Please be patient as the page loads
|
S12A5_HUMAN
|
||||||
| SwissProt Accessions | Q9H2X9, Q5VZ41, Q9H4Z0, Q9ULP4 | Gene names | SLC12A5, KCC2, KIAA1176 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (Erythroid K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (hKCC2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
S12A4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994714 (rank : 6) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UP95, O60632, O75893, Q13953, Q96LD5 | Gene names | SLC12A4, KCC1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 4 (Electroneutral potassium-chloride cotransporter 1) (Erythroid K-Cl cotransporter 1) (hKCC1). | |||||
|
S12A4_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.994771 (rank : 5) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JIS8, O55069, Q9ET57 | Gene names | Slc12a4, Kcc1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 4 (Electroneutral potassium-chloride cotransporter 1) (Erythroid K-Cl cotransporter 1) (mKCC1). | |||||
|
S12A5_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9H2X9, Q5VZ41, Q9H4Z0, Q9ULP4 | Gene names | SLC12A5, KCC2, KIAA1176 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (Erythroid K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (hKCC2). | |||||
|
S12A5_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.998885 (rank : 2) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q91V14, Q9Z0M7 | Gene names | Slc12a5, Kcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (mKCC2). | |||||
|
S12A6_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.993246 (rank : 7) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UHW9, Q8TDD4, Q9UFR2, Q9Y642, Q9Y665 | Gene names | SLC12A6, KCC3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
S12A6_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.993067 (rank : 8) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q924N4, Q924N3 | Gene names | Slc12a6, Kcc3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
S12A7_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.996581 (rank : 3) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y666, Q96I81, Q9H7I3, Q9H7I7, Q9UFW2 | Gene names | SLC12A7, KCC4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
S12A7_MOUSE
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.996431 (rank : 4) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9WVL3 | Gene names | Slc12a7, Kcc4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
S12A3_MOUSE
|
||||||
| θ value | 4.22625e-74 (rank : 9) | NC score | 0.838752 (rank : 9) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P59158 | Gene names | Slc12a3, Tsc | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 3 (Thiazide-sensitive sodium-chloride cotransporter) (Na-Cl symporter). | |||||
|
S12A3_HUMAN
|
||||||
| θ value | 1.36585e-56 (rank : 10) | NC score | 0.820717 (rank : 10) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P55017 | Gene names | SLC12A3, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 3 (Thiazide-sensitive sodium-chloride cotransporter) (Na-Cl symporter). | |||||
|
S12A2_HUMAN
|
||||||
| θ value | 5.73848e-55 (rank : 11) | NC score | 0.811584 (rank : 12) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P55011, Q8N713, Q8WWH7 | Gene names | SLC12A2, NKCC1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
|
S12A2_MOUSE
|
||||||
| θ value | 1.2784e-54 (rank : 12) | NC score | 0.812458 (rank : 11) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P55012 | Gene names | Slc12a2, Nkcc1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
|
S12A1_MOUSE
|
||||||
| θ value | 2.041e-52 (rank : 13) | NC score | 0.804343 (rank : 14) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P55014, O35938, Q91W55, Q9Z1E0 | Gene names | Slc12a1, Nkcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 1 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 2) (BSC1) (Kidney-specific Na-K-Cl symporter). | |||||
|
S12A1_HUMAN
|
||||||
| θ value | 3.48141e-52 (rank : 14) | NC score | 0.804567 (rank : 13) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13621 | Gene names | SLC12A1, NKCC2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 1 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 2) (Kidney-specific Na-K-Cl symporter). | |||||
|
LAT2_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 15) | NC score | 0.122608 (rank : 22) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UHI5, Q9UKQ6, Q9UKQ7, Q9UKQ8, Q9Y445 | Gene names | SLC7A8, LAT2 | |||
|
Domain Architecture |
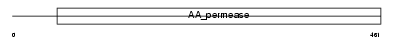
|
|||||
| Description | Large neutral amino acids transporter small subunit 2 (L-type amino acid transporter 2) (hLAT2). | |||||
|
LAT2_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 16) | NC score | 0.122407 (rank : 23) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QXW9 | Gene names | Slc7a8, Lat2 | |||
|
Domain Architecture |
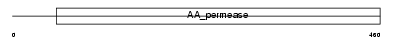
|
|||||
| Description | Large neutral amino acids transporter small subunit 2 (L-type amino acid transporter 2). | |||||
|
BAT1_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 17) | NC score | 0.133506 (rank : 21) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QXA6 | Gene names | Slc7a9, Bat1 | |||
|
Domain Architecture |
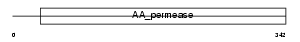
|
|||||
| Description | B(0,+)-type amino acid transporter 1 (B(0,+)AT) (Glycoprotein- associated amino acid transporter b0,+AT1). | |||||
|
CTR1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 18) | NC score | 0.175881 (rank : 17) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q09143, P30824 | Gene names | Slc7a1, Atrc1, Rec-1 | |||
|
Domain Architecture |
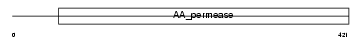
|
|||||
| Description | High-affinity cationic amino acid transporter 1 (CAT-1) (CAT1) (System Y+ basic amino acid transporter) (Ecotropic retroviral leukemia receptor) (ERR) (Ecotropic retrovirus receptor). | |||||
|
CTR2_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 19) | NC score | 0.209022 (rank : 15) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P18581 | Gene names | Slc7a2, Atrc2, Tea | |||
|
Domain Architecture |

|
|||||
| Description | Low-affinity cationic amino acid transporter 2 (CAT-2) (CAT2) (TEA protein) (T-cell early activation protein) (20.5). | |||||
|
BAT1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 20) | NC score | 0.115754 (rank : 24) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P82251 | Gene names | SLC7A9, BAT1 | |||
|
Domain Architecture |
|
|||||
| Description | B(0,+)-type amino acid transporter 1 (B(0,+)AT) (Glycoprotein- associated amino acid transporter b0,+AT1). | |||||
|
CTR1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 21) | NC score | 0.158728 (rank : 18) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30825 | Gene names | SLC7A1, ATRC1, REC1L | |||
|
Domain Architecture |
|
|||||
| Description | High-affinity cationic amino acid transporter 1 (CAT-1) (CAT1) (System Y+ basic amino acid transporter) (Ecotropic retroviral leukemia receptor homolog) (ERR) (Ecotropic retrovirus receptor homolog). | |||||
|
CTR3_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 22) | NC score | 0.144789 (rank : 19) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WY07, Q8N185, Q8NCA7, Q96SZ7 | Gene names | SLC7A3, ATRC3, CAT3 | |||
|
Domain Architecture |
|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 23) | NC score | 0.021370 (rank : 36) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
CTR2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 24) | NC score | 0.185171 (rank : 16) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52569 | Gene names | SLC7A2, ATRC2 | |||
|
Domain Architecture |
|
|||||
| Description | Low-affinity cationic amino acid transporter 2 (CAT-2) (CAT2). | |||||
|
CTR4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.102716 (rank : 25) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43246, Q3KNQ6, Q4VC45, Q6IBY8, Q96H88 | Gene names | SLC7A4 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 4 (CAT-4) (CAT4). | |||||
|
CTR3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.139303 (rank : 20) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70423 | Gene names | Slc7a3, Atrc3, Cat3 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
YLA1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.070352 (rank : 28) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UM01, O95984, Q9P2V5 | Gene names | SLC7A7 | |||
|
Domain Architecture |
|
|||||
| Description | Y+L amino acid transporter 1 (y(+)L-type amino acid transporter 1) (y+LAT-1) (Y+LAT1) (Monocyte amino acid permease 2) (MOP-2). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.001393 (rank : 66) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
RTN1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.021496 (rank : 35) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16799, Q16800, Q16801 | Gene names | RTN1, NSP | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
O1052_MOUSE
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.000746 (rank : 67) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 684 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VGR8 | Gene names | Olfr1052, Mor172-1 | |||
|
Domain Architecture |
|
|||||
| Description | Olfactory receptor 1052 (Olfactory receptor 172-1). | |||||
|
SYM_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.022654 (rank : 34) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56192, Q14895, Q53H14, Q96A15, Q96BZ0, Q9NSE0 | Gene names | MARS | |||
|
Domain Architecture |

|
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
EVL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.014658 (rank : 39) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UI08, O95884, Q7Z522, Q8TBV1, Q9UF25, Q9UIC2 | Gene names | EVL, RNB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
CHIT1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.007422 (rank : 49) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D7Q1, Q6QJD2 | Gene names | Chit1 | |||
|
Domain Architecture |
|
|||||
| Description | Chitotriosidase-1 precursor (EC 3.2.1.14) (Chitinase-1). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.002027 (rank : 63) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
ADDG_MOUSE
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.010390 (rank : 41) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QYB5, Q9D2M5, Q9QYB6 | Gene names | Add3, Addl | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-adducin (Adducin-like protein 70). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.001787 (rank : 64) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.020222 (rank : 37) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
OR5J2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | -0.000458 (rank : 68) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 649 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NH18, Q6IEU5 | Gene names | OR5J2 | |||
|
Domain Architecture |
|
|||||
| Description | Olfactory receptor 5J2 (Olfactory receptor OR11-266). | |||||
|
ROM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.016537 (rank : 38) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03395 | Gene names | ROM1, TSPAN23 | |||
|
Domain Architecture |
|
|||||
| Description | Rod outer segment membrane protein 1 (ROSP1) (Tetraspanin-23) (Tspan- 23). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.001508 (rank : 65) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.006810 (rank : 51) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
CENPC_MOUSE
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.008781 (rank : 43) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49452 | Gene names | Cenpc1, Cenpc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C). | |||||
|
CTL4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.008469 (rank : 44) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91VA1, Q7TQ02, Q9CVA7 | Gene names | Slc44a4, Ctl4, Ng22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Choline transporter-like protein 4 (Solute carrier family 44 member 4). | |||||
|
EDG1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.005425 (rank : 55) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O08530, Q9R235 | Gene names | Edg1, Lpb1 | |||
|
Domain Architecture |
|
|||||
| Description | Sphingosine 1-phosphate receptor Edg-1 (Sphingosine 1-phosphate receptor 1) (S1P1) (Lysophospholipid receptor B1). | |||||
|
EVL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.011437 (rank : 40) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
G137B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.051648 (rank : 33) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60478 | Gene names | GPR137B, TM7SF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integral membrane protein GPR137B (Transmembrane 7 superfamily member 1). | |||||
|
TPM4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.002942 (rank : 60) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 703 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6IRU2 | Gene names | Tpm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4). | |||||
|
ARI4A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.006241 (rank : 53) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
EPHA5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | -0.001916 (rank : 73) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54756 | Gene names | EPHA5, EHK1, HEK7 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 5 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EHK-1) (EPH homology kinase 1) (Receptor protein- tyrosine kinase HEK7). | |||||
|
EPHA5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | -0.001881 (rank : 72) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60629 | Gene names | Epha5, Bsk, Cek7, Ehk1 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 5 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EHK-1) (EPH homology kinase 1) (Brain-specific kinase) (CEK-7). | |||||
|
EYA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.004047 (rank : 57) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99502 | Gene names | EYA1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.006120 (rank : 54) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
GPR84_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.008138 (rank : 45) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NQS5 | Gene names | GPR84, EX33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 84 (Inflammation-related G-protein coupled receptor EX33). | |||||
|
IPR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.007671 (rank : 48) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVK9, Q3UCV7, Q80V00 | Gene names | Ipr1, Ifi75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intracellular pathogen resistance protein 1. | |||||
|
LPHN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.003479 (rank : 58) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94910, Q96IE7, Q9BU07, Q9HAR3 | Gene names | LPHN1, KIAA0821, LEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-1 precursor (Calcium-independent alpha-latrotoxin receptor 1) (Lectomedin-2). | |||||
|
PP4R1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.006415 (rank : 52) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TF05, Q99774, Q9UNQ7 | Gene names | PPP4R1, MEG1, PP4R1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 regulatory subunit 1. | |||||
|
ZN403_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.007934 (rank : 46) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H3C7, Q96T90, Q9GZR8, Q9H767 | Gene names | ZNF403, LCRG1, LZK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF403 (Laryngeal carcinoma-related protein 1). | |||||
|
ACOD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.009450 (rank : 42) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00767, Q16150, Q53GR9, Q5W037, Q5W038, Q6GSS4, Q96KF6, Q9BS07, Q9Y695 | Gene names | SCD | |||
|
Domain Architecture |
|
|||||
| Description | Acyl-CoA desaturase (EC 1.14.19.1) (Stearoyl-CoA desaturase) (Fatty acid desaturase) (Delta(9)-desaturase). | |||||
|
GPR97_MOUSE
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.003001 (rank : 59) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R0T6 | Gene names | Gpr97 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 97 precursor. | |||||
|
HYALP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.006917 (rank : 50) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48794, Q7TSD6, Q812F4, Q9DAQ1 | Gene names | Spam1, Ph20, Spam | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronidase PH-20 precursor (EC 3.2.1.35) (Hyal-PH20) (Sperm surface protein PH-20) (Sperm adhesion molecule 1). | |||||
|
MASP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | -0.000761 (rank : 69) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91WP0, Q9QXA4, Q9QXD2, Q9QXD5, Q9Z338 | Gene names | Masp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.002242 (rank : 62) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
OR5K1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | -0.001403 (rank : 71) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 782 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NHB7 | Gene names | OR5K1 | |||
|
Domain Architecture |
|
|||||
| Description | Olfactory receptor 5K1 (HTPCRX10). | |||||
|
PEG3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | -0.002251 (rank : 74) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | -0.001255 (rank : 70) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SYF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.007800 (rank : 47) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95926, Q5TH73 | Gene names | SYF2, CBPIN, GCIPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor SYF2 (CCNDBP1-interactor) (p29). | |||||
|
TPM4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.002454 (rank : 61) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P67936, P07226, Q15659, Q9BU85, Q9H8Q3 | Gene names | TPM4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4) (TM30p1). | |||||
|
V2R1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.004494 (rank : 56) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6TAC4, O70409 | Gene names | V2r1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vomeronasal type-2 receptor 1b precursor (Putative pheromone receptor V2R1b). | |||||
|
AAA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.089411 (rank : 27) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NS82 | Gene names | SLC7A10, ASC1 | |||
|
Domain Architecture |
|
|||||
| Description | Asc-type amino acid transporter 1 (Asc-1). | |||||
|
AAA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.096175 (rank : 26) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P63115, Q9CW25, Q9JMH8 | Gene names | Slc7a10, Asc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Asc-type amino acid transporter 1 (Asc-1) (D-serine transporter). | |||||
|
LAT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.069642 (rank : 29) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q01650, Q9UBN8, Q9UP15, Q9UQC0 | Gene names | SLC7A5, CD98LC, LAT1, MPE16 | |||
|
Domain Architecture |

|
|||||
| Description | Large neutral amino acids transporter small subunit 1 (L-type amino acid transporter 1) (4F2 light chain) (4F2 LC) (4F2LC) (CD98 light chain) (Integral membrane protein E16) (hLAT1). | |||||
|
LAT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.065983 (rank : 31) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z127, Q9JMI4 | Gene names | Slc7a5, Lat1 | |||
|
Domain Architecture |
|
|||||
| Description | Large neutral amino acids transporter small subunit 1 (L-type amino acid transporter 1) (4F2 light chain) (4F2 LC) (4F2LC). | |||||
|
XCT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.062895 (rank : 32) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UPY5 | Gene names | SLC7A11 | |||
|
Domain Architecture |
|
|||||
| Description | Cystine/glutamate transporter (Amino acid transport system xc-) (xCT) (Calcium channel blocker resistance protein CCBR1). | |||||
|
XCT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.068962 (rank : 30) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WTR6 | Gene names | Slc7a11 | |||
|
Domain Architecture |
|
|||||
| Description | Cystine/glutamate transporter (Amino acid transport system xc-) (xCT). | |||||
|
S12A5_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9H2X9, Q5VZ41, Q9H4Z0, Q9ULP4 | Gene names | SLC12A5, KCC2, KIAA1176 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (Erythroid K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (hKCC2). | |||||
|
S12A5_MOUSE
|
||||||
| NC score | 0.998885 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q91V14, Q9Z0M7 | Gene names | Slc12a5, Kcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (mKCC2). | |||||
|
S12A7_HUMAN
|
||||||
| NC score | 0.996581 (rank : 3) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y666, Q96I81, Q9H7I3, Q9H7I7, Q9UFW2 | Gene names | SLC12A7, KCC4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
S12A7_MOUSE
|
||||||
| NC score | 0.996431 (rank : 4) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9WVL3 | Gene names | Slc12a7, Kcc4 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 7 (Electroneutral potassium-chloride cotransporter 4) (K-Cl cotransporter 4). | |||||
|
S12A4_MOUSE
|
||||||
| NC score | 0.994771 (rank : 5) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JIS8, O55069, Q9ET57 | Gene names | Slc12a4, Kcc1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 4 (Electroneutral potassium-chloride cotransporter 1) (Erythroid K-Cl cotransporter 1) (mKCC1). | |||||
|
S12A4_HUMAN
|
||||||
| NC score | 0.994714 (rank : 6) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UP95, O60632, O75893, Q13953, Q96LD5 | Gene names | SLC12A4, KCC1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 4 (Electroneutral potassium-chloride cotransporter 1) (Erythroid K-Cl cotransporter 1) (hKCC1). | |||||
|
S12A6_HUMAN
|
||||||
| NC score | 0.993246 (rank : 7) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UHW9, Q8TDD4, Q9UFR2, Q9Y642, Q9Y665 | Gene names | SLC12A6, KCC3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
S12A6_MOUSE
|
||||||
| NC score | 0.993067 (rank : 8) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q924N4, Q924N3 | Gene names | Slc12a6, Kcc3 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 6 (Electroneutral potassium-chloride cotransporter 3) (K-Cl cotransporter 3). | |||||
|
S12A3_MOUSE
|
||||||
| NC score | 0.838752 (rank : 9) | θ value | 4.22625e-74 (rank : 9) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P59158 | Gene names | Slc12a3, Tsc | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 3 (Thiazide-sensitive sodium-chloride cotransporter) (Na-Cl symporter). | |||||
|
S12A3_HUMAN
|
||||||
| NC score | 0.820717 (rank : 10) | θ value | 1.36585e-56 (rank : 10) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P55017 | Gene names | SLC12A3, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 3 (Thiazide-sensitive sodium-chloride cotransporter) (Na-Cl symporter). | |||||
|
S12A2_MOUSE
|
||||||
| NC score | 0.812458 (rank : 11) | θ value | 1.2784e-54 (rank : 12) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P55012 | Gene names | Slc12a2, Nkcc1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
|
S12A2_HUMAN
|
||||||
| NC score | 0.811584 (rank : 12) | θ value | 5.73848e-55 (rank : 11) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P55011, Q8N713, Q8WWH7 | Gene names | SLC12A2, NKCC1 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 2 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 1) (Basolateral Na-K-Cl symporter). | |||||
|
S12A1_HUMAN
|
||||||
| NC score | 0.804567 (rank : 13) | θ value | 3.48141e-52 (rank : 14) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13621 | Gene names | SLC12A1, NKCC2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 1 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 2) (Kidney-specific Na-K-Cl symporter). | |||||
|
S12A1_MOUSE
|
||||||
| NC score | 0.804343 (rank : 14) | θ value | 2.041e-52 (rank : 13) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P55014, O35938, Q91W55, Q9Z1E0 | Gene names | Slc12a1, Nkcc2 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 1 (Bumetanide-sensitive sodium- (potassium)-chloride cotransporter 2) (BSC1) (Kidney-specific Na-K-Cl symporter). | |||||
|
CTR2_MOUSE
|
||||||
| NC score | 0.209022 (rank : 15) | θ value | 0.0113563 (rank : 19) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P18581 | Gene names | Slc7a2, Atrc2, Tea | |||
|
Domain Architecture |

|
|||||
| Description | Low-affinity cationic amino acid transporter 2 (CAT-2) (CAT2) (TEA protein) (T-cell early activation protein) (20.5). | |||||
|
CTR2_HUMAN
|
||||||
| NC score | 0.185171 (rank : 16) | θ value | 0.21417 (rank : 24) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52569 | Gene names | SLC7A2, ATRC2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-affinity cationic amino acid transporter 2 (CAT-2) (CAT2). | |||||
|
CTR1_MOUSE
|
||||||
| NC score | 0.175881 (rank : 17) | θ value | 0.00869519 (rank : 18) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q09143, P30824 | Gene names | Slc7a1, Atrc1, Rec-1 | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cationic amino acid transporter 1 (CAT-1) (CAT1) (System Y+ basic amino acid transporter) (Ecotropic retroviral leukemia receptor) (ERR) (Ecotropic retrovirus receptor). | |||||
|
CTR1_HUMAN
|
||||||
| NC score | 0.158728 (rank : 18) | θ value | 0.0736092 (rank : 21) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30825 | Gene names | SLC7A1, ATRC1, REC1L | |||
|
Domain Architecture |

|
|||||
| Description | High-affinity cationic amino acid transporter 1 (CAT-1) (CAT1) (System Y+ basic amino acid transporter) (Ecotropic retroviral leukemia receptor homolog) (ERR) (Ecotropic retrovirus receptor homolog). | |||||
|
CTR3_HUMAN
|
||||||
| NC score | 0.144789 (rank : 19) | θ value | 0.0736092 (rank : 22) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WY07, Q8N185, Q8NCA7, Q96SZ7 | Gene names | SLC7A3, ATRC3, CAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
CTR3_MOUSE
|
||||||
| NC score | 0.139303 (rank : 20) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70423 | Gene names | Slc7a3, Atrc3, Cat3 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 3 (CAT-3) (Solute carrier family 7 member 3) (Cationic amino acid transporter y+). | |||||
|
BAT1_MOUSE
|
||||||
| NC score | 0.133506 (rank : 21) | θ value | 0.00390308 (rank : 17) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QXA6 | Gene names | Slc7a9, Bat1 | |||
|
Domain Architecture |

|
|||||
| Description | B(0,+)-type amino acid transporter 1 (B(0,+)AT) (Glycoprotein- associated amino acid transporter b0,+AT1). | |||||
|
LAT2_HUMAN
|
||||||
| NC score | 0.122608 (rank : 22) | θ value | 0.000270298 (rank : 15) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UHI5, Q9UKQ6, Q9UKQ7, Q9UKQ8, Q9Y445 | Gene names | SLC7A8, LAT2 | |||
|
Domain Architecture |

|
|||||
| Description | Large neutral amino acids transporter small subunit 2 (L-type amino acid transporter 2) (hLAT2). | |||||
|
LAT2_MOUSE
|
||||||
| NC score | 0.122407 (rank : 23) | θ value | 0.00035302 (rank : 16) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QXW9 | Gene names | Slc7a8, Lat2 | |||
|
Domain Architecture |

|
|||||
| Description | Large neutral amino acids transporter small subunit 2 (L-type amino acid transporter 2). | |||||
|
BAT1_HUMAN
|
||||||
| NC score | 0.115754 (rank : 24) | θ value | 0.0148317 (rank : 20) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P82251 | Gene names | SLC7A9, BAT1 | |||
|
Domain Architecture |

|
|||||
| Description | B(0,+)-type amino acid transporter 1 (B(0,+)AT) (Glycoprotein- associated amino acid transporter b0,+AT1). | |||||
|
CTR4_HUMAN
|
||||||
| NC score | 0.102716 (rank : 25) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43246, Q3KNQ6, Q4VC45, Q6IBY8, Q96H88 | Gene names | SLC7A4 | |||
|
Domain Architecture |

|
|||||
| Description | Cationic amino acid transporter 4 (CAT-4) (CAT4). | |||||
|
AAA1_MOUSE
|
||||||
| NC score | 0.096175 (rank : 26) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P63115, Q9CW25, Q9JMH8 | Gene names | Slc7a10, Asc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Asc-type amino acid transporter 1 (Asc-1) (D-serine transporter). | |||||
|
AAA1_HUMAN
|
||||||
| NC score | 0.089411 (rank : 27) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NS82 | Gene names | SLC7A10, ASC1 | |||
|
Domain Architecture |

|
|||||
| Description | Asc-type amino acid transporter 1 (Asc-1). | |||||
|
YLA1_HUMAN
|
||||||
| NC score | 0.070352 (rank : 28) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UM01, O95984, Q9P2V5 | Gene names | SLC7A7 | |||
|
Domain Architecture |

|
|||||
| Description | Y+L amino acid transporter 1 (y(+)L-type amino acid transporter 1) (y+LAT-1) (Y+LAT1) (Monocyte amino acid permease 2) (MOP-2). | |||||
|
LAT1_HUMAN
|
||||||
| NC score | 0.069642 (rank : 29) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q01650, Q9UBN8, Q9UP15, Q9UQC0 | Gene names | SLC7A5, CD98LC, LAT1, MPE16 | |||
|
Domain Architecture |

|
|||||
| Description | Large neutral amino acids transporter small subunit 1 (L-type amino acid transporter 1) (4F2 light chain) (4F2 LC) (4F2LC) (CD98 light chain) (Integral membrane protein E16) (hLAT1). | |||||
|
XCT_MOUSE
|
||||||
| NC score | 0.068962 (rank : 30) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WTR6 | Gene names | Slc7a11 | |||
|
Domain Architecture |

|
|||||
| Description | Cystine/glutamate transporter (Amino acid transport system xc-) (xCT). | |||||
|
LAT1_MOUSE
|
||||||
| NC score | 0.065983 (rank : 31) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z127, Q9JMI4 | Gene names | Slc7a5, Lat1 | |||
|
Domain Architecture |

|
|||||
| Description | Large neutral amino acids transporter small subunit 1 (L-type amino acid transporter 1) (4F2 light chain) (4F2 LC) (4F2LC). | |||||
|
XCT_HUMAN
|
||||||
| NC score | 0.062895 (rank : 32) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UPY5 | Gene names | SLC7A11 | |||
|
Domain Architecture |

|
|||||
| Description | Cystine/glutamate transporter (Amino acid transport system xc-) (xCT) (Calcium channel blocker resistance protein CCBR1). | |||||
|
G137B_HUMAN
|
||||||
| NC score | 0.051648 (rank : 33) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60478 | Gene names | GPR137B, TM7SF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integral membrane protein GPR137B (Transmembrane 7 superfamily member 1). | |||||
|
SYM_HUMAN
|
||||||
| NC score | 0.022654 (rank : 34) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P56192, Q14895, Q53H14, Q96A15, Q96BZ0, Q9NSE0 | Gene names | MARS | |||
|
Domain Architecture |

|
|||||
| Description | Methionyl-tRNA synthetase (EC 6.1.1.10) (Methionine--tRNA ligase) (MetRS). | |||||
|
RTN1_HUMAN
|
||||||
| NC score | 0.021496 (rank : 35) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16799, Q16800, Q16801 | Gene names | RTN1, NSP | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.021370 (rank : 36) | θ value | 0.0736092 (rank : 23) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.020222 (rank : 37) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
ROM1_HUMAN
|
||||||
| NC score | 0.016537 (rank : 38) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03395 | Gene names | ROM1, TSPAN23 | |||
|
Domain Architecture |

|
|||||
| Description | Rod outer segment membrane protein 1 (ROSP1) (Tetraspanin-23) (Tspan- 23). | |||||
|
EVL_HUMAN
|
||||||
| NC score | 0.014658 (rank : 39) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UI08, O95884, Q7Z522, Q8TBV1, Q9UF25, Q9UIC2 | Gene names | EVL, RNB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
EVL_MOUSE
|
||||||
| NC score | 0.011437 (rank : 40) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
ADDG_MOUSE
|
||||||
| NC score | 0.010390 (rank : 41) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QYB5, Q9D2M5, Q9QYB6 | Gene names | Add3, Addl | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-adducin (Adducin-like protein 70). | |||||
|
ACOD_HUMAN
|
||||||
| NC score | 0.009450 (rank : 42) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00767, Q16150, Q53GR9, Q5W037, Q5W038, Q6GSS4, Q96KF6, Q9BS07, Q9Y695 | Gene names | SCD | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-CoA desaturase (EC 1.14.19.1) (Stearoyl-CoA desaturase) (Fatty acid desaturase) (Delta(9)-desaturase). | |||||
|
CENPC_MOUSE
|
||||||
| NC score | 0.008781 (rank : 43) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49452 | Gene names | Cenpc1, Cenpc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C). | |||||
|
CTL4_MOUSE
|
||||||
| NC score | 0.008469 (rank : 44) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91VA1, Q7TQ02, Q9CVA7 | Gene names | Slc44a4, Ctl4, Ng22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Choline transporter-like protein 4 (Solute carrier family 44 member 4). | |||||
|
GPR84_HUMAN
|
||||||
| NC score | 0.008138 (rank : 45) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NQS5 | Gene names | GPR84, EX33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 84 (Inflammation-related G-protein coupled receptor EX33). | |||||
|
ZN403_HUMAN
|
||||||
| NC score | 0.007934 (rank : 46) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H3C7, Q96T90, Q9GZR8, Q9H767 | Gene names | ZNF403, LCRG1, LZK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF403 (Laryngeal carcinoma-related protein 1). | |||||
|
SYF2_HUMAN
|
||||||
| NC score | 0.007800 (rank : 47) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95926, Q5TH73 | Gene names | SYF2, CBPIN, GCIPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pre-mRNA-splicing factor SYF2 (CCNDBP1-interactor) (p29). | |||||
|
IPR1_MOUSE
|
||||||
| NC score | 0.007671 (rank : 48) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVK9, Q3UCV7, Q80V00 | Gene names | Ipr1, Ifi75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intracellular pathogen resistance protein 1. | |||||
|
CHIT1_MOUSE
|
||||||
| NC score | 0.007422 (rank : 49) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D7Q1, Q6QJD2 | Gene names | Chit1 | |||
|
Domain Architecture |
|
|||||
| Description | Chitotriosidase-1 precursor (EC 3.2.1.14) (Chitinase-1). | |||||
|
HYALP_MOUSE
|
||||||
| NC score | 0.006917 (rank : 50) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48794, Q7TSD6, Q812F4, Q9DAQ1 | Gene names | Spam1, Ph20, Spam | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronidase PH-20 precursor (EC 3.2.1.35) (Hyal-PH20) (Sperm surface protein PH-20) (Sperm adhesion molecule 1). | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.006810 (rank : 51) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
PP4R1_HUMAN
|
||||||
| NC score | 0.006415 (rank : 52) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TF05, Q99774, Q9UNQ7 | Gene names | PPP4R1, MEG1, PP4R1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 regulatory subunit 1. | |||||
|
ARI4A_HUMAN
|
||||||
| NC score | 0.006241 (rank : 53) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.006120 (rank : 54) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
EDG1_MOUSE
|
||||||
| NC score | 0.005425 (rank : 55) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O08530, Q9R235 | Gene names | Edg1, Lpb1 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosine 1-phosphate receptor Edg-1 (Sphingosine 1-phosphate receptor 1) (S1P1) (Lysophospholipid receptor B1). | |||||
|
V2R1B_MOUSE
|
||||||
| NC score | 0.004494 (rank : 56) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6TAC4, O70409 | Gene names | V2r1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vomeronasal type-2 receptor 1b precursor (Putative pheromone receptor V2R1b). | |||||
|
EYA1_HUMAN
|
||||||
| NC score | 0.004047 (rank : 57) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99502 | Gene names | EYA1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
LPHN1_HUMAN
|
||||||
| NC score | 0.003479 (rank : 58) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94910, Q96IE7, Q9BU07, Q9HAR3 | Gene names | LPHN1, KIAA0821, LEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-1 precursor (Calcium-independent alpha-latrotoxin receptor 1) (Lectomedin-2). | |||||
|
GPR97_MOUSE
|
||||||
| NC score | 0.003001 (rank : 59) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R0T6 | Gene names | Gpr97 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 97 precursor. | |||||
|
TPM4_MOUSE
|
||||||
| NC score | 0.002942 (rank : 60) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 703 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6IRU2 | Gene names | Tpm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4). | |||||
|
TPM4_HUMAN
|
||||||
| NC score | 0.002454 (rank : 61) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P67936, P07226, Q15659, Q9BU85, Q9H8Q3 | Gene names | TPM4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4) (TM30p1). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.002242 (rank : 62) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.002027 (rank : 63) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.001787 (rank : 64) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.001508 (rank : 65) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.001393 (rank : 66) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
O1052_MOUSE
|
||||||
| NC score | 0.000746 (rank : 67) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 684 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VGR8 | Gene names | Olfr1052, Mor172-1 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 1052 (Olfactory receptor 172-1). | |||||
|
OR5J2_HUMAN
|
||||||
| NC score | -0.000458 (rank : 68) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 649 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NH18, Q6IEU5 | Gene names | OR5J2 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 5J2 (Olfactory receptor OR11-266). | |||||
|
MASP2_MOUSE
|
||||||
| NC score | -0.000761 (rank : 69) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91WP0, Q9QXA4, Q9QXD2, Q9QXD5, Q9Z338 | Gene names | Masp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
SPEG_HUMAN
|
||||||
| NC score | -0.001255 (rank : 70) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
OR5K1_HUMAN
|
||||||
| NC score | -0.001403 (rank : 71) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 782 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NHB7 | Gene names | OR5K1 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 5K1 (HTPCRX10). | |||||
|
EPHA5_MOUSE
|
||||||
| NC score | -0.001881 (rank : 72) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60629 | Gene names | Epha5, Bsk, Cek7, Ehk1 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 5 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EHK-1) (EPH homology kinase 1) (Brain-specific kinase) (CEK-7). | |||||
|
EPHA5_HUMAN
|
||||||
| NC score | -0.001916 (rank : 73) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54756 | Gene names | EPHA5, EHK1, HEK7 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 5 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EHK-1) (EPH homology kinase 1) (Receptor protein- tyrosine kinase HEK7). | |||||
|
PEG3_MOUSE
|
||||||
| NC score | -0.002251 (rank : 74) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||