Please be patient as the page loads
|
PMS2_HUMAN
|
||||||
| SwissProt Accessions | P54278 | Gene names | PMS2, PMSL2 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PMS2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 131 | |
| SwissProt Accessions | P54278 | Gene names | PMS2, PMSL2 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
PMS2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.959278 (rank : 2) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P54279 | Gene names | Pms2 | |||
|
Domain Architecture |
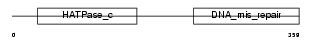
|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
PMS1_HUMAN
|
||||||
| θ value | 6.58091e-43 (rank : 3) | NC score | 0.700437 (rank : 5) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P54277 | Gene names | PMS1, PMSL1 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 1 (DNA mismatch repair protein PMS1). | |||||
|
MLH1_HUMAN
|
||||||
| θ value | 3.61106e-41 (rank : 4) | NC score | 0.743396 (rank : 3) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P40692 | Gene names | MLH1, COCA2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh1 (MutL protein homolog 1). | |||||
|
MLH1_MOUSE
|
||||||
| θ value | 6.15952e-41 (rank : 5) | NC score | 0.740578 (rank : 4) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JK91, Q62454 | Gene names | Mlh1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh1 (MutL protein homolog 1). | |||||
|
MLH3_HUMAN
|
||||||
| θ value | 6.85773e-16 (rank : 6) | NC score | 0.568280 (rank : 6) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UHC1, P49751, Q56DK9, Q9P292, Q9UHC0 | Gene names | MLH3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh3 (MutL protein homolog 3). | |||||
|
LPIN2_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 7) | NC score | 0.073685 (rank : 11) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99PI5, Q8C357, Q8C7I8, Q8CC85, Q8CHR7 | Gene names | Lpin2 | |||
|
Domain Architecture |
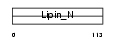
|
|||||
| Description | Lipin-2. | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 8) | NC score | 0.078164 (rank : 8) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 9) | NC score | 0.059218 (rank : 22) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 10) | NC score | 0.050872 (rank : 31) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
LPIN2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 11) | NC score | 0.077323 (rank : 9) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92539 | Gene names | LPIN2, KIAA0249 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-2. | |||||
|
UBP34_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 12) | NC score | 0.066808 (rank : 15) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
CD19_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 13) | NC score | 0.074530 (rank : 10) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P15391, Q96S68, Q9BRD6 | Gene names | CD19 | |||
|
Domain Architecture |
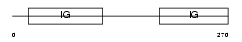
|
|||||
| Description | B-lymphocyte antigen CD19 precursor (Differentiation antigen CD19) (B- lymphocyte surface antigen B4) (Leu-12). | |||||
|
UBP34_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 14) | NC score | 0.065695 (rank : 16) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 15) | NC score | 0.081882 (rank : 7) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
TXLNG_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 16) | NC score | 0.028439 (rank : 60) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NUQ3, Q9P0X1 | Gene names | TXLNG, CXorf15, LSR5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-taxilin (Lipopolysaccharide-specific response protein 5). | |||||
|
ZCH14_MOUSE
|
||||||
| θ value | 0.125558 (rank : 17) | NC score | 0.060589 (rank : 20) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VIG0, Q80TX3, Q91VU4 | Gene names | Zcchc14, Kiaa0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 0.163984 (rank : 18) | NC score | 0.065364 (rank : 17) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
WASF3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.043538 (rank : 37) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VHI6 | Gene names | Wasf3, Wave3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 3 (WASP-family protein member 3) (Protein WAVE-3). | |||||
|
ZCPW1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.055883 (rank : 28) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.059350 (rank : 21) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
IP3KC_MOUSE
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.049297 (rank : 33) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TS72, Q3U384 | Gene names | Itpkc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol-trisphosphate 3-kinase C (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase C) (IP3K-C). | |||||
|
ZN592_MOUSE
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.013724 (rank : 102) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BHZ4, Q80XM1 | Gene names | Znf592, Kiaa0211, Zfp592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 592 (Zfp-592). | |||||
|
ENL_HUMAN
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.057230 (rank : 27) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.024416 (rank : 70) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.039215 (rank : 46) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
RAVR2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.042815 (rank : 38) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TPD6 | Gene names | Raver2, Kiaa1579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 2 (Protein raver-2). | |||||
|
T2FA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.073202 (rank : 12) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.013412 (rank : 104) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
BRWD1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.022430 (rank : 77) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
CD2L1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.012541 (rank : 107) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.027781 (rank : 62) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.063571 (rank : 18) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.034939 (rank : 53) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
ATX7_MOUSE
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.035353 (rank : 52) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.012326 (rank : 108) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.062247 (rank : 19) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.058383 (rank : 24) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
PP2CG_MOUSE
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.032070 (rank : 55) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61074 | Gene names | Ppm1g, Fin13, Ppm1c | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C) (Fibroblast growth factor-inducible protein 13) (FIN13). | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.042362 (rank : 40) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
TIAM1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.021499 (rank : 81) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60610 | Gene names | Tiam1, Tiam-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
E41L2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.022545 (rank : 76) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
NP1L3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.030451 (rank : 57) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99457, O60788 | Gene names | NAP1L3, BNAP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome assembly protein 1-like 3. | |||||
|
THOC2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.049956 (rank : 32) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
AXN2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.020073 (rank : 84) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2T1, Q9H3M6, Q9UH84 | Gene names | AXIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
ICAL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.040799 (rank : 42) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P20810, O95360, Q96D08, Q9H1Z5 | Gene names | CAST | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor) (Sperm BS-17 component). | |||||
|
REXO4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.030792 (rank : 56) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PAQ4, Q3UU17, Q8C524 | Gene names | Rexo4, Gm111, Pmc2, Xpmc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 4 (EC 3.1.-.-) (Exonuclease XPMC2) (Prevents mitotic catastrophe 2 protein homolog). | |||||
|
RHG06_MOUSE
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.022898 (rank : 74) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.039607 (rank : 45) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
CEP41_MOUSE
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.067360 (rank : 14) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99NF3, Q8BT53 | Gene names | Tsga14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 41 kDa (Cep41 protein) (Testis-specific gene A14 protein). | |||||
|
IRS2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.035493 (rank : 51) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.058451 (rank : 23) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
SH2D3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.019285 (rank : 88) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N5H7, Q5HYE5, Q6UY42, Q8N6X3, Q9Y2X5 | Gene names | SH2D3C, NSP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (Novel SH2-containing protein 3). | |||||
|
SLK_MOUSE
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.006896 (rank : 123) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.015662 (rank : 99) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
THYG_HUMAN
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.014670 (rank : 101) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01266, O15274, O43899, Q15593, Q15948, Q9NYR1, Q9NYR2, Q9UMZ0, Q9UNY3 | Gene names | TG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
ZC313_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.040368 (rank : 43) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
CEP41_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.057603 (rank : 25) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYV8, Q7Z496, Q86TM1, Q8NFU8, Q9H6A3, Q9NPV3 | Gene names | TSGA14, CEP41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 41 kDa (Cep41 protein) (Testis-specific gene A14 protein). | |||||
|
CL025_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.046608 (rank : 35) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IYM0, Q8TCP7, Q9H0L3 | Gene names | C12orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C12orf25. | |||||
|
IP3KC_HUMAN
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.036845 (rank : 49) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96DU7, Q9UE25, Q9Y475 | Gene names | ITPKC, IP3KC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol-trisphosphate 3-kinase C (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase C) (InsP 3-kinase C) (IP3K-C). | |||||
|
LZTS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.013558 (rank : 103) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y250, Q9Y5V7, Q9Y5V8, Q9Y5V9, Q9Y5W0, Q9Y5W1, Q9Y5W2 | Gene names | LZTS1, FEZ1 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.039801 (rank : 44) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.042624 (rank : 39) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.046147 (rank : 36) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.026030 (rank : 65) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CUBN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.004883 (rank : 126) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.067837 (rank : 13) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
FARP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.012135 (rank : 109) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
K1C20_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.007337 (rank : 121) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35900 | Gene names | KRT20 | |||
|
Domain Architecture |
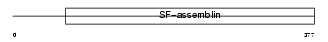
|
|||||
| Description | Keratin, type I cytoskeletal 20 (Cytokeratin-20) (CK-20) (Keratin-20) (K20) (Protein IT). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.057470 (rank : 26) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
R51A1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.035651 (rank : 50) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C551, O55219, Q8BP36, Q99L94, Q9D0J0 | Gene names | Rad51ap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein) (RAB22). | |||||
|
SAC3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.019878 (rank : 86) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91WF7, Q6A092 | Gene names | Sac3, Kiaa0274 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAC domain-containing protein 3. | |||||
|
TSKS_MOUSE
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.023191 (rank : 73) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54887 | Gene names | Stk22s1, Tsks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific serine kinase substrate (Testis-specific kinase substrate) (STK22 substrate 1). | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.024435 (rank : 69) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
WASF3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.033070 (rank : 54) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPY6, O94974 | Gene names | WASF3, KIAA0900, SCAR3, WAVE3 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein family member 3 (WASP-family protein member 3) (Protein WAVE-3) (Verprolin homology domain-containing protein 3). | |||||
|
ZN592_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.010086 (rank : 111) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 762 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92610 | Gene names | ZNF592, KIAA0211 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 592. | |||||
|
ARI5B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.021286 (rank : 82) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
BTBD7_MOUSE
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.021762 (rank : 80) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CFE5, Q80TC3, Q8BZY9 | Gene names | Btbd7, Kiaa1525 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 7. | |||||
|
CCD50_MOUSE
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.021869 (rank : 79) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q810U5, Q3TRW1, Q8BP82, Q9CZT1, Q9D436 | Gene names | Ccdc50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 50 (Protein Ymer). | |||||
|
CD2AP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.009502 (rank : 114) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLQ0, O88903, Q8K4Z1, Q8VCI9 | Gene names | Cd2ap, Mets1 | |||
|
Domain Architecture |
|
|||||
| Description | CD2-associated protein (Mesenchyme-to-epithelium transition protein with SH3 domains 1) (METS-1). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.008701 (rank : 118) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CGRE1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.028869 (rank : 59) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R1U2, Q9D1K1 | Gene names | Cgref1, Cgr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
EHMT1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.006600 (rank : 124) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H9B1, Q8TCN7, Q96F53, Q96JF1, Q96KH4 | Gene names | EHMT1, EUHMTASE1, KIAA1876 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Euchromatic histone-lysine N-methyltransferase 1) (Eu-HMTase1) (G9a- like protein 1) (GLP1). | |||||
|
IRS2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.030141 (rank : 58) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
PARM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.027527 (rank : 63) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6UWI2, Q96DV8, Q9Y4S1 | Gene names | ames=UNQ1879/PRO4322 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
RHG06_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.020343 (rank : 83) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.046618 (rank : 34) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
AIOL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.000066 (rank : 132) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UKT9, Q8WWQ9, Q8WWR0, Q8WWR1, Q8WWR2, Q8WWR3 | Gene names | IKZF3, ZNFN1A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein Aiolos (IKAROS family zinc finger protein 3). | |||||
|
BARH2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.003781 (rank : 129) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NY43, Q7Z4N7 | Gene names | BARHL2 | |||
|
Domain Architecture |

|
|||||
| Description | BarH-like 2 homeobox protein. | |||||
|
CEBPZ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.017900 (rank : 94) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q03701, Q8NE75 | Gene names | CEBPZ, CBF2 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein zeta (CCAAT-box-binding transcription factor) (CCAAT-binding factor) (CBF). | |||||
|
FNDC3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.007290 (rank : 122) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y2H6, Q5JVF9, Q6EVH3, Q6N020, Q6P9D5, Q9H1W1 | Gene names | FNDC3A, FNDC3, KIAA0970 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibronectin type-III domain-containing protein 3a. | |||||
|
HS90B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.025166 (rank : 66) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P08238, Q5T9W7, Q9NQW0 | Gene names | HSP90AB1, HSP90B, HSPC2, HSPCB | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (HSP 90). | |||||
|
HS90B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.024877 (rank : 67) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11499, O89078 | Gene names | Hsp90ab1, Hsp84, Hsp84-1, Hspcb | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (Tumor-specific transplantation 84 kDa antigen) (TSTA). | |||||
|
LIPB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 94) | NC score | 0.009497 (rank : 115) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35711, Q7TMG4, Q8CBS6, Q99KX6 | Gene names | Ppfibp2, Cclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2) (Coiled-coil-like protein 1). | |||||
|
RBM15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.018902 (rank : 90) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96T37, Q5D058, Q96PE4, Q96SC5, Q96SC6, Q96SC9, Q96SD0, Q96T38, Q9BRA5, Q9H6R8, Q9H9Y0 | Gene names | RBM15, OTT | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 15 (RNA-binding motif protein 15) (One- twenty two protein). | |||||
|
RTN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.018044 (rank : 92) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K0T0, Q8K4S4 | Gene names | Rtn1, Nsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
SRCA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.022700 (rank : 75) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
TRDN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.027819 (rank : 61) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
WASL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.019207 (rank : 89) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
ZCH10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.037478 (rank : 47) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
ZCH14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.040886 (rank : 41) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WYQ9, O60324, Q9UFP0 | Gene names | ZCCHC14, KIAA0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.036869 (rank : 48) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
CD008_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.019946 (rank : 85) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P78312, O43607, P78311, P78313, Q9UEG8 | Gene names | C4orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C4orf8 (Protein IT14). | |||||
|
DZIP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.012724 (rank : 106) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BMD2, Q6ZQ10, Q9CRK5 | Gene names | Dzip1, Kiaa0996 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein DZIP1 (DAZ-interacting protein 1 homolog). | |||||
|
FBX43_MOUSE
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.017624 (rank : 97) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CDI2, Q4G0C8, Q8R0R1 | Gene names | Fbxo43, Emi2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 43 (Endogenous meiotic inhibitor 2). | |||||
|
GAGD4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.013068 (rank : 105) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WTP9, Q5JS82, Q8WYS9 | Gene names | XAGE3, GAGED4, PLAC6 | |||
|
Domain Architecture |
|
|||||
| Description | G antigen family D member 4 (Placenta-specific gene 6 protein) (Protein XAGE-3). | |||||
|
HIPK3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.005697 (rank : 125) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H422, O14632, Q2PBG4, Q2PBG5, Q92632, Q9HAS2 | Gene names | HIPK3, DYRK6, FIST3, PKY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeodomain-interacting protein kinase 3 (EC 2.7.11.1) (Homolog of protein kinase YAK1) (Fas-interacting serine/threonine-protein kinase) (FIST) (Androgen receptor-interacting nuclear protein kinase) (ANPK). | |||||
|
HS90A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.024599 (rank : 68) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P07900, Q2PP14, Q9BVQ5 | Gene names | HSP90AA1, HSP90A, HSPC1, HSPCA | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (NY-REN-38 antigen). | |||||
|
HS90A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.024235 (rank : 72) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P07901 | Gene names | Hsp90aa1, Hsp86, Hsp86-1, Hspca | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (Tumor-specific transplantation 86 kDa antigen) (TSTA). | |||||
|
IRS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.024416 (rank : 71) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35569 | Gene names | Irs1, Irs-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
K0494_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.014902 (rank : 100) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BGQ6, Q3TQN6, Q8BJV7, Q8C0R1 | Gene names | Kiaa0494 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized calcium-binding protein KIAA0494. | |||||
|
MADCA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.017786 (rank : 96) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
MAST2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.002244 (rank : 131) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.026179 (rank : 64) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
S30BP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.019520 (rank : 87) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q02614, Q8VDJ5 | Gene names | Sap30bp, Hcngp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAP30-binding protein (Transcriptional regulator protein HCNGP). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.022078 (rank : 78) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
WASL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.018194 (rank : 91) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
ZN403_MOUSE
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.016399 (rank : 98) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5SV77, Q5SV75, Q5SV76, Q5SV78, Q6GVH6, Q6P9J4, Q920N4 | Gene names | Znf403, Ggnbp2, Zfp403 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF403 (Gametogenetin-binding protein 2) (Dioxin-inducible factor 3) (DIF-3). | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.017886 (rank : 95) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.017909 (rank : 93) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.008469 (rank : 119) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
CCNB3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.009907 (rank : 112) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
DYH5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.007627 (rank : 120) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
LMBTL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.009316 (rank : 116) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y468, Q5H8Y8, Q8IUV7, Q9H1E6, Q9H1G5, Q9UG06, Q9UJB9, Q9Y4C9 | Gene names | L3MBTL, KIAA0681, L3MBT | |||
|
Domain Architecture |

|
|||||
| Description | Lethal(3)malignant brain tumor-like protein (L(3)mbt-like) (L(3)mbt protein homolog) (H-l(3)mbt protein) (H-L(3)MBT) (L3MBTL1). | |||||
|
M3K3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.002532 (rank : 130) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 876 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61084 | Gene names | Map3k3, Mekk3 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 3 (EC 2.7.11.25) (MAPK/ERK kinase kinase 3) (MEK kinase 3) (MEKK 3). | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.004387 (rank : 127) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
MYCD_MOUSE
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.009502 (rank : 113) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VIM5, Q8C3W6, Q8VIL4 | Gene names | Myocd, Bsac2, Mycd, Srfcp | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin (SRF cofactor protein) (Basic SAP coiled-coil transcription activator 2). | |||||
|
NUCL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.011939 (rank : 110) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
TACC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.009199 (rank : 117) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.003805 (rank : 128) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
ZBT40_HUMAN
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | -0.001015 (rank : 133) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NUA8, O75066, Q5TFU5, Q8N1R1 | Gene names | ZBTB40, KIAA0478 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 40. | |||||
|
IWS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.050917 (rank : 30) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.051561 (rank : 29) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
PMS2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 131 | |
| SwissProt Accessions | P54278 | Gene names | PMS2, PMSL2 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
PMS2_MOUSE
|
||||||
| NC score | 0.959278 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P54279 | Gene names | Pms2 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
MLH1_HUMAN
|
||||||
| NC score | 0.743396 (rank : 3) | θ value | 3.61106e-41 (rank : 4) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P40692 | Gene names | MLH1, COCA2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh1 (MutL protein homolog 1). | |||||
|
MLH1_MOUSE
|
||||||
| NC score | 0.740578 (rank : 4) | θ value | 6.15952e-41 (rank : 5) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JK91, Q62454 | Gene names | Mlh1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh1 (MutL protein homolog 1). | |||||
|
PMS1_HUMAN
|
||||||
| NC score | 0.700437 (rank : 5) | θ value | 6.58091e-43 (rank : 3) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P54277 | Gene names | PMS1, PMSL1 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 1 (DNA mismatch repair protein PMS1). | |||||
|
MLH3_HUMAN
|
||||||
| NC score | 0.568280 (rank : 6) | θ value | 6.85773e-16 (rank : 6) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UHC1, P49751, Q56DK9, Q9P292, Q9UHC0 | Gene names | MLH3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh3 (MutL protein homolog 3). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.081882 (rank : 7) | θ value | 0.0330416 (rank : 15) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.078164 (rank : 8) | θ value | 0.00869519 (rank : 8) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
LPIN2_HUMAN
|
||||||
| NC score | 0.077323 (rank : 9) | θ value | 0.0193708 (rank : 11) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92539 | Gene names | LPIN2, KIAA0249 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-2. | |||||
|
CD19_HUMAN
|
||||||
| NC score | 0.074530 (rank : 10) | θ value | 0.0252991 (rank : 13) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P15391, Q96S68, Q9BRD6 | Gene names | CD19 | |||
|
Domain Architecture |

|
|||||
| Description | B-lymphocyte antigen CD19 precursor (Differentiation antigen CD19) (B- lymphocyte surface antigen B4) (Leu-12). | |||||
|
LPIN2_MOUSE
|
||||||
| NC score | 0.073685 (rank : 11) | θ value | 0.00390308 (rank : 7) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99PI5, Q8C357, Q8C7I8, Q8CC85, Q8CHR7 | Gene names | Lpin2 | |||
|
Domain Architecture |

|
|||||
| Description | Lipin-2. | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.073202 (rank : 12) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.067837 (rank : 13) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
CEP41_MOUSE
|
||||||
| NC score | 0.067360 (rank : 14) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99NF3, Q8BT53 | Gene names | Tsga14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 41 kDa (Cep41 protein) (Testis-specific gene A14 protein). | |||||
|
UBP34_MOUSE
|
||||||
| NC score | 0.066808 (rank : 15) | θ value | 0.0193708 (rank : 12) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
UBP34_HUMAN
|
||||||
| NC score | 0.065695 (rank : 16) | θ value | 0.0252991 (rank : 14) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.065364 (rank : 17) | θ value | 0.163984 (rank : 18) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.063571 (rank : 18) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.062247 (rank : 19) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
ZCH14_MOUSE
|
||||||
| NC score | 0.060589 (rank : 20) | θ value | 0.125558 (rank : 17) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VIG0, Q80TX3, Q91VU4 | Gene names | Zcchc14, Kiaa0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.059350 (rank : 21) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.059218 (rank : 22) | θ value | 0.0113563 (rank : 9) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.058451 (rank : 23) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.058383 (rank : 24) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
CEP41_HUMAN
|
||||||
| NC score | 0.057603 (rank : 25) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYV8, Q7Z496, Q86TM1, Q8NFU8, Q9H6A3, Q9NPV3 | Gene names | TSGA14, CEP41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 41 kDa (Cep41 protein) (Testis-specific gene A14 protein). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.057470 (rank : 26) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.057230 (rank : 27) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
ZCPW1_MOUSE
|
||||||
| NC score | 0.055883 (rank : 28) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.051561 (rank : 29) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.050917 (rank : 30) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.050872 (rank : 31) | θ value | 0.0113563 (rank : 10) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
THOC2_HUMAN
|
||||||
| NC score | 0.049956 (rank : 32) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
IP3KC_MOUSE
|
||||||
| NC score | 0.049297 (rank : 33) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TS72, Q3U384 | Gene names | Itpkc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol-trisphosphate 3-kinase C (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase C) (IP3K-C). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.046618 (rank : 34) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
CL025_HUMAN
|
||||||
| NC score | 0.046608 (rank : 35) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IYM0, Q8TCP7, Q9H0L3 | Gene names | C12orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C12orf25. | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.046147 (rank : 36) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
WASF3_MOUSE
|
||||||
| NC score | 0.043538 (rank : 37) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VHI6 | Gene names | Wasf3, Wave3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 3 (WASP-family protein member 3) (Protein WAVE-3). | |||||
|
RAVR2_MOUSE
|
||||||
| NC score | 0.042815 (rank : 38) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TPD6 | Gene names | Raver2, Kiaa1579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 2 (Protein raver-2). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.042624 (rank : 39) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.042362 (rank : 40) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
ZCH14_HUMAN
|
||||||
| NC score | 0.040886 (rank : 41) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WYQ9, O60324, Q9UFP0 | Gene names | ZCCHC14, KIAA0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
ICAL_HUMAN
|
||||||
| NC score | 0.040799 (rank : 42) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P20810, O95360, Q96D08, Q9H1Z5 | Gene names | CAST | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor) (Sperm BS-17 component). | |||||
|
ZC313_HUMAN
|
||||||
| NC score | 0.040368 (rank : 43) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.039801 (rank : 44) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.039607 (rank : 45) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.039215 (rank : 46) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
ZCH10_HUMAN
|
||||||
| NC score | 0.037478 (rank : 47) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.036869 (rank : 48) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
IP3KC_HUMAN
|
||||||
| NC score | 0.036845 (rank : 49) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96DU7, Q9UE25, Q9Y475 | Gene names | ITPKC, IP3KC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol-trisphosphate 3-kinase C (EC 2.7.1.127) (Inositol 1,4,5- trisphosphate 3-kinase C) (InsP 3-kinase C) (IP3K-C). | |||||
|
R51A1_MOUSE
|
||||||
| NC score | 0.035651 (rank : 50) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C551, O55219, Q8BP36, Q99L94, Q9D0J0 | Gene names | Rad51ap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein) (RAB22). | |||||
|
IRS2_MOUSE
|
||||||
| NC score | 0.035493 (rank : 51) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
ATX7_MOUSE
|
||||||
| NC score | 0.035353 (rank : 52) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.034939 (rank : 53) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
WASF3_HUMAN
|
||||||
| NC score | 0.033070 (rank : 54) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UPY6, O94974 | Gene names | WASF3, KIAA0900, SCAR3, WAVE3 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein family member 3 (WASP-family protein member 3) (Protein WAVE-3) (Verprolin homology domain-containing protein 3). | |||||
|
PP2CG_MOUSE
|
||||||
| NC score | 0.032070 (rank : 55) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61074 | Gene names | Ppm1g, Fin13, Ppm1c | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 2C isoform gamma (EC 3.1.3.16) (PP2C-gamma) (Protein phosphatase magnesium-dependent 1 gamma) (Protein phosphatase 1C) (Fibroblast growth factor-inducible protein 13) (FIN13). | |||||
|
REXO4_MOUSE
|
||||||
| NC score | 0.030792 (rank : 56) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PAQ4, Q3UU17, Q8C524 | Gene names | Rexo4, Gm111, Pmc2, Xpmc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 4 (EC 3.1.-.-) (Exonuclease XPMC2) (Prevents mitotic catastrophe 2 protein homolog). | |||||
|
NP1L3_HUMAN
|
||||||
| NC score | 0.030451 (rank : 57) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99457, O60788 | Gene names | NAP1L3, BNAP | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome assembly protein 1-like 3. | |||||
|
IRS2_HUMAN
|
||||||
| NC score | 0.030141 (rank : 58) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
CGRE1_MOUSE
|
||||||
| NC score | 0.028869 (rank : 59) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R1U2, Q9D1K1 | Gene names | Cgref1, Cgr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
TXLNG_HUMAN
|
||||||
| NC score | 0.028439 (rank : 60) | θ value | 0.0563607 (rank : 16) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NUQ3, Q9P0X1 | Gene names | TXLNG, CXorf15, LSR5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-taxilin (Lipopolysaccharide-specific response protein 5). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.027819 (rank : 61) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.027781 (rank : 62) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PARM1_HUMAN
|
||||||
| NC score | 0.027527 (rank : 63) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6UWI2, Q96DV8, Q9Y4S1 | Gene names | ames=UNQ1879/PRO4322 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.026179 (rank : 64) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.026030 (rank : 65) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
HS90B_HUMAN
|
||||||
| NC score | 0.025166 (rank : 66) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P08238, Q5T9W7, Q9NQW0 | Gene names | HSP90AB1, HSP90B, HSPC2, HSPCB | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (HSP 90). | |||||
|
HS90B_MOUSE
|
||||||
| NC score | 0.024877 (rank : 67) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11499, O89078 | Gene names | Hsp90ab1, Hsp84, Hsp84-1, Hspcb | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (Tumor-specific transplantation 84 kDa antigen) (TSTA). | |||||
|
HS90A_HUMAN
|
||||||
| NC score | 0.024599 (rank : 68) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P07900, Q2PP14, Q9BVQ5 | Gene names | HSP90AA1, HSP90A, HSPC1, HSPCA | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (NY-REN-38 antigen). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.024435 (rank : 69) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.024416 (rank : 70) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
IRS1_MOUSE
|
||||||
| NC score | 0.024416 (rank : 71) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35569 | Gene names | Irs1, Irs-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
HS90A_MOUSE
|
||||||
| NC score | 0.024235 (rank : 72) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P07901 | Gene names | Hsp90aa1, Hsp86, Hsp86-1, Hspca | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (Tumor-specific transplantation 86 kDa antigen) (TSTA). | |||||
|
TSKS_MOUSE
|
||||||
| NC score | 0.023191 (rank : 73) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54887 | Gene names | Stk22s1, Tsks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific serine kinase substrate (Testis-specific kinase substrate) (STK22 substrate 1). | |||||
|
RHG06_MOUSE
|
||||||
| NC score | 0.022898 (rank : 74) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
SRCA_MOUSE
|
||||||
| NC score | 0.022700 (rank : 75) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
E41L2_MOUSE
|
||||||
| NC score | 0.022545 (rank : 76) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
BRWD1_MOUSE
|
||||||
| NC score | 0.022430 (rank : 77) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.022078 (rank : 78) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
CCD50_MOUSE
|
||||||
| NC score | 0.021869 (rank : 79) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q810U5, Q3TRW1, Q8BP82, Q9CZT1, Q9D436 | Gene names | Ccdc50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 50 (Protein Ymer). | |||||
|
BTBD7_MOUSE
|
||||||
| NC score | 0.021762 (rank : 80) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CFE5, Q80TC3, Q8BZY9 | Gene names | Btbd7, Kiaa1525 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 7. | |||||
|
TIAM1_MOUSE
|
||||||
| NC score | 0.021499 (rank : 81) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60610 | Gene names | Tiam1, Tiam-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
ARI5B_MOUSE
|
||||||
| NC score | 0.021286 (rank : 82) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
RHG06_HUMAN
|
||||||
| NC score | 0.020343 (rank : 83) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43182, O43437, Q9P1B3, Q9UK81, Q9UK82 | Gene names | ARHGAP6, RHOGAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
AXN2_HUMAN
|
||||||
| NC score | 0.020073 (rank : 84) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2T1, Q9H3M6, Q9UH84 | Gene names | AXIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
CD008_HUMAN
|
||||||
| NC score | 0.019946 (rank : 85) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P78312, O43607, P78311, P78313, Q9UEG8 | Gene names | C4orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C4orf8 (Protein IT14). | |||||
|
SAC3_MOUSE
|
||||||
| NC score | 0.019878 (rank : 86) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91WF7, Q6A092 | Gene names | Sac3, Kiaa0274 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAC domain-containing protein 3. | |||||
|
S30BP_MOUSE
|
||||||
| NC score | 0.019520 (rank : 87) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q02614, Q8VDJ5 | Gene names | Sap30bp, Hcngp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAP30-binding protein (Transcriptional regulator protein HCNGP). | |||||
|
SH2D3_HUMAN
|
||||||
| NC score | 0.019285 (rank : 88) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N5H7, Q5HYE5, Q6UY42, Q8N6X3, Q9Y2X5 | Gene names | SH2D3C, NSP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (Novel SH2-containing protein 3). | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.019207 (rank : 89) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
RBM15_HUMAN
|
||||||
| NC score | 0.018902 (rank : 90) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96T37, Q5D058, Q96PE4, Q96SC5, Q96SC6, Q96SC9, Q96SD0, Q96T38, Q9BRA5, Q9H6R8, Q9H9Y0 | Gene names | RBM15, OTT | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 15 (RNA-binding motif protein 15) (One- twenty two protein). | |||||
|
WASL_HUMAN
|
||||||
| NC score | 0.018194 (rank : 91) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
RTN1_MOUSE
|
||||||
| NC score | 0.018044 (rank : 92) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K0T0, Q8K4S4 | Gene names | Rtn1, Nsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.017909 (rank : 93) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CEBPZ_HUMAN
|
||||||
| NC score | 0.017900 (rank : 94) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q03701, Q8NE75 | Gene names | CEBPZ, CBF2 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein zeta (CCAAT-box-binding transcription factor) (CCAAT-binding factor) (CBF). | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.017886 (rank : 95) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
MADCA_HUMAN
|
||||||
| NC score | 0.017786 (rank : 96) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
FBX43_MOUSE
|
||||||
| NC score | 0.017624 (rank : 97) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CDI2, Q4G0C8, Q8R0R1 | Gene names | Fbxo43, Emi2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 43 (Endogenous meiotic inhibitor 2). | |||||
|
ZN403_MOUSE
|
||||||
| NC score | 0.016399 (rank : 98) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5SV77, Q5SV75, Q5SV76, Q5SV78, Q6GVH6, Q6P9J4, Q920N4 | Gene names | Znf403, Ggnbp2, Zfp403 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF403 (Gametogenetin-binding protein 2) (Dioxin-inducible factor 3) (DIF-3). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.015662 (rank : 99) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
K0494_MOUSE
|
||||||
| NC score | 0.014902 (rank : 100) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BGQ6, Q3TQN6, Q8BJV7, Q8C0R1 | Gene names | Kiaa0494 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized calcium-binding protein KIAA0494. | |||||
|
THYG_HUMAN
|
||||||
| NC score | 0.014670 (rank : 101) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01266, O15274, O43899, Q15593, Q15948, Q9NYR1, Q9NYR2, Q9UMZ0, Q9UNY3 | Gene names | TG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
ZN592_MOUSE
|
||||||
| NC score | 0.013724 (rank : 102) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BHZ4, Q80XM1 | Gene names | Znf592, Kiaa0211, Zfp592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 592 (Zfp-592). | |||||
|
LZTS1_HUMAN
|
||||||
| NC score | 0.013558 (rank : 103) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y250, Q9Y5V7, Q9Y5V8, Q9Y5V9, Q9Y5W0, Q9Y5W1, Q9Y5W2 | Gene names | LZTS1, FEZ1 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 1 (F37/esophageal cancer- related gene-coding leucine-zipper motif) (Fez1). | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | 0.013412 (rank : 104) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
GAGD4_HUMAN
|
||||||
| NC score | 0.013068 (rank : 105) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WTP9, Q5JS82, Q8WYS9 | Gene names | XAGE3, GAGED4, PLAC6 | |||
|
Domain Architecture |

|
|||||
| Description | G antigen family D member 4 (Placenta-specific gene 6 protein) (Protein XAGE-3). | |||||
|
DZIP1_MOUSE
|
||||||
| NC score | 0.012724 (rank : 106) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BMD2, Q6ZQ10, Q9CRK5 | Gene names | Dzip1, Kiaa0996 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein DZIP1 (DAZ-interacting protein 1 homolog). | |||||
|
CD2L1_MOUSE
|
||||||
| NC score | 0.012541 (rank : 107) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.012326 (rank : 108) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
FARP1_HUMAN
|
||||||
| NC score | 0.012135 (rank : 109) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y4F1, Q6IQ29 | Gene names | FARP1, CDEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 1 (Chondrocyte- derived ezrin-like protein). | |||||
|
NUCL_MOUSE
|
||||||
| NC score | 0.011939 (rank : 110) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
ZN592_HUMAN
|
||||||
| NC score | 0.010086 (rank : 111) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 762 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92610 | Gene names | ZNF592, KIAA0211 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 592. | |||||
|
CCNB3_MOUSE
|
||||||
| NC score | 0.009907 (rank : 112) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
MYCD_MOUSE
|
||||||
| NC score | 0.009502 (rank : 113) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VIM5, Q8C3W6, Q8VIL4 | Gene names | Myocd, Bsac2, Mycd, Srfcp | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin (SRF cofactor protein) (Basic SAP coiled-coil transcription activator 2). | |||||
|
CD2AP_MOUSE
|
||||||
| NC score | 0.009502 (rank : 114) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLQ0, O88903, Q8K4Z1, Q8VCI9 | Gene names | Cd2ap, Mets1 | |||
|
Domain Architecture |

|
|||||
| Description | CD2-associated protein (Mesenchyme-to-epithelium transition protein with SH3 domains 1) (METS-1). | |||||
|
LIPB2_MOUSE
|
||||||
| NC score | 0.009497 (rank : 115) | θ value | 5.27518 (rank : 94) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35711, Q7TMG4, Q8CBS6, Q99KX6 | Gene names | Ppfibp2, Cclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2) (Coiled-coil-like protein 1). | |||||
|
LMBTL_HUMAN
|
||||||
| NC score | 0.009316 (rank : 116) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y468, Q5H8Y8, Q8IUV7, Q9H1E6, Q9H1G5, Q9UG06, Q9UJB9, Q9Y4C9 | Gene names | L3MBTL, KIAA0681, L3MBT | |||
|
Domain Architecture |

|
|||||
| Description | Lethal(3)malignant brain tumor-like protein (L(3)mbt-like) (L(3)mbt protein homolog) (H-l(3)mbt protein) (H-L(3)MBT) (L3MBTL1). | |||||
|
TACC3_HUMAN
|
||||||
| NC score | 0.009199 (rank : 117) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.008701 (rank : 118) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.008469 (rank : 119) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
DYH5_MOUSE
|
||||||
| NC score | 0.007627 (rank : 120) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
K1C20_HUMAN
|
||||||
| NC score | 0.007337 (rank : 121) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35900 | Gene names | KRT20 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 20 (Cytokeratin-20) (CK-20) (Keratin-20) (K20) (Protein IT). | |||||
|
FNDC3_HUMAN
|
||||||
| NC score | 0.007290 (rank : 122) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y2H6, Q5JVF9, Q6EVH3, Q6N020, Q6P9D5, Q9H1W1 | Gene names | FNDC3A, FNDC3, KIAA0970 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibronectin type-III domain-containing protein 3a. | |||||
|
SLK_MOUSE
|
||||||
| NC score | 0.006896 (rank : 123) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
EHMT1_HUMAN
|
||||||
| NC score | 0.006600 (rank : 124) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H9B1, Q8TCN7, Q96F53, Q96JF1, Q96KH4 | Gene names | EHMT1, EUHMTASE1, KIAA1876 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Euchromatic histone-lysine N-methyltransferase 1) (Eu-HMTase1) (G9a- like protein 1) (GLP1). | |||||
|
HIPK3_HUMAN
|
||||||
| NC score | 0.005697 (rank : 125) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H422, O14632, Q2PBG4, Q2PBG5, Q92632, Q9HAS2 | Gene names | HIPK3, DYRK6, FIST3, PKY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeodomain-interacting protein kinase 3 (EC 2.7.11.1) (Homolog of protein kinase YAK1) (Fas-interacting serine/threonine-protein kinase) (FIST) (Androgen receptor-interacting nuclear protein kinase) (ANPK). | |||||
|
CUBN_MOUSE
|
||||||
| NC score | 0.004883 (rank : 126) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JLB4 | Gene names | Cubn, Ifcr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor). | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.004387 (rank : 127) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
WNK1_MOUSE
|
||||||
| NC score | 0.003805 (rank : 128) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
BARH2_HUMAN
|
||||||
| NC score | 0.003781 (rank : 129) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NY43, Q7Z4N7 | Gene names | BARHL2 | |||
|
Domain Architecture |

|
|||||
| Description | BarH-like 2 homeobox protein. | |||||
|
M3K3_MOUSE
|
||||||
| NC score | 0.002532 (rank : 130) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 876 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61084 | Gene names | Map3k3, Mekk3 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 3 (EC 2.7.11.25) (MAPK/ERK kinase kinase 3) (MEK kinase 3) (MEKK 3). | |||||
|
MAST2_MOUSE
|
||||||
| NC score | 0.002244 (rank : 131) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
AIOL_HUMAN
|
||||||
| NC score | 0.000066 (rank : 132) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UKT9, Q8WWQ9, Q8WWR0, Q8WWR1, Q8WWR2, Q8WWR3 | Gene names | IKZF3, ZNFN1A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein Aiolos (IKAROS family zinc finger protein 3). | |||||
|
ZBT40_HUMAN
|
||||||
| NC score | -0.001015 (rank : 133) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 131 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NUA8, O75066, Q5TFU5, Q8N1R1 | Gene names | ZBTB40, KIAA0478 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 40. | |||||