Please be patient as the page loads
|
MLH1_HUMAN
|
||||||
| SwissProt Accessions | P40692 | Gene names | MLH1, COCA2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh1 (MutL protein homolog 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MLH1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P40692 | Gene names | MLH1, COCA2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh1 (MutL protein homolog 1). | |||||
|
MLH1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.992120 (rank : 2) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JK91, Q62454 | Gene names | Mlh1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh1 (MutL protein homolog 1). | |||||
|
PMS2_HUMAN
|
||||||
| θ value | 3.61106e-41 (rank : 3) | NC score | 0.743396 (rank : 3) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P54278 | Gene names | PMS2, PMSL2 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
PMS2_MOUSE
|
||||||
| θ value | 1.9192e-34 (rank : 4) | NC score | 0.739299 (rank : 4) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54279 | Gene names | Pms2 | |||
|
Domain Architecture |
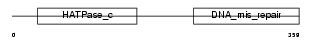
|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
PMS1_HUMAN
|
||||||
| θ value | 2.19584e-30 (rank : 5) | NC score | 0.678292 (rank : 5) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54277 | Gene names | PMS1, PMSL1 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 1 (DNA mismatch repair protein PMS1). | |||||
|
MLH3_HUMAN
|
||||||
| θ value | 3.07116e-24 (rank : 6) | NC score | 0.651832 (rank : 6) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UHC1, P49751, Q56DK9, Q9P292, Q9UHC0 | Gene names | MLH3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh3 (MutL protein homolog 3). | |||||
|
EMSY_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 7) | NC score | 0.062653 (rank : 8) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
CCM2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 8) | NC score | 0.066981 (rank : 7) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K2Y9 | Gene names | Ccm2, Osm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Malcavernin (Cerebral cavernous malformations protein 2 homolog) (Osmosensing scaffold for MEKK3). | |||||
|
EMSY_MOUSE
|
||||||
| θ value | 1.38821 (rank : 9) | NC score | 0.051128 (rank : 9) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
CA106_HUMAN
|
||||||
| θ value | 1.81305 (rank : 10) | NC score | 0.025942 (rank : 13) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3KP66, Q9NV65, Q9NVI0 | Gene names | C1orf106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106. | |||||
|
FYCO1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 11) | NC score | 0.007920 (rank : 26) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
HCFC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 12) | NC score | 0.017382 (rank : 16) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5Z7 | Gene names | HCFC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor 2 (HCF-2) (C2 factor). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.019121 (rank : 14) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
CCM2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.042840 (rank : 10) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BSQ5, Q71RE5, Q8TAT4 | Gene names | CCM2, C7orf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Malcavernin (Cerebral cavernous malformations 2 protein). | |||||
|
GLE1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.018010 (rank : 15) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q53GS7, O75458, Q53GT9, Q5VVU1, Q8NCP6, Q9UFL6 | Gene names | GLE1L, GLE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein) (hGLE1). | |||||
|
G3BP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.013941 (rank : 17) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97855 | Gene names | G3bp, G3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 1 (EC 3.6.1.-) (ATP- dependent DNA helicase VIII) (GAP SH3-domain-binding protein 1) (G3BP- 1) (HDH-VIII). | |||||
|
PTN21_MOUSE
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.006118 (rank : 28) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62136 | Gene names | Ptpn21 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.013229 (rank : 20) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.009226 (rank : 24) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.013741 (rank : 18) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
GRIK5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.009691 (rank : 21) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16478 | Gene names | GRIK5, GRIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Excitatory amino acid receptor 2) (EAA2). | |||||
|
GRIK5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.009665 (rank : 22) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61626 | Gene names | Grik5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Glutamate receptor gamma-2) (GluR gamma-2). | |||||
|
ROBO2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.003627 (rank : 30) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TPD3, Q8BJ59 | Gene names | Robo2, Kiaa1568 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 2 precursor. | |||||
|
UBP24_HUMAN
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.009509 (rank : 23) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UPU5, Q6ZSY2, Q8N2Y4, Q9NXD1 | Gene names | USP24, KIAA1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 24 (EC 3.1.2.15) (Ubiquitin thioesterase 24) (Ubiquitin-specific-processing protease 24) (Deubiquitinating enzyme 24). | |||||
|
MKRN2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.028841 (rank : 11) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
RHG07_MOUSE
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.007141 (rank : 27) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0Z9, Q8R541 | Gene names | Dlc1, Arhgap7, Stard12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein homolog) (Dlc-1) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
CCNL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.008965 (rank : 25) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJA7, Q5XK66, Q60995, Q8C136, Q8CIJ8, Q99L73, Q9QXH5 | Gene names | Ccnl2, Ania6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Cyclin Ania-6b) (Paneth cell-enhanced expression protein) (PCEE). | |||||
|
COX10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.013447 (rank : 19) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CFY5 | Gene names | Cox10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protoheme IX farnesyltransferase, mitochondrial precursor (EC 2.5.1.-) (Heme O synthase). | |||||
|
FLNA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.005121 (rank : 29) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BTM8, O54934, Q7TQI1, Q8BLK1, Q8BTN7, Q8VHX5, Q8VHX8, Q99KQ2 | Gene names | Flna, Fln, Fln1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
MKRN2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.027424 (rank : 12) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
MLH1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P40692 | Gene names | MLH1, COCA2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh1 (MutL protein homolog 1). | |||||
|
MLH1_MOUSE
|
||||||
| NC score | 0.992120 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JK91, Q62454 | Gene names | Mlh1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh1 (MutL protein homolog 1). | |||||
|
PMS2_HUMAN
|
||||||
| NC score | 0.743396 (rank : 3) | θ value | 3.61106e-41 (rank : 3) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P54278 | Gene names | PMS2, PMSL2 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
PMS2_MOUSE
|
||||||
| NC score | 0.739299 (rank : 4) | θ value | 1.9192e-34 (rank : 4) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54279 | Gene names | Pms2 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
PMS1_HUMAN
|
||||||
| NC score | 0.678292 (rank : 5) | θ value | 2.19584e-30 (rank : 5) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54277 | Gene names | PMS1, PMSL1 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 1 (DNA mismatch repair protein PMS1). | |||||
|
MLH3_HUMAN
|
||||||
| NC score | 0.651832 (rank : 6) | θ value | 3.07116e-24 (rank : 6) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UHC1, P49751, Q56DK9, Q9P292, Q9UHC0 | Gene names | MLH3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh3 (MutL protein homolog 3). | |||||
|
CCM2_MOUSE
|
||||||
| NC score | 0.066981 (rank : 7) | θ value | 0.365318 (rank : 8) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K2Y9 | Gene names | Ccm2, Osm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Malcavernin (Cerebral cavernous malformations protein 2 homolog) (Osmosensing scaffold for MEKK3). | |||||
|
EMSY_HUMAN
|
||||||
| NC score | 0.062653 (rank : 8) | θ value | 0.0736092 (rank : 7) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
EMSY_MOUSE
|
||||||
| NC score | 0.051128 (rank : 9) | θ value | 1.38821 (rank : 9) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
CCM2_HUMAN
|
||||||
| NC score | 0.042840 (rank : 10) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BSQ5, Q71RE5, Q8TAT4 | Gene names | CCM2, C7orf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Malcavernin (Cerebral cavernous malformations 2 protein). | |||||
|
MKRN2_HUMAN
|
||||||
| NC score | 0.028841 (rank : 11) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
MKRN2_MOUSE
|
||||||
| NC score | 0.027424 (rank : 12) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
CA106_HUMAN
|
||||||
| NC score | 0.025942 (rank : 13) | θ value | 1.81305 (rank : 10) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3KP66, Q9NV65, Q9NVI0 | Gene names | C1orf106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf106. | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.019121 (rank : 14) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
GLE1_HUMAN
|
||||||
| NC score | 0.018010 (rank : 15) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q53GS7, O75458, Q53GT9, Q5VVU1, Q8NCP6, Q9UFL6 | Gene names | GLE1L, GLE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein) (hGLE1). | |||||
|
HCFC2_HUMAN
|
||||||
| NC score | 0.017382 (rank : 16) | θ value | 2.36792 (rank : 12) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5Z7 | Gene names | HCFC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor 2 (HCF-2) (C2 factor). | |||||
|
G3BP_MOUSE
|
||||||
| NC score | 0.013941 (rank : 17) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97855 | Gene names | G3bp, G3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 1 (EC 3.6.1.-) (ATP- dependent DNA helicase VIII) (GAP SH3-domain-binding protein 1) (G3BP- 1) (HDH-VIII). | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.013741 (rank : 18) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
COX10_MOUSE
|
||||||
| NC score | 0.013447 (rank : 19) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CFY5 | Gene names | Cox10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protoheme IX farnesyltransferase, mitochondrial precursor (EC 2.5.1.-) (Heme O synthase). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.013229 (rank : 20) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
GRIK5_HUMAN
|
||||||
| NC score | 0.009691 (rank : 21) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16478 | Gene names | GRIK5, GRIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Excitatory amino acid receptor 2) (EAA2). | |||||
|
GRIK5_MOUSE
|
||||||
| NC score | 0.009665 (rank : 22) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61626 | Gene names | Grik5 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor, ionotropic kainate 5 precursor (Glutamate receptor KA-2) (KA2) (Glutamate receptor gamma-2) (GluR gamma-2). | |||||
|
UBP24_HUMAN
|
||||||
| NC score | 0.009509 (rank : 23) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UPU5, Q6ZSY2, Q8N2Y4, Q9NXD1 | Gene names | USP24, KIAA1057 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 24 (EC 3.1.2.15) (Ubiquitin thioesterase 24) (Ubiquitin-specific-processing protease 24) (Deubiquitinating enzyme 24). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.009226 (rank : 24) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
CCNL2_MOUSE
|
||||||
| NC score | 0.008965 (rank : 25) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJA7, Q5XK66, Q60995, Q8C136, Q8CIJ8, Q99L73, Q9QXH5 | Gene names | Ccnl2, Ania6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Cyclin Ania-6b) (Paneth cell-enhanced expression protein) (PCEE). | |||||
|
FYCO1_MOUSE
|
||||||
| NC score | 0.007920 (rank : 26) | θ value | 2.36792 (rank : 11) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
RHG07_MOUSE
|
||||||
| NC score | 0.007141 (rank : 27) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0Z9, Q8R541 | Gene names | Dlc1, Arhgap7, Stard12 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 7 (Rho-type GTPase-activating protein 7) (Deleted in liver cancer 1 protein homolog) (Dlc-1) (StAR-related lipid transfer protein 12) (StARD12) (START domain-containing protein 12). | |||||
|
PTN21_MOUSE
|
||||||
| NC score | 0.006118 (rank : 28) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62136 | Gene names | Ptpn21 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 21 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-RL10). | |||||
|
FLNA_MOUSE
|
||||||
| NC score | 0.005121 (rank : 29) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BTM8, O54934, Q7TQI1, Q8BLK1, Q8BTN7, Q8VHX5, Q8VHX8, Q99KQ2 | Gene names | Flna, Fln, Fln1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-A (Alpha-filamin) (Filamin-1) (Endothelial actin-binding protein) (Actin-binding protein 280) (ABP-280) (Nonmuscle filamin). | |||||
|
ROBO2_MOUSE
|
||||||
| NC score | 0.003627 (rank : 30) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 30 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TPD3, Q8BJ59 | Gene names | Robo2, Kiaa1568 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 2 precursor. | |||||