Please be patient as the page loads
|
S12A6_HUMAN
|
||||||
| SwissProt Accessions | Q9UHW9, Q8TDD4, Q9UFR2, Q9Y642, Q9Y665 | Gene names | SLC12A6, KCC3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
S12A4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.996905 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UP95, O60632, O75893, Q13953, Q96LD5 | Gene names | SLC12A4, KCC1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 4 (Electroneutral potassium-chloride cotransporter 1) (Erythroid K-Cl cotransporter 1) (hKCC1). | |||||
|
S12A4_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.996957 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JIS8, O55069, Q9ET57 | Gene names | Slc12a4, Kcc1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 4 (Electroneutral potassium-chloride cotransporter 1) (Erythroid K-Cl cotransporter 1) (mKCC1). | |||||
|
S12A5_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.993246 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H2X9, Q5VZ41, Q9H4Z0, Q9ULP4 | Gene names | SLC12A5, KCC2, KIAA1176 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (Erythroid K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (hKCC2). | |||||
|
S12A5_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.993142 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91V14, Q9Z0M7 | Gene names | Slc12a5, Kcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (mKCC2). | |||||
|
S12A6_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UHW9, Q8TDD4, Q9UFR2, Q9Y642, Q9Y665 | Gene names | SLC12A6, KCC3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
S12A6_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.999225 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q924N4, Q924N3 | Gene names | Slc12a6, Kcc3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
S12A7_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.993834 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y666, Q96I81, Q9H7I3, Q9H7I7, Q9UFW2 | Gene names | SLC12A7, KCC4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
S12A7_MOUSE
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.994345 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9WVL3 | Gene names | Slc12a7, Kcc4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
S12A3_MOUSE
|
||||||
| θ value | 1.7238e-59 (rank : 9) | NC score | 0.833241 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P59158 | Gene names | Slc12a3, Tsc | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 3 (Thiazide-sensitive sodium-chloride cotransporter) (Na-Cl symporter). | |||||
|
S12A3_HUMAN
|
||||||
| θ value | 3.974e-56 (rank : 10) | NC score | 0.816516 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P55017 | Gene names | SLC12A3, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 3 (Thiazide-sensitive sodium-chloride cotransporter) (Na-Cl symporter). | |||||
|
S12A2_HUMAN
|
||||||
| θ value | 2.18062e-54 (rank : 11) | NC score | 0.807786 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55011, Q8N713, Q8WWH7 | Gene names | SLC12A2, NKCC1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
|
S12A2_MOUSE
|
||||||
| θ value | 2.84797e-54 (rank : 12) | NC score | 0.809632 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P55012 | Gene names | Slc12a2, Nkcc1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
|
S12A1_HUMAN
|
||||||
| θ value | 3.84914e-51 (rank : 13) | NC score | 0.801784 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13621 | Gene names | SLC12A1, NKCC2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 1 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 2) (Kidney-specific Na-K-Cl symporter). | |||||
|
S12A1_MOUSE
|
||||||
| θ value | 1.46268e-50 (rank : 14) | NC score | 0.801674 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P55014, O35938, Q91W55, Q9Z1E0 | Gene names | Slc12a1, Nkcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 1 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 2) (BSC1) (Kidney-specific Na-K-Cl symporter). | |||||
|
CTR2_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 15) | NC score | 0.212208 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P18581 | Gene names | Slc7a2, Atrc2, Tea | |||
|
Domain Architecture |

|
|||||
| Description | Low-affinity cationic amino acid transporter 2 (CAT-2) (CAT2) (TEA protein) (T-cell early activation protein) (20.5). | |||||
|
CTR2_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 16) | NC score | 0.188325 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52569 | Gene names | SLC7A2, ATRC2 | |||
|
Domain Architecture |
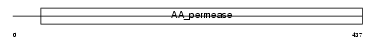
|
|||||
| Description | Low-affinity cationic amino acid transporter 2 (CAT-2) (CAT2). | |||||
|
CTR1_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 17) | NC score | 0.162649 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P30825 | Gene names | SLC7A1, ATRC1, REC1L | |||
|
Domain Architecture |
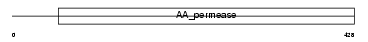
|
|||||
| Description | High-affinity cationic amino acid transporter 1 (CAT-1) (CAT1) (System Y+ basic amino acid transporter) (Ecotropic retroviral leukemia receptor homolog) (ERR) (Ecotropic retrovirus receptor homolog). | |||||
|
CTR1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 18) | NC score | 0.177353 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q09143, P30824 | Gene names | Slc7a1, Atrc1, Rec-1 | |||
|
Domain Architecture |
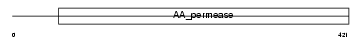
|
|||||
| Description | High-affinity cationic amino acid transporter 1 (CAT-1) (CAT1) (System Y+ basic amino acid transporter) (Ecotropic retroviral leukemia receptor) (ERR) (Ecotropic retrovirus receptor). | |||||
|
AAA1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 19) | NC score | 0.080837 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NS82 | Gene names | SLC7A10, ASC1 | |||
|
Domain Architecture |
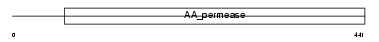
|
|||||
| Description | Asc-type amino acid transporter 1 (Asc-1). | |||||
|
DNMT1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 20) | NC score | 0.033405 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P26358, Q9UHG5, Q9ULA2, Q9UMZ6 | Gene names | DNMT1, AIM, DNMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI). | |||||
|
BAT1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 21) | NC score | 0.122116 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QXA6 | Gene names | Slc7a9, Bat1 | |||
|
Domain Architecture |
|
|||||
| Description | B(0,+)-type amino acid transporter 1 (B(0,+)AT) (Glycoprotein- associated amino acid transporter b0,+AT1). | |||||
|
AAA1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 22) | NC score | 0.087389 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P63115, Q9CW25, Q9JMH8 | Gene names | Slc7a10, Asc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Asc-type amino acid transporter 1 (Asc-1) (D-serine transporter). | |||||
|
CTR3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.147021 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WY07, Q8N185, Q8NCA7, Q96SZ7 | Gene names | SLC7A3, ATRC3, CAT3 | |||
|
Domain Architecture |
|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
BAT1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 24) | NC score | 0.104275 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P82251 | Gene names | SLC7A9, BAT1 | |||
|
Domain Architecture |
|
|||||
| Description | B(0,+)-type amino acid transporter 1 (B(0,+)AT) (Glycoprotein- associated amino acid transporter b0,+AT1). | |||||
|
CTR3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 25) | NC score | 0.142090 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70423 | Gene names | Slc7a3, Atrc3, Cat3 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
KCC2B_MOUSE
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.001116 (rank : 61) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 841 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28652, Q5SVH9 | Gene names | Camk2b, Camk2d | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent protein kinase type II beta chain (EC 2.7.11.17) (CaM-kinase II beta chain) (CaM kinase II subunit beta) (CaMK-II subunit beta). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.008359 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
LAT1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.061590 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q01650, Q9UBN8, Q9UP15, Q9UQC0 | Gene names | SLC7A5, CD98LC, LAT1, MPE16 | |||
|
Domain Architecture |

|
|||||
| Description | Large neutral amino acids transporter small subunit 1 (L-type amino acid transporter 1) (4F2 light chain) (4F2 LC) (4F2LC) (CD98 light chain) (Integral membrane protein E16) (hLAT1). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.018165 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.012088 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
HS90A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.018456 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P07901 | Gene names | Hsp90aa1, Hsp86, Hsp86-1, Hspca | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (Tumor-specific transplantation 86 kDa antigen) (TSTA). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.004147 (rank : 59) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
PMS2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.014602 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54279 | Gene names | Pms2 | |||
|
Domain Architecture |
|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
S36A1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.033868 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z2H8, Q7Z7C0, Q96M74 | Gene names | SLC36A1, PAT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proton-coupled amino acid transporter 1 (Proton/amino acid transporter 1) (Solute carrier family 36 member 1). | |||||
|
COX10_MOUSE
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.024969 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CFY5 | Gene names | Cox10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protoheme IX farnesyltransferase, mitochondrial precursor (EC 2.5.1.-) (Heme O synthase). | |||||
|
RM21_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.020263 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z2W9 | Gene names | MRPL21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 39S ribosomal protein L21, mitochondrial precursor (L21mt) (MRP-L21). | |||||
|
TAOK3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.000247 (rank : 63) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1458 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BYC6, Q3V3B3 | Gene names | Taok3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO3 (EC 2.7.11.1) (Thousand and one amino acid protein 3). | |||||
|
DGAT1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.026871 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75907, Q96BB8 | Gene names | DGAT1, AGRP1, DGAT | |||
|
Domain Architecture |
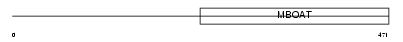
|
|||||
| Description | Diacylglycerol O-acyltransferase 1 (EC 2.3.1.20) (Diglyceride acyltransferase) (ACAT-related gene product 1). | |||||
|
EDG1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.004953 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P21453 | Gene names | EDG1 | |||
|
Domain Architecture |
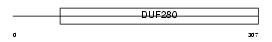
|
|||||
| Description | Sphingosine 1-phosphate receptor Edg-1 (Sphingosine 1-phosphate receptor 1) (S1P1). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.016520 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
NASP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.016847 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
DGAT1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.025089 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z2A7, Q9D7Q5 | Gene names | Dgat1, Dgat | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol O-acyltransferase 1 (EC 2.3.1.20) (Diglyceride acyltransferase). | |||||
|
LAT1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.057773 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z127, Q9JMI4 | Gene names | Slc7a5, Lat1 | |||
|
Domain Architecture |
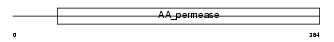
|
|||||
| Description | Large neutral amino acids transporter small subunit 1 (L-type amino acid transporter 1) (4F2 light chain) (4F2 LC) (4F2LC). | |||||
|
RHG09_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.005821 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
KIF4A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.002705 (rank : 60) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
HS90B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.013577 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08238, Q5T9W7, Q9NQW0 | Gene names | HSP90AB1, HSP90B, HSPC2, HSPCB | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (HSP 90). | |||||
|
IF3A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.005078 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
OR5T1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.000653 (rank : 62) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 654 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NG75 | Gene names | OR5T1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 5T1 (Olfactory receptor OR11-179). | |||||
|
OR7A2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | -0.001616 (rank : 64) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 782 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NGA2, Q96R95 | Gene names | OR7A2, OR7A7 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 7A2. | |||||
|
PRP6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.016021 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94906, O95109, Q5VXS5, Q9H3Z1, Q9H4T9, Q9H4U8, Q9NTE6 | Gene names | PRPF6, C20orf14 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 6 homolog (U5 snRNP-associated 102 kDa protein) (U5-102 kDa protein). | |||||
|
PRP6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.016012 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91YR7, Q8CIK9, Q8R3M8, Q99JN1, Q9CSZ0 | Gene names | Prpf6 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 6 homolog (U5 snRNP-associated 102 kDa protein) (U5-102 kDa protein). | |||||
|
SOAT2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.018231 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75908, Q96TD4, Q9UNR2 | Gene names | SOAT2, ACACT2, ACAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol O-acyltransferase 2 (EC 2.3.1.26) (Cholesterol acyltransferase 2) (Acyl coenzyme A:cholesterol acyltransferase 2) (ACAT-2). | |||||
|
EDG1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.006512 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08530, Q9R235 | Gene names | Edg1, Lpb1 | |||
|
Domain Architecture |
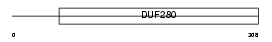
|
|||||
| Description | Sphingosine 1-phosphate receptor Edg-1 (Sphingosine 1-phosphate receptor 1) (S1P1) (Lysophospholipid receptor B1). | |||||
|
NPT2B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.008410 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBP0, Q9Z290 | Gene names | Slc34a2, Npt2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium-dependent phosphate transport protein 2B (Sodium/phosphate cotransporter 2B) (Na(+)/Pi cotransporter 2B) (Sodium-phosphate transport protein 2B) (Na(+)-dependent phosphate cotransporter 2B) (NaPi-2b) (Solute carrier family 34 member 2). | |||||
|
G137B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.049166 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60478 | Gene names | GPR137B, TM7SF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integral membrane protein GPR137B (Transmembrane 7 superfamily member 1). | |||||
|
P52K_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.006162 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
SNIP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.010854 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9C0H9, Q8N4W8 | Gene names | P140, KIAA1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
STX12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.004432 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86Y82, O95564 | Gene names | STX12 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-12. | |||||
|
CTR4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.102070 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43246, Q3KNQ6, Q4VC45, Q6IBY8, Q96H88 | Gene names | SLC7A4 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 4 (CAT-4) (CAT4). | |||||
|
LAT2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.106637 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UHI5, Q9UKQ6, Q9UKQ7, Q9UKQ8, Q9Y445 | Gene names | SLC7A8, LAT2 | |||
|
Domain Architecture |
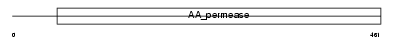
|
|||||
| Description | Large neutral amino acids transporter small subunit 2 (L-type amino acid transporter 2) (hLAT2). | |||||
|
LAT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.107253 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QXW9 | Gene names | Slc7a8, Lat2 | |||
|
Domain Architecture |
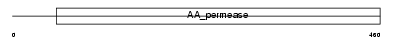
|
|||||
| Description | Large neutral amino acids transporter small subunit 2 (L-type amino acid transporter 2). | |||||
|
XCT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.052742 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UPY5 | Gene names | SLC7A11 | |||
|
Domain Architecture |
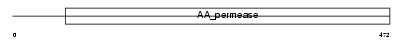
|
|||||
| Description | Cystine/glutamate transporter (Amino acid transport system xc-) (xCT) (Calcium channel blocker resistance protein CCBR1). | |||||
|
XCT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.058520 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WTR6 | Gene names | Slc7a11 | |||
|
Domain Architecture |
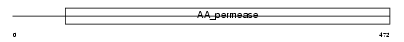
|
|||||
| Description | Cystine/glutamate transporter (Amino acid transport system xc-) (xCT). | |||||
|
YLA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.057343 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UM01, O95984, Q9P2V5 | Gene names | SLC7A7 | |||
|
Domain Architecture |
|
|||||
| Description | Y+L amino acid transporter 1 (y(+)L-type amino acid transporter 1) (y+LAT-1) (Y+LAT1) (Monocyte amino acid permease 2) (MOP-2). | |||||
|
S12A6_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UHW9, Q8TDD4, Q9UFR2, Q9Y642, Q9Y665 | Gene names | SLC12A6, KCC3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
S12A6_MOUSE
|
||||||
| NC score | 0.999225 (rank : 2) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q924N4, Q924N3 | Gene names | Slc12a6, Kcc3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
S12A4_MOUSE
|
||||||
| NC score | 0.996957 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JIS8, O55069, Q9ET57 | Gene names | Slc12a4, Kcc1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 4 (Electroneutral potassium-chloride cotransporter 1) (Erythroid K-Cl cotransporter 1) (mKCC1). | |||||
|
S12A4_HUMAN
|
||||||
| NC score | 0.996905 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UP95, O60632, O75893, Q13953, Q96LD5 | Gene names | SLC12A4, KCC1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 4 (Electroneutral potassium-chloride cotransporter 1) (Erythroid K-Cl cotransporter 1) (hKCC1). | |||||
|
S12A7_MOUSE
|
||||||
| NC score | 0.994345 (rank : 5) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9WVL3 | Gene names | Slc12a7, Kcc4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
S12A7_HUMAN
|
||||||
| NC score | 0.993834 (rank : 6) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y666, Q96I81, Q9H7I3, Q9H7I7, Q9UFW2 | Gene names | SLC12A7, KCC4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
S12A5_HUMAN
|
||||||
| NC score | 0.993246 (rank : 7) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H2X9, Q5VZ41, Q9H4Z0, Q9ULP4 | Gene names | SLC12A5, KCC2, KIAA1176 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (Erythroid K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (hKCC2). | |||||
|
S12A5_MOUSE
|
||||||
| NC score | 0.993142 (rank : 8) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91V14, Q9Z0M7 | Gene names | Slc12a5, Kcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (mKCC2). | |||||
|
S12A3_MOUSE
|
||||||
| NC score | 0.833241 (rank : 9) | θ value | 1.7238e-59 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P59158 | Gene names | Slc12a3, Tsc | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 3 (Thiazide-sensitive sodium-chloride cotransporter) (Na-Cl symporter). | |||||
|
S12A3_HUMAN
|
||||||
| NC score | 0.816516 (rank : 10) | θ value | 3.974e-56 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P55017 | Gene names | SLC12A3, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 3 (Thiazide-sensitive sodium-chloride cotransporter) (Na-Cl symporter). | |||||
|
S12A2_MOUSE
|
||||||
| NC score | 0.809632 (rank : 11) | θ value | 2.84797e-54 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P55012 | Gene names | Slc12a2, Nkcc1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
|
S12A2_HUMAN
|
||||||
| NC score | 0.807786 (rank : 12) | θ value | 2.18062e-54 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P55011, Q8N713, Q8WWH7 | Gene names | SLC12A2, NKCC1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
|
S12A1_HUMAN
|
||||||
| NC score | 0.801784 (rank : 13) | θ value | 3.84914e-51 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13621 | Gene names | SLC12A1, NKCC2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 1 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 2) (Kidney-specific Na-K-Cl symporter). | |||||
|
S12A1_MOUSE
|
||||||
| NC score | 0.801674 (rank : 14) | θ value | 1.46268e-50 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P55014, O35938, Q91W55, Q9Z1E0 | Gene names | Slc12a1, Nkcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 1 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 2) (BSC1) (Kidney-specific Na-K-Cl symporter). | |||||
|
CTR2_MOUSE
|
||||||
| NC score | 0.212208 (rank : 15) | θ value | 0.000270298 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P18581 | Gene names | Slc7a2, Atrc2, Tea | |||
|
Domain Architecture |

|
|||||
| Description | Low-affinity cationic amino acid transporter 2 (CAT-2) (CAT2) (TEA protein) (T-cell early activation protein) (20.5). | |||||
|
CTR2_HUMAN
|
||||||
| NC score | 0.188325 (rank : 16) | θ value | 0.000461057 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52569 | Gene names | SLC7A2, ATRC2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-affinity cationic amino acid transporter 2 (CAT-2) (CAT2). | |||||
|
CTR1_MOUSE
|
||||||
| NC score | 0.177353 (rank : 17) | θ value | 0.0252991 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q09143, P30824 | Gene names | Slc7a1, Atrc1, Rec-1 | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cationic amino acid transporter 1 (CAT-1) (CAT1) (System Y+ basic amino acid transporter) (Ecotropic retroviral leukemia receptor) (ERR) (Ecotropic retrovirus receptor). | |||||
|
CTR1_HUMAN
|
||||||
| NC score | 0.162649 (rank : 18) | θ value | 0.00509761 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P30825 | Gene names | SLC7A1, ATRC1, REC1L | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cationic amino acid transporter 1 (CAT-1) (CAT1) (System Y+ basic amino acid transporter) (Ecotropic retroviral leukemia receptor homolog) (ERR) (Ecotropic retrovirus receptor homolog). | |||||
|
CTR3_HUMAN
|
||||||
| NC score | 0.147021 (rank : 19) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WY07, Q8N185, Q8NCA7, Q96SZ7 | Gene names | SLC7A3, ATRC3, CAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
CTR3_MOUSE
|
||||||
| NC score | 0.142090 (rank : 20) | θ value | 0.47712 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70423 | Gene names | Slc7a3, Atrc3, Cat3 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
BAT1_MOUSE
|
||||||
| NC score | 0.122116 (rank : 21) | θ value | 0.125558 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QXA6 | Gene names | Slc7a9, Bat1 | |||
|
Domain Architecture |

|
|||||
| Description | B(0,+)-type amino acid transporter 1 (B(0,+)AT) (Glycoprotein- associated amino acid transporter b0,+AT1). | |||||
|
LAT2_MOUSE
|
||||||
| NC score | 0.107253 (rank : 22) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QXW9 | Gene names | Slc7a8, Lat2 | |||
|
Domain Architecture |

|
|||||
| Description | Large neutral amino acids transporter small subunit 2 (L-type amino acid transporter 2). | |||||
|
LAT2_HUMAN
|
||||||
| NC score | 0.106637 (rank : 23) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UHI5, Q9UKQ6, Q9UKQ7, Q9UKQ8, Q9Y445 | Gene names | SLC7A8, LAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Large neutral amino acids transporter small subunit 2 (L-type amino acid transporter 2) (hLAT2). | |||||
|
BAT1_HUMAN
|
||||||
| NC score | 0.104275 (rank : 24) | θ value | 0.21417 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P82251 | Gene names | SLC7A9, BAT1 | |||
|
Domain Architecture |

|
|||||
| Description | B(0,+)-type amino acid transporter 1 (B(0,+)AT) (Glycoprotein- associated amino acid transporter b0,+AT1). | |||||
|
CTR4_HUMAN
|
||||||
| NC score | 0.102070 (rank : 25) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43246, Q3KNQ6, Q4VC45, Q6IBY8, Q96H88 | Gene names | SLC7A4 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 4 (CAT-4) (CAT4). | |||||
|
AAA1_MOUSE
|
||||||
| NC score | 0.087389 (rank : 26) | θ value | 0.163984 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P63115, Q9CW25, Q9JMH8 | Gene names | Slc7a10, Asc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Asc-type amino acid transporter 1 (Asc-1) (D-serine transporter). | |||||
|
AAA1_HUMAN
|
||||||
| NC score | 0.080837 (rank : 27) | θ value | 0.0431538 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NS82 | Gene names | SLC7A10, ASC1 | |||
|
Domain Architecture |

|
|||||
| Description | Asc-type amino acid transporter 1 (Asc-1). | |||||
|
LAT1_HUMAN
|
||||||
| NC score | 0.061590 (rank : 28) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q01650, Q9UBN8, Q9UP15, Q9UQC0 | Gene names | SLC7A5, CD98LC, LAT1, MPE16 | |||
|
Domain Architecture |

|
|||||
| Description | Large neutral amino acids transporter small subunit 1 (L-type amino acid transporter 1) (4F2 light chain) (4F2 LC) (4F2LC) (CD98 light chain) (Integral membrane protein E16) (hLAT1). | |||||
|
XCT_MOUSE
|
||||||
| NC score | 0.058520 (rank : 29) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WTR6 | Gene names | Slc7a11 | |||
|
Domain Architecture |

|
|||||
| Description | Cystine/glutamate transporter (Amino acid transport system xc-) (xCT). | |||||
|
LAT1_MOUSE
|
||||||
| NC score | 0.057773 (rank : 30) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z127, Q9JMI4 | Gene names | Slc7a5, Lat1 | |||
|
Domain Architecture |

|
|||||
| Description | Large neutral amino acids transporter small subunit 1 (L-type amino acid transporter 1) (4F2 light chain) (4F2 LC) (4F2LC). | |||||
|
YLA1_HUMAN
|
||||||
| NC score | 0.057343 (rank : 31) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UM01, O95984, Q9P2V5 | Gene names | SLC7A7 | |||
|
Domain Architecture |

|
|||||
| Description | Y+L amino acid transporter 1 (y(+)L-type amino acid transporter 1) (y+LAT-1) (Y+LAT1) (Monocyte amino acid permease 2) (MOP-2). | |||||
|
XCT_HUMAN
|
||||||
| NC score | 0.052742 (rank : 32) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UPY5 | Gene names | SLC7A11 | |||
|
Domain Architecture |

|
|||||
| Description | Cystine/glutamate transporter (Amino acid transport system xc-) (xCT) (Calcium channel blocker resistance protein CCBR1). | |||||
|
G137B_HUMAN
|
||||||
| NC score | 0.049166 (rank : 33) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60478 | Gene names | GPR137B, TM7SF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integral membrane protein GPR137B (Transmembrane 7 superfamily member 1). | |||||
|
S36A1_HUMAN
|
||||||
| NC score | 0.033868 (rank : 34) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z2H8, Q7Z7C0, Q96M74 | Gene names | SLC36A1, PAT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proton-coupled amino acid transporter 1 (Proton/amino acid transporter 1) (Solute carrier family 36 member 1). | |||||
|
DNMT1_HUMAN
|
||||||
| NC score | 0.033405 (rank : 35) | θ value | 0.0961366 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P26358, Q9UHG5, Q9ULA2, Q9UMZ6 | Gene names | DNMT1, AIM, DNMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI). | |||||
|
DGAT1_HUMAN
|
||||||
| NC score | 0.026871 (rank : 36) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75907, Q96BB8 | Gene names | DGAT1, AGRP1, DGAT | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol O-acyltransferase 1 (EC 2.3.1.20) (Diglyceride acyltransferase) (ACAT-related gene product 1). | |||||
|
DGAT1_MOUSE
|
||||||
| NC score | 0.025089 (rank : 37) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z2A7, Q9D7Q5 | Gene names | Dgat1, Dgat | |||
|
Domain Architecture |

|
|||||
| Description | Diacylglycerol O-acyltransferase 1 (EC 2.3.1.20) (Diglyceride acyltransferase). | |||||
|
COX10_MOUSE
|
||||||
| NC score | 0.024969 (rank : 38) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CFY5 | Gene names | Cox10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protoheme IX farnesyltransferase, mitochondrial precursor (EC 2.5.1.-) (Heme O synthase). | |||||
|
RM21_HUMAN
|
||||||
| NC score | 0.020263 (rank : 39) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z2W9 | Gene names | MRPL21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 39S ribosomal protein L21, mitochondrial precursor (L21mt) (MRP-L21). | |||||
|
HS90A_MOUSE
|
||||||
| NC score | 0.018456 (rank : 40) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P07901 | Gene names | Hsp90aa1, Hsp86, Hsp86-1, Hspca | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-alpha (HSP 86) (Tumor-specific transplantation 86 kDa antigen) (TSTA). | |||||
|
SOAT2_HUMAN
|
||||||
| NC score | 0.018231 (rank : 41) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75908, Q96TD4, Q9UNR2 | Gene names | SOAT2, ACACT2, ACAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol O-acyltransferase 2 (EC 2.3.1.26) (Cholesterol acyltransferase 2) (Acyl coenzyme A:cholesterol acyltransferase 2) (ACAT-2). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.018165 (rank : 42) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.016847 (rank : 43) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.016520 (rank : 44) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
PRP6_HUMAN
|
||||||
| NC score | 0.016021 (rank : 45) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94906, O95109, Q5VXS5, Q9H3Z1, Q9H4T9, Q9H4U8, Q9NTE6 | Gene names | PRPF6, C20orf14 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 6 homolog (U5 snRNP-associated 102 kDa protein) (U5-102 kDa protein). | |||||
|
PRP6_MOUSE
|
||||||
| NC score | 0.016012 (rank : 46) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91YR7, Q8CIK9, Q8R3M8, Q99JN1, Q9CSZ0 | Gene names | Prpf6 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-mRNA-processing factor 6 homolog (U5 snRNP-associated 102 kDa protein) (U5-102 kDa protein). | |||||
|
PMS2_MOUSE
|
||||||
| NC score | 0.014602 (rank : 47) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54279 | Gene names | Pms2 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
HS90B_HUMAN
|
||||||
| NC score | 0.013577 (rank : 48) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08238, Q5T9W7, Q9NQW0 | Gene names | HSP90AB1, HSP90B, HSPC2, HSPCB | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein HSP 90-beta (HSP 84) (HSP 90). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.012088 (rank : 49) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
SNIP_HUMAN
|
||||||
| NC score | 0.010854 (rank : 50) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9C0H9, Q8N4W8 | Gene names | P140, KIAA1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
NPT2B_MOUSE
|
||||||
| NC score | 0.008410 (rank : 51) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBP0, Q9Z290 | Gene names | Slc34a2, Npt2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium-dependent phosphate transport protein 2B (Sodium/phosphate cotransporter 2B) (Na(+)/Pi cotransporter 2B) (Sodium-phosphate transport protein 2B) (Na(+)-dependent phosphate cotransporter 2B) (NaPi-2b) (Solute carrier family 34 member 2). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.008359 (rank : 52) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
EDG1_MOUSE
|
||||||
| NC score | 0.006512 (rank : 53) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08530, Q9R235 | Gene names | Edg1, Lpb1 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosine 1-phosphate receptor Edg-1 (Sphingosine 1-phosphate receptor 1) (S1P1) (Lysophospholipid receptor B1). | |||||
|
P52K_HUMAN
|
||||||
| NC score | 0.006162 (rank : 54) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
RHG09_HUMAN
|
||||||
| NC score | 0.005821 (rank : 55) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BRR9, Q8TCJ3, Q8WYR0, Q96EZ2, Q96S74 | Gene names | ARHGAP9 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 9. | |||||
|
IF3A_HUMAN
|
||||||
| NC score | 0.005078 (rank : 56) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
EDG1_HUMAN
|
||||||
| NC score | 0.004953 (rank : 57) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P21453 | Gene names | EDG1 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosine 1-phosphate receptor Edg-1 (Sphingosine 1-phosphate receptor 1) (S1P1). | |||||
|
STX12_HUMAN
|
||||||
| NC score | 0.004432 (rank : 58) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86Y82, O95564 | Gene names | STX12 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-12. | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.004147 (rank : 59) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
KIF4A_MOUSE
|
||||||
| NC score | 0.002705 (rank : 60) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
KCC2B_MOUSE
|
||||||
| NC score | 0.001116 (rank : 61) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 841 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28652, Q5SVH9 | Gene names | Camk2b, Camk2d | |||
|
Domain Architecture |

|
|||||
| Description | Calcium/calmodulin-dependent protein kinase type II beta chain (EC 2.7.11.17) (CaM-kinase II beta chain) (CaM kinase II subunit beta) (CaMK-II subunit beta). | |||||
|
OR5T1_HUMAN
|
||||||
| NC score | 0.000653 (rank : 62) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 654 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NG75 | Gene names | OR5T1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 5T1 (Olfactory receptor OR11-179). | |||||
|
TAOK3_MOUSE
|
||||||
| NC score | 0.000247 (rank : 63) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1458 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BYC6, Q3V3B3 | Gene names | Taok3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO3 (EC 2.7.11.1) (Thousand and one amino acid protein 3). | |||||
|
OR7A2_HUMAN
|
||||||
| NC score | -0.001616 (rank : 64) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 782 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NGA2, Q96R95 | Gene names | OR7A2, OR7A7 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 7A2. | |||||