA1, HSP90A, HSPC1, HSPCA | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (NY-REN-38 antigen). | |||||
|
HS90A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.979765 (rank : 3) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P07901 | Gene names | Hsp90aa1, Hsp86, Hsp86-1, Hspca | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (Tumor-specific transplantation 86 kDa antigen) (TSTA). | |||||
|
HS90B_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.998135 (rank : 2) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P08238, Q5T9W7, Q9NQW0 | Gene names | HSP90AB1, HSP90B, HSPC2, HSPCB | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (HSP 90). | |||||
|
HS90B_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P11499, O89078 | Gene names | Hsp90ab1, Hsp84, Hsp84-1, Hspcb | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (Tumor-specific transplantation 84 kDa antigen) (TSTA). | |||||
|
ENPL_HUMAN
|
||||||
| θ value | 1.0648e-109 (rank : 5) | NC score | 0.933307 (rank : 5) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P14625, Q96A97 | Gene names | HSP90B1, TRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmin precursor (Heat shock protein 90 kDa beta member 1) (94 kDa glucose-regulated protein) (GRP94) (gp96 homolog) (Tumor rejection antigen 1). | |||||
|
ENPL_MOUSE
|
||||||
| θ value | 6.90186e-109 (rank : 6) | NC score | 0.931581 (rank : 6) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P08113, P11427 | Gene names | Hsp90b1, Tra-1, Tra1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmin precursor (Heat shock protein 90 kDa beta member 1) (94 kDa glucose-regulated protein) (GRP94) (ERP99) (Polymorphic tumor rejection antigen 1) (Tumor rejection antigen gp96). | |||||
|
TRAP1_HUMAN
|
||||||
| θ value | 3.60269e-49 (rank : 7) | NC score | 0.855428 (rank : 7) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12931, O43642, O75235, Q9UHL5 | Gene names | TRAP1, HSP75 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein 75 kDa, mitochondrial precursor (HSP 75) (Tumor necrosis factor type 1 receptor-associated protein) (TRAP-1) (TNFR- associated protein 1). | |||||
|
TRAP1_MOUSE
|
||||||
| θ value | 1.36902e-48 (rank : 8) | NC score | 0.854343 (rank : 8) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CQN1, Q542I4 | Gene names | Trap1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein 75 kDa, mitochondrial precursor (HSP 75) (Tumor necrosis factor type 1 receptor-associated protein) (TRAP-1) (TNFR- associated protein 1). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 9) | NC score | 0.107818 (rank : 10) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 10) | NC score | 0.101448 (rank : 11) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 11) | NC score | 0.038145 (rank : 20) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
KI67_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 12) | NC score | 0.058169 (rank : 14) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
CND3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 13) | NC score | 0.080262 (rank : 13) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BPX3, Q96SV9, Q9BUR3, Q9BVY1, Q9H914, Q9H9Z6, Q9HBI9 | Gene names | NCAPG, CAPG, NYMEL3 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 3 (Non-SMC condensin I complex subunit G) (Chromosome-associated protein G) (Condensin subunit CAP-G) (hCAP-G) (XCAP-G homolog) (Antigen NY-MEL-3). | |||||
|
RELN_HUMAN
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.046467 (rank : 15) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78509, Q86UJ0, Q86UJ8, Q8NDV0, Q9UDQ2 | Gene names | RELN | |||
|
Domain Architecture |
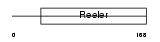
|
|||||
| Description | Reelin precursor (EC 3.4.21.-). | |||||
|
NUF2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.037940 (rank : 21) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BZD4, Q8WU69, Q96HJ4, Q96Q78 | Gene names | CDCA1, NUF2, NUF2R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (hNuf2) (hNuf2R) (hsNuf2) (Cell division cycle-associated protein 1). | |||||
|
HSF2B_HUMAN
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.144171 (rank : 9) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75031 | Gene names | HSF2BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock factor 2-binding protein. | |||||
|
OXSR1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.012642 (rank : 81) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 865 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P9R2, Q8BVZ9, Q8C0B9 | Gene names | Oxsr1, Osr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase OSR1 (EC 2.7.11.1) (Oxidative stress- responsive 1 protein). | |||||
|
PRI1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.089966 (rank : 12) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P20664 | Gene names | Prim1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA primase small subunit (EC 2.7.7.-) (DNA primase 49 kDa subunit) (p49). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.028831 (rank : 33) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
LAMA2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.016738 (rank : 71) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.037353 (rank : 23) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
AT10A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.021567 (rank : 53) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60312, Q969I4 | Gene names | ATP10A, ATP10C, ATPVC, KIAA0566 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (ATPVA) (Aminophospholipid translocase VA). | |||||
|
KIF5A_MOUSE
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.019853 (rank : 58) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
OXSR1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.011295 (rank : 84) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95747, Q3LR53, Q7Z501, Q9UPQ1 | Gene names | OXSR1, KIAA1101, OSR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase OSR1 (EC 2.7.11.1) (Oxidative stress- responsive 1 protein). | |||||
|
PHF3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.040352 (rank : 18) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
CCD46_MOUSE
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.023852 (rank : 48) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
I5P2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.014264 (rank : 77) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P32019, Q5VSG9, Q5VSH0, Q5VSH1, Q658Q5, Q6P6D4, Q6PD53, Q86YE1 | Gene names | INPP5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-1,4,5-trisphosphate 5-phosphatase precursor (EC 3.1.3.36) (Phosphoinositide 5-phosphatase) (5PTase) (75 kDa inositol polyphosphate-5-phosphatase). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.033895 (rank : 26) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
DCDC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.023484 (rank : 49) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UHG0, Q5VTR8, Q5VTR9, Q86W35, Q9UFD1, Q9ULR6 | Gene names | DCDC2, KIAA1154 | |||
|
Domain Architecture |
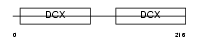
|
|||||
| Description | Doublecortin domain-containing protein 2 (RU2S protein). | |||||
|
DYH11_HUMAN
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.025208 (rank : 43) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96DT5, Q9UJ82 | Gene names | DNAH11 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 11 (Axonemal beta dynein heavy chain 11). | |||||
|
RELN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.035908 (rank : 24) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60841, Q9CUA6 | Gene names | Reln, Rl | |||
|
Domain Architecture |
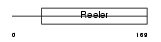
|
|||||
| Description | Reelin precursor (EC 3.4.21.-) (Reeler protein). | |||||
|
CEP55_MOUSE
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.027082 (rank : 35) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BT07, Q8C2J0, Q8K2I8, Q8R2Y4, Q9DBZ8 | Gene names | Cep55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55. | |||||
|
E2F6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.017440 (rank : 67) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75461, O60544, Q7Z2H6 | Gene names | E2F6 | |||
|
Domain Architecture |
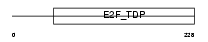
|
|||||
| Description | Transcription factor E2F6 (E2F-6). | |||||
|
NAF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.025371 (rank : 42) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
CLC6A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.012691 (rank : 80) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6EIG7 | Gene names | CLEC6A, CLECSF10, DECTIN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 6 member A (C-type lectin superfamily member 10) (Dendritic cell-associated lectin 2) (DC-associated C-type lectin-2) (DECTIN-2). | |||||
|
EVI1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.001986 (rank : 95) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
MPIP3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.031126 (rank : 28) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30307, Q96PL3, Q9H168, Q9H2E8, Q9H2E9, Q9H2F1 | Gene names | CDC25C | |||
|
Domain Architecture |

|
|||||
| Description | M-phase inducer phosphatase 3 (EC 3.1.3.48) (Dual specificity phosphatase Cdc25C). | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.025803 (rank : 41) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
NOC2L_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.026067 (rank : 39) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y3T9, Q5SVA3, Q9BTN6 | Gene names | NOC2L | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar complex protein 2 homolog (Protein NOC2 homolog) (NOC2- like). | |||||
|
P52K_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.024497 (rank : 46) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.030608 (rank : 30) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.029602 (rank : 31) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
SMC2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.037901 (rank : 22) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
ST1E1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.023162 (rank : 50) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49888, Q8N6X5 | Gene names | SULT1E1, STE | |||
|
Domain Architecture |

|
|||||
| Description | Estrogen sulfotransferase (EC 2.8.2.4) (Sulfotransferase, estrogen- preferring) (EST-1). | |||||
|
ZA2G_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.007748 (rank : 87) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64726 | Gene names | Azgp1 | |||
|
Domain Architecture |
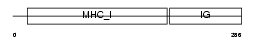
|
|||||
| Description | Zinc-alpha-2-glycoprotein precursor (Zn-alpha-2-glycoprotein) (Zn- alpha-2-GP). | |||||
|
ZN318_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.039485 (rank : 19) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.019115 (rank : 60) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.021429 (rank : 55) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.021492 (rank : 54) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.022404 (rank : 51) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CJ092_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.031197 (rank : 27) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IYW2, Q5JSF7, Q9NTQ5 | Gene names | C10orf92 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf92. | |||||
|
RGS6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.011854 (rank : 83) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49758, O75576, O75577, Q8TE13, Q8TE14, Q8TE15, Q8TE16, Q8TE17, Q8TE18, Q8TE19, Q8TE20, Q8TE21, Q8TE22, Q9Y245, Q9Y647 | Gene names | RGS6 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 6 (RGS6) (S914). | |||||
|
STK39_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.007281 (rank : 89) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UEW8, O14774, Q9UER4 | Gene names | STK39, SPAK | |||
|
Domain Architecture |
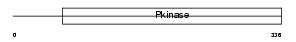
|
|||||
| Description | STE20/SPS1-related proline-alanine-rich protein kinase (EC 2.7.11.1) (Ste-20-related kinase) (Serine/threonine-protein kinase 39) (DCHT). | |||||
|
STK39_MOUSE
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.007902 (rank : 86) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 915 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z1W9, Q80W13 | Gene names | Stk39, Spak | |||
|
Domain Architecture |
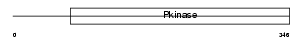
|
|||||
| Description | STE20/SPS1-related proline-alanine-rich protein kinase (EC 2.7.11.1) (Ste-20-related kinase) (Serine/threonine-protein kinase 39). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.031000 (rank : 29) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
ADA30_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.014636 (rank : 75) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UKF2, Q9UKF1 | Gene names | ADAM30 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 30 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 30). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.017632 (rank : 66) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
E2F6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.021672 (rank : 52) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54917 | Gene names | E2f6 | |||
|
Domain Architecture |
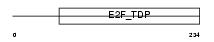
|
|||||
| Description | Transcription factor E2F6 (E2F-6) (E2F-binding site-modulating activity protein) (EMA). | |||||
|
IF4G3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.017248 (rank : 68) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
IF4G3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.017163 (rank : 69) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
KIF5C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.016105 (rank : 72) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
MRIP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.025966 (rank : 40) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
MRIP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.026268 (rank : 38) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.034181 (rank : 25) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
PMS2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.024877 (rank : 44) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54278 | Gene names | PMS2, PMSL2 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.027003 (rank : 36) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
REST_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.016852 (rank : 70) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
TARA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.024778 (rank : 45) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
AT10A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.012125 (rank : 82) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54827, Q8R3B8 | Gene names | Atp10a, Atpc5, Pfatp | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (P-locus fat-associated ATPase). | |||||
|
DYH9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.023947 (rank : 47) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NYC9, O95494, Q9NQ28 | Gene names | DNAH9, DNAH17L, DNAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 9 (Axonemal beta dynein heavy chain 9). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.040673 (rank : 17) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.043806 (rank : 16) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LMO7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.019363 (rank : 59) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
NCOAT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.010321 (rank : 85) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60502, O75166, Q86WV0, Q8IV98, Q9BVA5, Q9HAR0 | Gene names | MGEA5, HEXC, KIAA0679, MEA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
|
NOL10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.019944 (rank : 57) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BSC4, Q53RC9, Q96TA5, Q9H7Y7, Q9H855 | Gene names | NOL10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 10. | |||||
|
SALL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.000641 (rank : 96) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 826 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ER74, Q920R5 | Gene names | Sall1, Sal3 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 1 (Zinc finger protein Spalt-3) (Sal-3) (MSal-3). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.029313 (rank : 32) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TEX14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.007666 (rank : 88) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1333 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7M6U3, Q3UT36, Q5NC10, Q5NC11, Q8CGK1, Q8CGK2, Q99MV8 | Gene names | Tex14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
TGON1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.018318 (rank : 62) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
ANLN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.026535 (rank : 37) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
CDKL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | -0.000134 (rank : 97) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QUK0, Q3UL60, Q3V3X7, Q9QYI1, Q9QYI2 | Gene names | Cdkl2, Kkiamre, Kkm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-dependent kinase-like 2 (EC 2.7.11.22) (Serine/threonine- protein kinase KKIAMRE). | |||||
|
CP135_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.017911 (rank : 64) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
DESP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.018298 (rank : 63) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
DNJC8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.015714 (rank : 73) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6NZB0, Q6PG72, Q8C2M6 | Gene names | Dnajc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 8. | |||||
|
E41L2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.006628 (rank : 91) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.014273 (rank : 76) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.014830 (rank : 74) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
KIF5A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.014205 (rank : 78) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q12840, Q4LE26 | Gene names | KIF5A, NKHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
MX1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.005529 (rank : 92) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P20591 | Gene names | MX1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx1 (Interferon-regulated resistance GTP-binding protein MxA) (Interferon-induced protein p78) (IFI-78K). | |||||
|
PEG3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.007189 (rank : 90) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
PLXD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.003262 (rank : 94) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y4D7, Q6PJS9, Q8IZJ2, Q9BTQ2 | Gene names | PLXND1, KIAA0620 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-D1 precursor. | |||||
|
REST_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.017733 (rank : 65) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
RXINP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.019063 (rank : 61) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96RL1, Q5XKQ1, Q7Z3W7, Q8N5B9, Q9BZR1, Q9BZR5, Q9UHX7 | Gene names | RXRIP110, RAP80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoid X receptor-interacting protein 110 (Receptor-associated protein 80) (Nuclear zinc finger protein RAP80). | |||||
|
SGOL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.012846 (rank : 79) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5FBB7, Q588H5, Q5FBB4, Q5FBB5, Q5FBB6, Q5FBB8, Q8N579, Q8WVL0, Q9BVA8, Q9H275 | Gene names | SGOL1, SGO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 1 (hSgo1) (Serologically defined breast cancer antigen NY-BR-85). | |||||
|
SHOX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.004828 (rank : 93) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15266, O00412, O00413, O15267 | Gene names | SHOX, PHOG | |||
|
Domain Architecture |

|
|||||
| Description | Short stature homeobox protein (Short stature homeobox-containing protein) (Pseudoautosomal homeobox-containing osteogenic protein). | |||||
|
SMRC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.020982 (rank : 56) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
TSH1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.027689 (rank : 34) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
ZBT24_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | -0.000813 (rank : 98) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43167, Q5TED5, Q8N455 | Gene names | ZBTB24, KIAA0441, ZNF450 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 24 (Zinc finger protein 450). | |||||
|
HS90B_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P11499, O89078 | Gene names | Hsp90ab1, Hsp84, Hsp84-1, Hspcb | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (Tumor-specific transplantation 84 kDa antigen) (TSTA). | |||||
|
HS90B_HUMAN
|
||||||
| NC score | 0.998135 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P08238, Q5T9W7, Q9NQW0 | Gene names | HSP90AB1, HSP90B, HSPC2, HSPCB | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (HSP 90). | |||||
|
HS90A_MOUSE
|
||||||
| NC score | 0.979765 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P07901 | Gene names | Hsp90aa1, Hsp86, Hsp86-1, Hspca | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (Tumor-specific transplantation 86 kDa antigen) (TSTA). | |||||
|
HS90A_HUMAN
|
||||||
| NC score | 0.977869 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P07900, Q2PP14, Q9BVQ5 | Gene names | HSP90AA1, HSP90A, HSPC1, HSPCA | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (NY-REN-38 antigen). | |||||
|
ENPL_HUMAN
|
||||||
| NC score | 0.933307 (rank : 5) | θ value | 1.0648e-109 (rank : 5) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P14625, Q96A97 | Gene names | HSP90B1, TRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmin precursor (Heat shock protein 90 kDa beta member 1) (94 kDa glucose-regulated protein) (GRP94) (gp96 homolog) (Tumor rejection antigen 1). | |||||
|
ENPL_MOUSE
|
||||||
| NC score | 0.931581 (rank : 6) | θ value | 6.90186e-109 (rank : 6) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P08113, P11427 | Gene names | Hsp90b1, Tra-1, Tra1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmin precursor (Heat shock protein 90 kDa beta member 1) (94 kDa glucose-regulated protein) (GRP94) (ERP99) (Polymorphic tumor rejection antigen 1) (Tumor rejection antigen gp96). | |||||
|
TRAP1_HUMAN
|
||||||
| NC score | 0.855428 (rank : 7) | θ value | 3.60269e-49 (rank : 7) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12931, O43642, O75235, Q9UHL5 | Gene names | TRAP1, HSP75 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein 75 kDa, mitochondrial precursor (HSP 75) (Tumor necrosis factor type 1 receptor-associated protein) (TRAP-1) (TNFR- associated protein 1). | |||||
|
TRAP1_MOUSE
|
||||||
| NC score | 0.854343 (rank : 8) | θ value | 1.36902e-48 (rank : 8) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CQN1, Q542I4 | Gene names | Trap1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein 75 kDa, mitochondrial precursor (HSP 75) (Tumor necrosis factor type 1 receptor-associated protein) (TRAP-1) (TNFR- associated protein 1). | |||||
|
HSF2B_HUMAN
|
||||||
| NC score | 0.144171 (rank : 9) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75031 | Gene names | HSF2BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heat shock factor 2-binding protein. | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.107818 (rank : 10) | θ value | 0.0148317 (rank : 9) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.101448 (rank : 11) | θ value | 0.0330416 (rank : 10) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
PRI1_MOUSE
|
||||||
| NC score | 0.089966 (rank : 12) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P20664 | Gene names | Prim1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA primase small subunit (EC 2.7.7.-) (DNA primase 49 kDa subunit) (p49). | |||||
|
CND3_HUMAN
|
||||||
| NC score | 0.080262 (rank : 13) | θ value | 0.0961366 (rank : 13) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BPX3, Q96SV9, Q9BUR3, Q9BVY1, Q9H914, Q9H9Z6, Q9HBI9 | Gene names | NCAPG, CAPG, NYMEL3 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 3 (Non-SMC condensin I complex subunit G) (Chromosome-associated protein G) (Condensin subunit CAP-G) (hCAP-G) (XCAP-G homolog) (Antigen NY-MEL-3). | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.058169 (rank : 14) | θ value | 0.0563607 (rank : 12) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
RELN_HUMAN
|
||||||
| NC score | 0.046467 (rank : 15) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78509, Q86UJ0, Q86UJ8, Q8NDV0, Q9UDQ2 | Gene names | RELN | |||
|
Domain Architecture |

|
|||||
| Description | Reelin precursor (EC 3.4.21.-). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.043806 (rank : 16) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.040673 (rank : 17) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
PHF3_HUMAN
|
||||||
| NC score | 0.040352 (rank : 18) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
ZN318_MOUSE
|
||||||
| NC score | 0.039485 (rank : 19) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.038145 (rank : 20) | θ value | 0.0431538 (rank : 11) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
NUF2_HUMAN
|
||||||
| NC score | 0.037940 (rank : 21) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BZD4, Q8WU69, Q96HJ4, Q96Q78 | Gene names | CDCA1, NUF2, NUF2R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (hNuf2) (hNuf2R) (hsNuf2) (Cell division cycle-associated protein 1). | |||||
|
SMC2_MOUSE
|
||||||
| NC score | 0.037901 (rank : 22) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.037353 (rank : 23) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
RELN_MOUSE
|
||||||
| NC score | 0.035908 (rank : 24) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60841, Q9CUA6 | Gene names | Reln, Rl | |||
|
Domain Architecture |

|
|||||
| Description | Reelin precursor (EC 3.4.21.-) (Reeler protein). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.034181 (rank : 25) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.033895 (rank : 26) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
CJ092_HUMAN
|
||||||
| NC score | 0.031197 (rank : 27) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IYW2, Q5JSF7, Q9NTQ5 | Gene names | C10orf92 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf92. | |||||
|
MPIP3_HUMAN
|
||||||
| NC score | 0.031126 (rank : 28) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30307, Q96PL3, Q9H168, Q9H2E8, Q9H2E9, Q9H2F1 | Gene names | CDC25C | |||
|
Domain Architecture |

|
|||||
| Description | M-phase inducer phosphatase 3 (EC 3.1.3.48) (Dual specificity phosphatase Cdc25C). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.031000 (rank : 29) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.030608 (rank : 30) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.029602 (rank : 31) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.029313 (rank : 32) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.028831 (rank : 33) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
TSH1_MOUSE
|
||||||
| NC score | 0.027689 (rank : 34) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5DTH5, Q6PE60, Q8VD19, Q9JLD5 | Gene names | Tshz1, Kiaa4206, Sdccag33, Tsh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 1 (Serologically defined colon cancer antigen 3 homolog). | |||||
|
CEP55_MOUSE
|
||||||
| NC score | 0.027082 (rank : 35) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BT07, Q8C2J0, Q8K2I8, Q8R2Y4, Q9DBZ8 | Gene names | Cep55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55. | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.027003 (rank : 36) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
ANLN_HUMAN
|
||||||
| NC score | 0.026535 (rank : 37) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
MRIP_MOUSE
|
||||||
| NC score | 0.026268 (rank : 38) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
NOC2L_HUMAN
|
||||||
| NC score | 0.026067 (rank : 39) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y3T9, Q5SVA3, Q9BTN6 | Gene names | NOC2L | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar complex protein 2 homolog (Protein NOC2 homolog) (NOC2- like). | |||||
|
MRIP_HUMAN
|
||||||
| NC score | 0.025966 (rank : 40) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.025803 (rank : 41) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
NAF1_HUMAN
|
||||||
| NC score | 0.025371 (rank : 42) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
DYH11_HUMAN
|
||||||
| NC score | 0.025208 (rank : 43) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96DT5, Q9UJ82 | Gene names | DNAH11 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 11 (Axonemal beta dynein heavy chain 11). | |||||
|
PMS2_HUMAN
|
||||||
| NC score | 0.024877 (rank : 44) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54278 | Gene names | PMS2, PMSL2 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.024778 (rank : 45) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
P52K_HUMAN
|
||||||
| NC score | 0.024497 (rank : 46) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
DYH9_HUMAN
|
||||||
| NC score | 0.023947 (rank : 47) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NYC9, O95494, Q9NQ28 | Gene names | DNAH9, DNAH17L, DNAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 9 (Axonemal beta dynein heavy chain 9). | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.023852 (rank : 48) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
DCDC2_HUMAN
|
||||||
| NC score | 0.023484 (rank : 49) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UHG0, Q5VTR8, Q5VTR9, Q86W35, Q9UFD1, Q9ULR6 | Gene names | DCDC2, KIAA1154 | |||
|
Domain Architecture |

|
|||||
| Description | Doublecortin domain-containing protein 2 (RU2S protein). | |||||
|
ST1E1_HUMAN
|
||||||
| NC score | 0.023162 (rank : 50) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49888, Q8N6X5 | Gene names | SULT1E1, STE | |||
|
Domain Architecture |

|
|||||
| Description | Estrogen sulfotransferase (EC 2.8.2.4) (Sulfotransferase, estrogen- preferring) (EST-1). | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.022404 (rank : 51) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
E2F6_MOUSE
|
||||||
| NC score | 0.021672 (rank : 52) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54917 | Gene names | E2f6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F6 (E2F-6) (E2F-binding site-modulating activity protein) (EMA). | |||||
|
AT10A_HUMAN
|
||||||
| NC score | 0.021567 (rank : 53) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60312, Q969I4 | Gene names | ATP10A, ATP10C, ATPVC, KIAA0566 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (ATPVA) (Aminophospholipid translocase VA). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.021492 (rank : 54) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.021429 (rank : 55) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
SMRC2_MOUSE
|
||||||
| NC score | 0.020982 (rank : 56) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6PDG5, Q6P6P3 | Gene names | Smarcc2, Baf170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
NOL10_HUMAN
|
||||||
| NC score | 0.019944 (rank : 57) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BSC4, Q53RC9, Q96TA5, Q9H7Y7, Q9H855 | Gene names | NOL10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 10. | |||||
|
KIF5A_MOUSE
|
||||||
| NC score | 0.019853 (rank : 58) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
LMO7_HUMAN
|
||||||
| NC score | 0.019363 (rank : 59) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.019115 (rank : 60) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
RXINP_HUMAN
|
||||||
| NC score | 0.019063 (rank : 61) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96RL1, Q5XKQ1, Q7Z3W7, Q8N5B9, Q9BZR1, Q9BZR5, Q9UHX7 | Gene names | RXRIP110, RAP80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoid X receptor-interacting protein 110 (Receptor-associated protein 80) (Nuclear zinc finger protein RAP80). | |||||
|
TGON1_MOUSE
|
||||||
| NC score | 0.018318 (rank : 62) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.018298 (rank : 63) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.017911 (rank : 64) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.017733 (rank : 65) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.017632 (rank : 66) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
E2F6_HUMAN
|
||||||
| NC score | 0.017440 (rank : 67) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75461, O60544, Q7Z2H6 | Gene names | E2F6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F6 (E2F-6). | |||||
|
IF4G3_HUMAN
|
||||||
| NC score | 0.017248 (rank : 68) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
IF4G3_MOUSE
|
||||||
| NC score | 0.017163 (rank : 69) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.016852 (rank : 70) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
LAMA2_MOUSE
|
||||||
| NC score | 0.016738 (rank : 71) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
KIF5C_HUMAN
|
||||||
| NC score | 0.016105 (rank : 72) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
DNJC8_MOUSE
|
||||||
| NC score | 0.015714 (rank : 73) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6NZB0, Q6PG72, Q8C2M6 | Gene names | Dnajc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 8. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.014830 (rank : 74) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
ADA30_HUMAN
|
||||||
| NC score | 0.014636 (rank : 75) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UKF2, Q9UKF1 | Gene names | ADAM30 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 30 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 30). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.014273 (rank : 76) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
I5P2_HUMAN
|
||||||
| NC score | 0.014264 (rank : 77) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P32019, Q5VSG9, Q5VSH0, Q5VSH1, Q658Q5, Q6P6D4, Q6PD53, Q86YE1 | Gene names | INPP5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type II inositol-1,4,5-trisphosphate 5-phosphatase precursor (EC 3.1.3.36) (Phosphoinositide 5-phosphatase) (5PTase) (75 kDa inositol polyphosphate-5-phosphatase). | |||||
|
KIF5A_HUMAN
|
||||||
| NC score | 0.014205 (rank : 78) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q12840, Q4LE26 | Gene names | KIF5A, NKHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
SGOL1_HUMAN
|
||||||
| NC score | 0.012846 (rank : 79) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5FBB7, Q588H5, Q5FBB4, Q5FBB5, Q5FBB6, Q5FBB8, Q8N579, Q8WVL0, Q9BVA8, Q9H275 | Gene names | SGOL1, SGO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 1 (hSgo1) (Serologically defined breast cancer antigen NY-BR-85). | |||||
|
CLC6A_HUMAN
|
||||||
| NC score | 0.012691 (rank : 80) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6EIG7 | Gene names | CLEC6A, CLECSF10, DECTIN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 6 member A (C-type lectin superfamily member 10) (Dendritic cell-associated lectin 2) (DC-associated C-type lectin-2) (DECTIN-2). | |||||
|
OXSR1_MOUSE
|
||||||
| NC score | 0.012642 (rank : 81) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 865 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6P9R2, Q8BVZ9, Q8C0B9 | Gene names | Oxsr1, Osr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase OSR1 (EC 2.7.11.1) (Oxidative stress- responsive 1 protein). | |||||
|
AT10A_MOUSE
|
||||||
| NC score | 0.012125 (rank : 82) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54827, Q8R3B8 | Gene names | Atp10a, Atpc5, Pfatp | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VA (EC 3.6.3.1) (P-locus fat-associated ATPase). | |||||
|
RGS6_HUMAN
|
||||||
| NC score | 0.011854 (rank : 83) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49758, O75576, O75577, Q8TE13, Q8TE14, Q8TE15, Q8TE16, Q8TE17, Q8TE18, Q8TE19, Q8TE20, Q8TE21, Q8TE22, Q9Y245, Q9Y647 | Gene names | RGS6 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 6 (RGS6) (S914). | |||||
|
OXSR1_HUMAN
|
||||||
| NC score | 0.011295 (rank : 84) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95747, Q3LR53, Q7Z501, Q9UPQ1 | Gene names | OXSR1, KIAA1101, OSR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase OSR1 (EC 2.7.11.1) (Oxidative stress- responsive 1 protein). | |||||
|
NCOAT_HUMAN
|
||||||
| NC score | 0.010321 (rank : 85) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60502, O75166, Q86WV0, Q8IV98, Q9BVA5, Q9HAR0 | Gene names | MGEA5, HEXC, KIAA0679, MEA5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional protein NCOAT (Nuclear cytoplasmic O-GlcNAcase and acetyltransferase) (Meningioma-expressed antigen 5) [Includes: Beta- hexosaminidase (EC 3.2.1.52) (N-acetyl-beta-glucosaminidase) (Beta-N- acetylhexosaminidase) (Hexosaminidase C) (N-acetyl-beta-D- glucosaminidase) (O-GlcNAcase); Histone acetyltransferase (EC 2.3.1.48) (HAT)]. | |||||
|
STK39_MOUSE
|
||||||
| NC score | 0.007902 (rank : 86) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 915 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z1W9, Q80W13 | Gene names | Stk39, Spak | |||
|
Domain Architecture |

|
|||||
| Description | STE20/SPS1-related proline-alanine-rich protein kinase (EC 2.7.11.1) (Ste-20-related kinase) (Serine/threonine-protein kinase 39). | |||||
|
ZA2G_MOUSE
|
||||||
| NC score | 0.007748 (rank : 87) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64726 | Gene names | Azgp1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-alpha-2-glycoprotein precursor (Zn-alpha-2-glycoprotein) (Zn- alpha-2-GP). | |||||
|
TEX14_MOUSE
|
||||||
| NC score | 0.007666 (rank : 88) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1333 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7M6U3, Q3UT36, Q5NC10, Q5NC11, Q8CGK1, Q8CGK2, Q99MV8 | Gene names | Tex14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
STK39_HUMAN
|
||||||
| NC score | 0.007281 (rank : 89) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UEW8, O14774, Q9UER4 | Gene names | STK39, SPAK | |||
|
Domain Architecture |

|
|||||
| Description | STE20/SPS1-related proline-alanine-rich protein kinase (EC 2.7.11.1) (Ste-20-related kinase) (Serine/threonine-protein kinase 39) (DCHT). | |||||
|
PEG3_MOUSE
|
||||||
| NC score | 0.007189 (rank : 90) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
E41L2_MOUSE
|
||||||
| NC score | 0.006628 (rank : 91) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
MX1_HUMAN
|
||||||
| NC score | 0.005529 (rank : 92) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P20591 | Gene names | MX1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced GTP-binding protein Mx1 (Interferon-regulated resistance GTP-binding protein MxA) (Interferon-induced protein p78) (IFI-78K). | |||||
|
SHOX_HUMAN
|
||||||
| NC score | 0.004828 (rank : 93) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15266, O00412, O00413, O15267 | Gene names | SHOX, PHOG | |||
|
Domain Architecture |

|
|||||
| Description | Short stature homeobox protein (Short stature homeobox-containing protein) (Pseudoautosomal homeobox-containing osteogenic protein). | |||||
|
PLXD1_HUMAN
|
||||||
| NC score | 0.003262 (rank : 94) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y4D7, Q6PJS9, Q8IZJ2, Q9BTQ2 | Gene names | PLXND1, KIAA0620 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-D1 precursor. | |||||
|
EVI1_HUMAN
|
||||||
| NC score | 0.001986 (rank : 95) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
SALL1_MOUSE
|
||||||
| NC score | 0.000641 (rank : 96) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 826 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ER74, Q920R5 | Gene names | Sall1, Sal3 | |||
|
Domain Architecture |

|
|||||
| Description | Sal-like protein 1 (Zinc finger protein Spalt-3) (Sal-3) (MSal-3). | |||||
|
CDKL2_MOUSE
|
||||||
| NC score | -0.000134 (rank : 97) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QUK0, Q3UL60, Q3V3X7, Q9QYI1, Q9QYI2 | Gene names | Cdkl2, Kkiamre, Kkm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-dependent kinase-like 2 (EC 2.7.11.22) (Serine/threonine- protein kinase KKIAMRE). | |||||
|
ZBT24_HUMAN
|
||||||
| NC score | -0.000813 (rank : 98) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43167, Q5TED5, Q8N455 | Gene names | ZBTB24, KIAA0441, ZNF450 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and BTB domain-containing protein 24 (Zinc finger protein 450). | |||||