Please be patient as the page loads
|
PMS2_MOUSE
|
||||||
| SwissProt Accessions | P54279 | Gene names | Pms2 | |||
|
Domain Architecture |
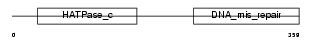
|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PMS2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.959278 (rank : 2) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P54278 | Gene names | PMS2, PMSL2 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
PMS2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P54279 | Gene names | Pms2 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
PMS1_HUMAN
|
||||||
| θ value | 1.42001e-37 (rank : 3) | NC score | 0.705067 (rank : 5) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P54277 | Gene names | PMS1, PMSL1 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 1 (DNA mismatch repair protein PMS1). | |||||
|
MLH1_HUMAN
|
||||||
| θ value | 1.9192e-34 (rank : 4) | NC score | 0.739299 (rank : 3) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P40692 | Gene names | MLH1, COCA2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh1 (MutL protein homolog 1). | |||||
|
MLH1_MOUSE
|
||||||
| θ value | 6.17384e-33 (rank : 5) | NC score | 0.735128 (rank : 4) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JK91, Q62454 | Gene names | Mlh1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh1 (MutL protein homolog 1). | |||||
|
MLH3_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 6) | NC score | 0.567305 (rank : 6) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UHC1, P49751, Q56DK9, Q9P292, Q9UHC0 | Gene names | MLH3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh3 (MutL protein homolog 3). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 7) | NC score | 0.037861 (rank : 14) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
ACM5_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 8) | NC score | 0.011915 (rank : 34) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q920H4 | Gene names | Chrm5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscarinic acetylcholine receptor M5. | |||||
|
C8AP2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 9) | NC score | 0.049078 (rank : 10) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
ICAL_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 10) | NC score | 0.056990 (rank : 9) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51125, Q9EQV4, Q9EQV5, Q9QXQ3, Q9QXQ4, Q9R0N1 | Gene names | Cast | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor). | |||||
|
CEP41_MOUSE
|
||||||
| θ value | 0.47712 (rank : 11) | NC score | 0.078516 (rank : 7) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99NF3, Q8BT53 | Gene names | Tsga14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 41 kDa (Cep41 protein) (Testis-specific gene A14 protein). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 12) | NC score | 0.026057 (rank : 19) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
LRMP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 13) | NC score | 0.039908 (rank : 12) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60664, Q7TMU8 | Gene names | Lrmp, Jaw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphoid-restricted membrane protein (Protein Jaw1). | |||||
|
GLIS3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.003920 (rank : 47) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 758 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NEA6 | Gene names | GLIS3, ZNF515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3) (Zinc finger protein 515). | |||||
|
MAST3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.003558 (rank : 49) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1128 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60307, Q7LDZ8, Q9UPI0 | Gene names | MAST3, KIAA0561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 3 (EC 2.7.11.1). | |||||
|
RAVR2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.038822 (rank : 13) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TPD6 | Gene names | Raver2, Kiaa1579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 2 (Protein raver-2). | |||||
|
S12A6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.014602 (rank : 29) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UHW9, Q8TDD4, Q9UFR2, Q9Y642, Q9Y665 | Gene names | SLC12A6, KCC3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 18) | NC score | 0.028084 (rank : 18) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
CEP41_HUMAN
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.068158 (rank : 8) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYV8, Q7Z496, Q86TM1, Q8NFU8, Q9H6A3, Q9NPV3 | Gene names | TSGA14, CEP41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 41 kDa (Cep41 protein) (Testis-specific gene A14 protein). | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.009614 (rank : 39) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
RT31_HUMAN
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.035970 (rank : 15) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92665, Q5VYC8, Q8WTV8 | Gene names | MRPS31, IMOGN38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 28S ribosomal protein S31, mitochondrial precursor (S31mt) (MRP-S31) (Imogen 38). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.012961 (rank : 32) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
ANC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.033121 (rank : 16) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H1A4, Q9BSE6, Q9H8D0 | Gene names | ANAPC1, TSG24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 1 (APC1) (Cyclosome subunit 1) (Protein Tsg24) (Mitotic checkpoint regulator). | |||||
|
LIPA2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.008211 (rank : 42) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
ZEP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.005984 (rank : 44) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P31629, Q02646, Q5THT5, Q9NS05 | Gene names | HIVEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 (HIV- EP2) (MHC-binding protein 2) (MBP-2). | |||||
|
CHCH3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.020551 (rank : 22) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NX63 | Gene names | CHCHD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil-helix-coiled-coil-helix domain-containing protein 3. | |||||
|
CL025_HUMAN
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.042986 (rank : 11) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IYM0, Q8TCP7, Q9H0L3 | Gene names | C12orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C12orf25. | |||||
|
DHSA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.021597 (rank : 21) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K2B3, Q3UH25 | Gene names | Sdha | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial precursor (EC 1.3.5.1) (Fp) (Flavoprotein subunit of complex II). | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.004872 (rank : 45) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
K1C20_HUMAN
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.007911 (rank : 43) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35900 | Gene names | KRT20 | |||
|
Domain Architecture |
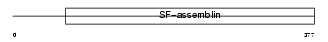
|
|||||
| Description | Keratin, type I cytoskeletal 20 (Cytokeratin-20) (CK-20) (Keratin-20) (K20) (Protein IT). | |||||
|
KSR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.004137 (rank : 46) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61097, Q61648, Q78DX8 | Gene names | Ksr1, Ksr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras) (mKSR1) (Hb protein). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.019837 (rank : 23) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
PERQ1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.025318 (rank : 20) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75420 | Gene names | PERQ1 | |||
|
Domain Architecture |

|
|||||
| Description | PERQ amino acid rich with GYF domain protein 1. | |||||
|
RGL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.013136 (rank : 31) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60695 | Gene names | Rgl1, Rgl | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 1 (RalGDS-like 1). | |||||
|
ANC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.029523 (rank : 17) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53995, Q8BP33, Q8C772 | Gene names | Anapc1, Mcpr, Tsg24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 1 (APC1) (Cyclosome subunit 1) (Protein Tsg24) (Mitotic checkpoint regulator). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.014081 (rank : 30) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.018749 (rank : 26) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
SOX4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.011415 (rank : 36) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q06831 | Gene names | Sox4, Sox-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.019790 (rank : 24) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TOIP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.017748 (rank : 27) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921T2, Q3U7A4 | Gene names | Tor1aip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 1 (Lamina-associated polypeptide 1B). | |||||
|
UBP43_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.009388 (rank : 41) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
4ET_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.015091 (rank : 28) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NRA8, Q8NCF2, Q9H708 | Gene names | EIF4ENIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
DTD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.018832 (rank : 25) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TEA8, Q496D1, Q5W184, Q8WXU8, Q9BW67, Q9H464, Q9H474 | Gene names | HARS2, C20orf88 | |||
|
Domain Architecture |

|
|||||
| Description | Probable D-tyrosyl-tRNA(Tyr) deacylase (EC 3.1.-.-). | |||||
|
EST1A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.012762 (rank : 33) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P61406, Q5NC64 | Gene names | Est1a, Kiaa0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A). | |||||
|
MYCB2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.011881 (rank : 35) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
MYO9B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.003661 (rank : 48) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.010712 (rank : 38) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
STK36_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.001066 (rank : 50) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NRP7, Q8TC32, Q9H9N9, Q9UF35, Q9ULE2 | Gene names | STK36, KIAA1278 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 36 (EC 2.7.11.1) (Fused homolog). | |||||
|
TAOK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.001026 (rank : 51) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1720 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5F2E8, Q69ZL2, Q8JZX2, Q91VG7 | Gene names | Taok1, Kiaa1361 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO1 (EC 2.7.11.1) (Thousand and one amino acid protein 1). | |||||
|
DEAF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.010995 (rank : 37) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75398, O15152, O75399, O75510, O75511, O75512, O75513, Q9UET1 | Gene names | DEAF1, SPN, ZMYND5 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR) (Suppressin) (Zinc finger MYND domain-containing protein 5). | |||||
|
MARE3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.009423 (rank : 40) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPY8, O00265, Q6FI15, Q9BZP7, Q9BZP8 | Gene names | MAPRE3 | |||
|
Domain Architecture |
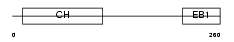
|
|||||
| Description | Microtubule-associated protein RP/EB family member 3 (End-binding protein 3) (EB3) (EB1 protein family member 3) (EBF3) (RP3). | |||||
|
PMS2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P54279 | Gene names | Pms2 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
PMS2_HUMAN
|
||||||
| NC score | 0.959278 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P54278 | Gene names | PMS2, PMSL2 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
MLH1_HUMAN
|
||||||
| NC score | 0.739299 (rank : 3) | θ value | 1.9192e-34 (rank : 4) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P40692 | Gene names | MLH1, COCA2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh1 (MutL protein homolog 1). | |||||
|
MLH1_MOUSE
|
||||||
| NC score | 0.735128 (rank : 4) | θ value | 6.17384e-33 (rank : 5) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JK91, Q62454 | Gene names | Mlh1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh1 (MutL protein homolog 1). | |||||
|
PMS1_HUMAN
|
||||||
| NC score | 0.705067 (rank : 5) | θ value | 1.42001e-37 (rank : 3) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P54277 | Gene names | PMS1, PMSL1 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 1 (DNA mismatch repair protein PMS1). | |||||
|
MLH3_HUMAN
|
||||||
| NC score | 0.567305 (rank : 6) | θ value | 2.88119e-14 (rank : 6) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UHC1, P49751, Q56DK9, Q9P292, Q9UHC0 | Gene names | MLH3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Mlh3 (MutL protein homolog 3). | |||||
|
CEP41_MOUSE
|
||||||
| NC score | 0.078516 (rank : 7) | θ value | 0.47712 (rank : 11) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99NF3, Q8BT53 | Gene names | Tsga14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 41 kDa (Cep41 protein) (Testis-specific gene A14 protein). | |||||
|
CEP41_HUMAN
|
||||||
| NC score | 0.068158 (rank : 8) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYV8, Q7Z496, Q86TM1, Q8NFU8, Q9H6A3, Q9NPV3 | Gene names | TSGA14, CEP41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 41 kDa (Cep41 protein) (Testis-specific gene A14 protein). | |||||
|
ICAL_MOUSE
|
||||||
| NC score | 0.056990 (rank : 9) | θ value | 0.0961366 (rank : 10) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51125, Q9EQV4, Q9EQV5, Q9QXQ3, Q9QXQ4, Q9R0N1 | Gene names | Cast | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor). | |||||
|
C8AP2_MOUSE
|
||||||
| NC score | 0.049078 (rank : 10) | θ value | 0.0961366 (rank : 9) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
CL025_HUMAN
|
||||||
| NC score | 0.042986 (rank : 11) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IYM0, Q8TCP7, Q9H0L3 | Gene names | C12orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C12orf25. | |||||
|
LRMP_MOUSE
|
||||||
| NC score | 0.039908 (rank : 12) | θ value | 0.813845 (rank : 13) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60664, Q7TMU8 | Gene names | Lrmp, Jaw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphoid-restricted membrane protein (Protein Jaw1). | |||||
|
RAVR2_MOUSE
|
||||||
| NC score | 0.038822 (rank : 13) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TPD6 | Gene names | Raver2, Kiaa1579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonucleoprotein PTB-binding 2 (Protein raver-2). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.037861 (rank : 14) | θ value | 0.0736092 (rank : 7) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
RT31_HUMAN
|
||||||
| NC score | 0.035970 (rank : 15) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q92665, Q5VYC8, Q8WTV8 | Gene names | MRPS31, IMOGN38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 28S ribosomal protein S31, mitochondrial precursor (S31mt) (MRP-S31) (Imogen 38). | |||||
|
ANC1_HUMAN
|
||||||
| NC score | 0.033121 (rank : 16) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H1A4, Q9BSE6, Q9H8D0 | Gene names | ANAPC1, TSG24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 1 (APC1) (Cyclosome subunit 1) (Protein Tsg24) (Mitotic checkpoint regulator). | |||||
|
ANC1_MOUSE
|
||||||
| NC score | 0.029523 (rank : 17) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53995, Q8BP33, Q8C772 | Gene names | Anapc1, Mcpr, Tsg24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 1 (APC1) (Cyclosome subunit 1) (Protein Tsg24) (Mitotic checkpoint regulator). | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.028084 (rank : 18) | θ value | 1.38821 (rank : 18) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.026057 (rank : 19) | θ value | 0.813845 (rank : 12) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
PERQ1_HUMAN
|
||||||
| NC score | 0.025318 (rank : 20) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75420 | Gene names | PERQ1 | |||
|
Domain Architecture |

|
|||||
| Description | PERQ amino acid rich with GYF domain protein 1. | |||||
|
DHSA_MOUSE
|
||||||
| NC score | 0.021597 (rank : 21) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K2B3, Q3UH25 | Gene names | Sdha | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial precursor (EC 1.3.5.1) (Fp) (Flavoprotein subunit of complex II). | |||||
|
CHCH3_HUMAN
|
||||||
| NC score | 0.020551 (rank : 22) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NX63 | Gene names | CHCHD3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil-helix-coiled-coil-helix domain-containing protein 3. | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.019837 (rank : 23) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.019790 (rank : 24) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
DTD1_HUMAN
|
||||||
| NC score | 0.018832 (rank : 25) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TEA8, Q496D1, Q5W184, Q8WXU8, Q9BW67, Q9H464, Q9H474 | Gene names | HARS2, C20orf88 | |||
|
Domain Architecture |

|
|||||
| Description | Probable D-tyrosyl-tRNA(Tyr) deacylase (EC 3.1.-.-). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.018749 (rank : 26) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
TOIP1_MOUSE
|
||||||
| NC score | 0.017748 (rank : 27) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921T2, Q3U7A4 | Gene names | Tor1aip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 1 (Lamina-associated polypeptide 1B). | |||||
|
4ET_HUMAN
|
||||||
| NC score | 0.015091 (rank : 28) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NRA8, Q8NCF2, Q9H708 | Gene names | EIF4ENIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
S12A6_HUMAN
|
||||||
| NC score | 0.014602 (rank : 29) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UHW9, Q8TDD4, Q9UFR2, Q9Y642, Q9Y665 | Gene names | SLC12A6, KCC3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.014081 (rank : 30) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
RGL1_MOUSE
|
||||||
| NC score | 0.013136 (rank : 31) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60695 | Gene names | Rgl1, Rgl | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator-like 1 (RalGDS-like 1). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.012961 (rank : 32) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
EST1A_MOUSE
|
||||||
| NC score | 0.012762 (rank : 33) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P61406, Q5NC64 | Gene names | Est1a, Kiaa0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A). | |||||
|
ACM5_MOUSE
|
||||||
| NC score | 0.011915 (rank : 34) | θ value | 0.0961366 (rank : 8) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q920H4 | Gene names | Chrm5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Muscarinic acetylcholine receptor M5. | |||||
|
MYCB2_HUMAN
|
||||||
| NC score | 0.011881 (rank : 35) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
SOX4_MOUSE
|
||||||
| NC score | 0.011415 (rank : 36) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q06831 | Gene names | Sox4, Sox-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-4. | |||||
|
DEAF1_HUMAN
|
||||||
| NC score | 0.010995 (rank : 37) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75398, O15152, O75399, O75510, O75511, O75512, O75513, Q9UET1 | Gene names | DEAF1, SPN, ZMYND5 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR) (Suppressin) (Zinc finger MYND domain-containing protein 5). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.010712 (rank : 38) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.009614 (rank : 39) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
MARE3_HUMAN
|
||||||
| NC score | 0.009423 (rank : 40) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPY8, O00265, Q6FI15, Q9BZP7, Q9BZP8 | Gene names | MAPRE3 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein RP/EB family member 3 (End-binding protein 3) (EB3) (EB1 protein family member 3) (EBF3) (RP3). | |||||
|
UBP43_HUMAN
|
||||||
| NC score | 0.009388 (rank : 41) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q70EL4, Q8N2C5, Q96DQ6 | Gene names | USP43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
LIPA2_HUMAN
|
||||||
| NC score | 0.008211 (rank : 42) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
K1C20_HUMAN
|
||||||
| NC score | 0.007911 (rank : 43) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35900 | Gene names | KRT20 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cytoskeletal 20 (Cytokeratin-20) (CK-20) (Keratin-20) (K20) (Protein IT). | |||||
|
ZEP2_HUMAN
|
||||||
| NC score | 0.005984 (rank : 44) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P31629, Q02646, Q5THT5, Q9NS05 | Gene names | HIVEP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 (HIV- EP2) (MHC-binding protein 2) (MBP-2). | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.004872 (rank : 45) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
KSR1_MOUSE
|
||||||
| NC score | 0.004137 (rank : 46) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61097, Q61648, Q78DX8 | Gene names | Ksr1, Ksr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras) (mKSR1) (Hb protein). | |||||
|
GLIS3_HUMAN
|
||||||
| NC score | 0.003920 (rank : 47) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 758 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NEA6 | Gene names | GLIS3, ZNF515 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3) (Zinc finger protein 515). | |||||
|
MYO9B_MOUSE
|
||||||
| NC score | 0.003661 (rank : 48) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
MAST3_HUMAN
|
||||||
| NC score | 0.003558 (rank : 49) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1128 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60307, Q7LDZ8, Q9UPI0 | Gene names | MAST3, KIAA0561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 3 (EC 2.7.11.1). | |||||
|
STK36_HUMAN
|
||||||
| NC score | 0.001066 (rank : 50) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NRP7, Q8TC32, Q9H9N9, Q9UF35, Q9ULE2 | Gene names | STK36, KIAA1278 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 36 (EC 2.7.11.1) (Fused homolog). | |||||
|
TAOK1_MOUSE
|
||||||
| NC score | 0.001026 (rank : 51) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 51 | Target Neighborhood Hits | 1720 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5F2E8, Q69ZL2, Q8JZX2, Q91VG7 | Gene names | Taok1, Kiaa1361 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO1 (EC 2.7.11.1) (Thousand and one amino acid protein 1). | |||||