Please be patient as the page loads
|
S12A2_MOUSE
|
||||||
| SwissProt Accessions | P55012 | Gene names | Slc12a2, Nkcc1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
S12A1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.993039 (rank : 4) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13621 | Gene names | SLC12A1, NKCC2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 1 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 2) (Kidney-specific Na-K-Cl symporter). | |||||
|
S12A1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.993195 (rank : 3) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P55014, O35938, Q91W55, Q9Z1E0 | Gene names | Slc12a1, Nkcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 1 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 2) (BSC1) (Kidney-specific Na-K-Cl symporter). | |||||
|
S12A2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.996600 (rank : 2) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P55011, Q8N713, Q8WWH7 | Gene names | SLC12A2, NKCC1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
|
S12A2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P55012 | Gene names | Slc12a2, Nkcc1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
|
S12A3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.988253 (rank : 5) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P55017 | Gene names | SLC12A3, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 3 (Thiazide-sensitive sodium-chloride cotransporter) (Na-Cl symporter). | |||||
|
S12A3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.985066 (rank : 6) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P59158 | Gene names | Slc12a3, Tsc | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 3 (Thiazide-sensitive sodium-chloride cotransporter) (Na-Cl symporter). | |||||
|
S12A7_MOUSE
|
||||||
| θ value | 6.55044e-59 (rank : 7) | NC score | 0.816293 (rank : 8) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WVL3 | Gene names | Slc12a7, Kcc4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
S12A7_HUMAN
|
||||||
| θ value | 3.59435e-57 (rank : 8) | NC score | 0.818899 (rank : 7) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y666, Q96I81, Q9H7I3, Q9H7I7, Q9UFW2 | Gene names | SLC12A7, KCC4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
S12A4_HUMAN
|
||||||
| θ value | 2.32979e-56 (rank : 9) | NC score | 0.812607 (rank : 10) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UP95, O60632, O75893, Q13953, Q96LD5 | Gene names | SLC12A4, KCC1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 4 (Electroneutral potassium-chloride cotransporter 1) (Erythroid K-Cl cotransporter 1) (hKCC1). | |||||
|
S12A4_MOUSE
|
||||||
| θ value | 2.32979e-56 (rank : 10) | NC score | 0.813453 (rank : 9) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JIS8, O55069, Q9ET57 | Gene names | Slc12a4, Kcc1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 4 (Electroneutral potassium-chloride cotransporter 1) (Erythroid K-Cl cotransporter 1) (mKCC1). | |||||
|
S12A5_HUMAN
|
||||||
| θ value | 1.2784e-54 (rank : 11) | NC score | 0.812458 (rank : 11) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H2X9, Q5VZ41, Q9H4Z0, Q9ULP4 | Gene names | SLC12A5, KCC2, KIAA1176 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (Erythroid K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (hKCC2). | |||||
|
S12A5_MOUSE
|
||||||
| θ value | 2.18062e-54 (rank : 12) | NC score | 0.812341 (rank : 12) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91V14, Q9Z0M7 | Gene names | Slc12a5, Kcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (mKCC2). | |||||
|
S12A6_HUMAN
|
||||||
| θ value | 2.84797e-54 (rank : 13) | NC score | 0.809632 (rank : 13) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UHW9, Q8TDD4, Q9UFR2, Q9Y642, Q9Y665 | Gene names | SLC12A6, KCC3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
S12A6_MOUSE
|
||||||
| θ value | 1.41344e-53 (rank : 14) | NC score | 0.808798 (rank : 14) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q924N4, Q924N3 | Gene names | Slc12a6, Kcc3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 15) | NC score | 0.024450 (rank : 32) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
CTR2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 16) | NC score | 0.145330 (rank : 16) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 20 | |
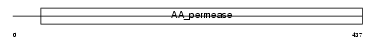
| SwissProt Accessions | P52569 | Gene names | SLC7A2, ATRC2 | |||
|
Domain Architecture |
|
|||||
| Description | Low-affinity cationic amino acid transporter 2 (CAT-2) (CAT2). | |||||
|
CTR2_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 17) | NC score | 0.159939 (rank : 15) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P18581 | Gene names | Slc7a2, Atrc2, Tea | |||
|
Domain Architecture |

|
|||||
| Description | Low-affinity cationic amino acid transporter 2 (CAT-2) (CAT2) (TEA protein) (T-cell early activation protein) (20.5). | |||||
|
AAA1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 18) | NC score | 0.080118 (rank : 21) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P63115, Q9CW25, Q9JMH8 | Gene names | Slc7a10, Asc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Asc-type amino acid transporter 1 (Asc-1) (D-serine transporter). | |||||
|
CTR1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 19) | NC score | 0.143395 (rank : 17) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 21 | |
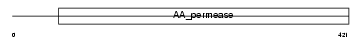
| SwissProt Accessions | Q09143, P30824 | Gene names | Slc7a1, Atrc1, Rec-1 | |||
|
Domain Architecture |
|
|||||
| Description | High-affinity cationic amino acid transporter 1 (CAT-1) (CAT1) (System Y+ basic amino acid transporter) (Ecotropic retroviral leukemia receptor) (ERR) (Ecotropic retrovirus receptor). | |||||
|
ZMAT2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.050051 (rank : 28) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96NC0 | Gene names | ZMAT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger matrin type 2. | |||||
|
ZMAT2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.050051 (rank : 29) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CPW7, Q5EBK0 | Gene names | Zmat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger matrin type 2. | |||||
|
CGRE1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.029656 (rank : 31) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
SNPC5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.038629 (rank : 30) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75971, Q96CF3 | Gene names | SNAPC5, SNAP19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 5 (SNAPc subunit 5) (snRNA- activating protein complex 19 kDa subunit) (SNAPc 19 kDa subunit) (Small nuclear RNA-activating complex polypeptide 5). | |||||
|
CNTP3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.006802 (rank : 42) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BZ76, Q9C0E9 | Gene names | CNTNAP3, CASPR3, KIAA1714 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 3 precursor (Cell recognition molecule Caspr3). | |||||
|
HIRP3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.017007 (rank : 33) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
LAT2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.071332 (rank : 22) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 13 | |
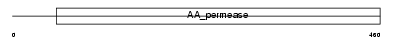
| SwissProt Accessions | Q9QXW9 | Gene names | Slc7a8, Lat2 | |||
|
Domain Architecture |
|
|||||
| Description | Large neutral amino acids transporter small subunit 2 (L-type amino acid transporter 2). | |||||
|
NSMA2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.014748 (rank : 34) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJY3, Q3UTQ5 | Gene names | smpd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sphingomyelin phosphodiesterase 3 (EC 3.1.4.12) (Neutral sphingomyelinase 2) (Neutral sphingomyelinase II) (nSMase2) (nSMase- 2). | |||||
|
PRC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.010782 (rank : 38) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43663, Q9BSB6 | Gene names | PRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein regulator of cytokinesis 1. | |||||
|
SDCG1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.013482 (rank : 35) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CCP0, Q66JX6, Q8C9R6, Q8CA65, Q8JZT9, Q8R072, Q9CW30, Q9CYB8, Q9D4A9 | Gene names | Sdccag1 | |||
|
Domain Architecture |

|
|||||
| Description | Serologically defined colon cancer antigen 1. | |||||
|
TBX18_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.005151 (rank : 45) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
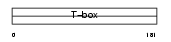
| SwissProt Accessions | O95935, Q7Z6U4, Q9UJI6 | Gene names | TBX18 | |||
|
Domain Architecture |
|
|||||
| Description | T-box transcription factor TBX18 (T-box protein 18). | |||||
|
CLMN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.009584 (rank : 39) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C5W0, Q91V71, Q91XT7, Q91XT8, Q91XU9 | Gene names | Clmn | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
CTR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.108272 (rank : 18) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P30825 | Gene names | SLC7A1, ATRC1, REC1L | |||
|
Domain Architecture |
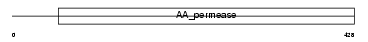
|
|||||
| Description | High-affinity cationic amino acid transporter 1 (CAT-1) (CAT1) (System Y+ basic amino acid transporter) (Ecotropic retroviral leukemia receptor homolog) (ERR) (Ecotropic retrovirus receptor homolog). | |||||
|
SORC3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.006652 (rank : 43) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPU3, Q5VXF9, Q9NQJ2 | Gene names | SORCS3, KIAA1059 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
|
E41L3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.004328 (rank : 48) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
LAT2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.068874 (rank : 23) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UHI5, Q9UKQ6, Q9UKQ7, Q9UKQ8, Q9Y445 | Gene names | SLC7A8, LAT2 | |||
|
Domain Architecture |
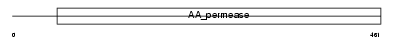
|
|||||
| Description | Large neutral amino acids transporter small subunit 2 (L-type amino acid transporter 2) (hLAT2). | |||||
|
TDRD5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.012036 (rank : 36) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NAT2, Q5VTV0 | Gene names | TDRD5 | |||
|
Domain Architecture |
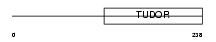
|
|||||
| Description | Tudor domain-containing protein 5. | |||||
|
TRI25_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.002764 (rank : 53) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14258 | Gene names | TRIM25, EFP, RNF147, ZNF147 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 25 (Zinc finger protein 147) (Estrogen-responsive finger protein) (Efp) (RING finger protein 147). | |||||
|
CLD6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.004369 (rank : 47) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56747 | Gene names | CLDN6 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-6 (Skullin 2). | |||||
|
CLD9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.004299 (rank : 49) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95484 | Gene names | CLDN9 | |||
|
Domain Architecture |
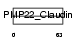
|
|||||
| Description | Claudin-9. | |||||
|
DOCK6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.004963 (rank : 46) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96HP0, Q9P2F2 | Gene names | DOCK6, KIAA1395 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 6. | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.011382 (rank : 37) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
NIBA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.008728 (rank : 41) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZQ8, Q5TEM8, Q8TEI5, Q9H593, Q9H9Y8, Q9HCB9 | Gene names | C1orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban protein. | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.006323 (rank : 44) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.004255 (rank : 51) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.002053 (rank : 57) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.002757 (rank : 54) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.003158 (rank : 52) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
TP53B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.009295 (rank : 40) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
FOXE2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.002086 (rank : 56) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99526 | Gene names | FOXE2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E2 (HNF-3/fork head-like protein 5) (HFKL5) (HFKH4). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.004293 (rank : 50) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
OLF15_MOUSE
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | -0.000742 (rank : 60) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 752 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23275 | Gene names | Olfr15 | |||
|
Domain Architecture |
|
|||||
| Description | Olfactory receptor 15 (Odorant receptor OR3). | |||||
|
PRP4B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | -0.002176 (rank : 61) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61136, O88378, Q8BND8, Q8R4Y5, Q9CTL9, Q9CTT0 | Gene names | Prpf4b, Cbp143, Prp4h, Prp4k, Prp4m, Prpk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (Pre-mRNA protein kinase). | |||||
|
TPM3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.001638 (rank : 59) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P06753, P12324, Q969Q2, Q9NQH8 | Gene names | TPM3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma) (hTM5). | |||||
|
TPM3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.001665 (rank : 58) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P21107, Q09021, Q60606, Q80SW6, Q9EPW3 | Gene names | Tpm3, Tpm-5, Tpm5 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma). | |||||
|
TS1R2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.002577 (rank : 55) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q925I4, Q923J8 | Gene names | Tas1r2, Gpr71, T1r2, Tr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 2 precursor (G-protein coupled receptor 71) (Sweet taste receptor T1R2). | |||||
|
BAT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.053909 (rank : 26) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P82251 | Gene names | SLC7A9, BAT1 | |||
|
Domain Architecture |
|
|||||
| Description | B(0,+)-type amino acid transporter 1 (B(0,+)AT) (Glycoprotein- associated amino acid transporter b0,+AT1). | |||||
|
BAT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.064352 (rank : 24) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QXA6 | Gene names | Slc7a9, Bat1 | |||
|
Domain Architecture |
|
|||||
| Description | B(0,+)-type amino acid transporter 1 (B(0,+)AT) (Glycoprotein- associated amino acid transporter b0,+AT1). | |||||
|
CTR3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.080280 (rank : 20) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WY07, Q8N185, Q8NCA7, Q96SZ7 | Gene names | SLC7A3, ATRC3, CAT3 | |||
|
Domain Architecture |
|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
CTR3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.098250 (rank : 19) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70423 | Gene names | Slc7a3, Atrc3, Cat3 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
CTR4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.056368 (rank : 25) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43246, Q3KNQ6, Q4VC45, Q6IBY8, Q96H88 | Gene names | SLC7A4 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 4 (CAT-4) (CAT4). | |||||
|
XCT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.052031 (rank : 27) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9WTR6 | Gene names | Slc7a11 | |||
|
Domain Architecture |
|
|||||
| Description | Cystine/glutamate transporter (Amino acid transport system xc-) (xCT). | |||||
|
S12A2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P55012 | Gene names | Slc12a2, Nkcc1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
|
S12A2_HUMAN
|
||||||
| NC score | 0.996600 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P55011, Q8N713, Q8WWH7 | Gene names | SLC12A2, NKCC1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
|
S12A1_MOUSE
|
||||||
| NC score | 0.993195 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P55014, O35938, Q91W55, Q9Z1E0 | Gene names | Slc12a1, Nkcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 1 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 2) (BSC1) (Kidney-specific Na-K-Cl symporter). | |||||
|
S12A1_HUMAN
|
||||||
| NC score | 0.993039 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13621 | Gene names | SLC12A1, NKCC2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 1 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 2) (Kidney-specific Na-K-Cl symporter). | |||||
|
S12A3_HUMAN
|
||||||
| NC score | 0.988253 (rank : 5) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P55017 | Gene names | SLC12A3, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 3 (Thiazide-sensitive sodium-chloride cotransporter) (Na-Cl symporter). | |||||
|
S12A3_MOUSE
|
||||||
| NC score | 0.985066 (rank : 6) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P59158 | Gene names | Slc12a3, Tsc | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 3 (Thiazide-sensitive sodium-chloride cotransporter) (Na-Cl symporter). | |||||
|
S12A7_HUMAN
|
||||||
| NC score | 0.818899 (rank : 7) | θ value | 3.59435e-57 (rank : 8) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y666, Q96I81, Q9H7I3, Q9H7I7, Q9UFW2 | Gene names | SLC12A7, KCC4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
S12A7_MOUSE
|
||||||
| NC score | 0.816293 (rank : 8) | θ value | 6.55044e-59 (rank : 7) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WVL3 | Gene names | Slc12a7, Kcc4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
S12A4_MOUSE
|
||||||
| NC score | 0.813453 (rank : 9) | θ value | 2.32979e-56 (rank : 10) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JIS8, O55069, Q9ET57 | Gene names | Slc12a4, Kcc1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 4 (Electroneutral potassium-chloride cotransporter 1) (Erythroid K-Cl cotransporter 1) (mKCC1). | |||||
|
S12A4_HUMAN
|
||||||
| NC score | 0.812607 (rank : 10) | θ value | 2.32979e-56 (rank : 9) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UP95, O60632, O75893, Q13953, Q96LD5 | Gene names | SLC12A4, KCC1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 4 (Electroneutral potassium-chloride cotransporter 1) (Erythroid K-Cl cotransporter 1) (hKCC1). | |||||
|
S12A5_HUMAN
|
||||||
| NC score | 0.812458 (rank : 11) | θ value | 1.2784e-54 (rank : 11) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H2X9, Q5VZ41, Q9H4Z0, Q9ULP4 | Gene names | SLC12A5, KCC2, KIAA1176 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (Erythroid K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (hKCC2). | |||||
|
S12A5_MOUSE
|
||||||
| NC score | 0.812341 (rank : 12) | θ value | 2.18062e-54 (rank : 12) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91V14, Q9Z0M7 | Gene names | Slc12a5, Kcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (mKCC2). | |||||
|
S12A6_HUMAN
|
||||||
| NC score | 0.809632 (rank : 13) | θ value | 2.84797e-54 (rank : 13) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UHW9, Q8TDD4, Q9UFR2, Q9Y642, Q9Y665 | Gene names | SLC12A6, KCC3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
S12A6_MOUSE
|
||||||
| NC score | 0.808798 (rank : 14) | θ value | 1.41344e-53 (rank : 14) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q924N4, Q924N3 | Gene names | Slc12a6, Kcc3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
CTR2_MOUSE
|
||||||
| NC score | 0.159939 (rank : 15) | θ value | 0.0193708 (rank : 17) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P18581 | Gene names | Slc7a2, Atrc2, Tea | |||
|
Domain Architecture |

|
|||||
| Description | Low-affinity cationic amino acid transporter 2 (CAT-2) (CAT2) (TEA protein) (T-cell early activation protein) (20.5). | |||||
|
CTR2_HUMAN
|
||||||
| NC score | 0.145330 (rank : 16) | θ value | 0.0193708 (rank : 16) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P52569 | Gene names | SLC7A2, ATRC2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-affinity cationic amino acid transporter 2 (CAT-2) (CAT2). | |||||
|
CTR1_MOUSE
|
||||||
| NC score | 0.143395 (rank : 17) | θ value | 0.0563607 (rank : 19) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q09143, P30824 | Gene names | Slc7a1, Atrc1, Rec-1 | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cationic amino acid transporter 1 (CAT-1) (CAT1) (System Y+ basic amino acid transporter) (Ecotropic retroviral leukemia receptor) (ERR) (Ecotropic retrovirus receptor). | |||||
|
CTR1_HUMAN
|
||||||
| NC score | 0.108272 (rank : 18) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P30825 | Gene names | SLC7A1, ATRC1, REC1L | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cationic amino acid transporter 1 (CAT-1) (CAT1) (System Y+ basic amino acid transporter) (Ecotropic retroviral leukemia receptor homolog) (ERR) (Ecotropic retrovirus receptor homolog). | |||||
|
CTR3_MOUSE
|
||||||
| NC score | 0.098250 (rank : 19) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70423 | Gene names | Slc7a3, Atrc3, Cat3 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
CTR3_HUMAN
|
||||||
| NC score | 0.080280 (rank : 20) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WY07, Q8N185, Q8NCA7, Q96SZ7 | Gene names | SLC7A3, ATRC3, CAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
AAA1_MOUSE
|
||||||
| NC score | 0.080118 (rank : 21) | θ value | 0.0252991 (rank : 18) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P63115, Q9CW25, Q9JMH8 | Gene names | Slc7a10, Asc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Asc-type amino acid transporter 1 (Asc-1) (D-serine transporter). | |||||
|
LAT2_MOUSE
|
||||||
| NC score | 0.071332 (rank : 22) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QXW9 | Gene names | Slc7a8, Lat2 | |||
|
Domain Architecture |

|
|||||
| Description | Large neutral amino acids transporter small subunit 2 (L-type amino acid transporter 2). | |||||
|
LAT2_HUMAN
|
||||||
| NC score | 0.068874 (rank : 23) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UHI5, Q9UKQ6, Q9UKQ7, Q9UKQ8, Q9Y445 | Gene names | SLC7A8, LAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Large neutral amino acids transporter small subunit 2 (L-type amino acid transporter 2) (hLAT2). | |||||
|
BAT1_MOUSE
|
||||||
| NC score | 0.064352 (rank : 24) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QXA6 | Gene names | Slc7a9, Bat1 | |||
|
Domain Architecture |

|
|||||
| Description | B(0,+)-type amino acid transporter 1 (B(0,+)AT) (Glycoprotein- associated amino acid transporter b0,+AT1). | |||||
|
CTR4_HUMAN
|
||||||
| NC score | 0.056368 (rank : 25) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43246, Q3KNQ6, Q4VC45, Q6IBY8, Q96H88 | Gene names | SLC7A4 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 4 (CAT-4) (CAT4). | |||||
|
BAT1_HUMAN
|
||||||
| NC score | 0.053909 (rank : 26) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P82251 | Gene names | SLC7A9, BAT1 | |||
|
Domain Architecture |

|
|||||
| Description | B(0,+)-type amino acid transporter 1 (B(0,+)AT) (Glycoprotein- associated amino acid transporter b0,+AT1). | |||||
|
XCT_MOUSE
|
||||||
| NC score | 0.052031 (rank : 27) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9WTR6 | Gene names | Slc7a11 | |||
|
Domain Architecture |

|
|||||
| Description | Cystine/glutamate transporter (Amino acid transport system xc-) (xCT). | |||||
|
ZMAT2_HUMAN
|
||||||
| NC score | 0.050051 (rank : 28) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96NC0 | Gene names | ZMAT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger matrin type 2. | |||||
|
ZMAT2_MOUSE
|
||||||
| NC score | 0.050051 (rank : 29) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CPW7, Q5EBK0 | Gene names | Zmat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger matrin type 2. | |||||
|
SNPC5_HUMAN
|
||||||
| NC score | 0.038629 (rank : 30) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75971, Q96CF3 | Gene names | SNAPC5, SNAP19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 5 (SNAPc subunit 5) (snRNA- activating protein complex 19 kDa subunit) (SNAPc 19 kDa subunit) (Small nuclear RNA-activating complex polypeptide 5). | |||||
|
CGRE1_HUMAN
|
||||||
| NC score | 0.029656 (rank : 31) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.024450 (rank : 32) | θ value | 0.0113563 (rank : 15) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
HIRP3_HUMAN
|
||||||
| NC score | 0.017007 (rank : 33) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
NSMA2_MOUSE
|
||||||
| NC score | 0.014748 (rank : 34) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJY3, Q3UTQ5 | Gene names | smpd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sphingomyelin phosphodiesterase 3 (EC 3.1.4.12) (Neutral sphingomyelinase 2) (Neutral sphingomyelinase II) (nSMase2) (nSMase- 2). | |||||
|
SDCG1_MOUSE
|
||||||
| NC score | 0.013482 (rank : 35) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CCP0, Q66JX6, Q8C9R6, Q8CA65, Q8JZT9, Q8R072, Q9CW30, Q9CYB8, Q9D4A9 | Gene names | Sdccag1 | |||
|
Domain Architecture |

|
|||||
| Description | Serologically defined colon cancer antigen 1. | |||||
|
TDRD5_HUMAN
|
||||||
| NC score | 0.012036 (rank : 36) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NAT2, Q5VTV0 | Gene names | TDRD5 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 5. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.011382 (rank : 37) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
PRC1_HUMAN
|
||||||
| NC score | 0.010782 (rank : 38) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43663, Q9BSB6 | Gene names | PRC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein regulator of cytokinesis 1. | |||||
|
CLMN_MOUSE
|
||||||
| NC score | 0.009584 (rank : 39) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C5W0, Q91V71, Q91XT7, Q91XT8, Q91XU9 | Gene names | Clmn | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.009295 (rank : 40) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
NIBA_HUMAN
|
||||||
| NC score | 0.008728 (rank : 41) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZQ8, Q5TEM8, Q8TEI5, Q9H593, Q9H9Y8, Q9HCB9 | Gene names | C1orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban protein. | |||||
|
CNTP3_HUMAN
|
||||||
| NC score | 0.006802 (rank : 42) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BZ76, Q9C0E9 | Gene names | CNTNAP3, CASPR3, KIAA1714 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 3 precursor (Cell recognition molecule Caspr3). | |||||
|
SORC3_HUMAN
|
||||||
| NC score | 0.006652 (rank : 43) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPU3, Q5VXF9, Q9NQJ2 | Gene names | SORCS3, KIAA1059 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.006323 (rank : 44) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
TBX18_HUMAN
|
||||||
| NC score | 0.005151 (rank : 45) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95935, Q7Z6U4, Q9UJI6 | Gene names | TBX18 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX18 (T-box protein 18). | |||||
|
DOCK6_HUMAN
|
||||||
| NC score | 0.004963 (rank : 46) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96HP0, Q9P2F2 | Gene names | DOCK6, KIAA1395 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 6. | |||||
|
CLD6_HUMAN
|
||||||
| NC score | 0.004369 (rank : 47) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56747 | Gene names | CLDN6 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-6 (Skullin 2). | |||||
|
E41L3_MOUSE
|
||||||
| NC score | 0.004328 (rank : 48) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
CLD9_HUMAN
|
||||||
| NC score | 0.004299 (rank : 49) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95484 | Gene names | CLDN9 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-9. | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.004293 (rank : 50) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.004255 (rank : 51) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.003158 (rank : 52) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
TRI25_HUMAN
|
||||||
| NC score | 0.002764 (rank : 53) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14258 | Gene names | TRIM25, EFP, RNF147, ZNF147 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 25 (Zinc finger protein 147) (Estrogen-responsive finger protein) (Efp) (RING finger protein 147). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.002757 (rank : 54) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
TS1R2_MOUSE
|
||||||
| NC score | 0.002577 (rank : 55) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q925I4, Q923J8 | Gene names | Tas1r2, Gpr71, T1r2, Tr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Taste receptor type 1 member 2 precursor (G-protein coupled receptor 71) (Sweet taste receptor T1R2). | |||||
|
FOXE2_HUMAN
|
||||||
| NC score | 0.002086 (rank : 56) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99526 | Gene names | FOXE2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E2 (HNF-3/fork head-like protein 5) (HFKL5) (HFKH4). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.002053 (rank : 57) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
TPM3_MOUSE
|
||||||
| NC score | 0.001665 (rank : 58) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P21107, Q09021, Q60606, Q80SW6, Q9EPW3 | Gene names | Tpm3, Tpm-5, Tpm5 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma). | |||||
|
TPM3_HUMAN
|
||||||
| NC score | 0.001638 (rank : 59) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P06753, P12324, Q969Q2, Q9NQH8 | Gene names | TPM3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma) (hTM5). | |||||
|
OLF15_MOUSE
|
||||||
| NC score | -0.000742 (rank : 60) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 752 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23275 | Gene names | Olfr15 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 15 (Odorant receptor OR3). | |||||
|
PRP4B_MOUSE
|
||||||
| NC score | -0.002176 (rank : 61) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61136, O88378, Q8BND8, Q8R4Y5, Q9CTL9, Q9CTT0 | Gene names | Prpf4b, Cbp143, Prp4h, Prp4k, Prp4m, Prpk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (Pre-mRNA protein kinase). | |||||