Please be patient as the page loads
|
EYA1_HUMAN
|
||||||
| SwissProt Accessions | Q99502 | Gene names | EYA1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
EYA1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q99502 | Gene names | EYA1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
EYA1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.989351 (rank : 2) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P97767, O08818 | Gene names | Eya1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
EYA4_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.970248 (rank : 6) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95677, O95464, O95679, Q8IW39, Q9NTR7 | Gene names | EYA4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
EYA4_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.975708 (rank : 3) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z191 | Gene names | Eya4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
EYA2_MOUSE
|
||||||
| θ value | 1.71888e-184 (rank : 5) | NC score | 0.975222 (rank : 4) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O08575, P97925 | Gene names | Eya2, Eab1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
EYA2_HUMAN
|
||||||
| θ value | 1.40939e-178 (rank : 6) | NC score | 0.974414 (rank : 5) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00167, Q5JSW8, Q96CV6, Q96H97, Q99503, Q99812, Q9BWF6, Q9H4S3, Q9H4S9, Q9NPZ4, Q9UIX7 | Gene names | EYA2, EAB1 | |||
|
Domain Architecture |
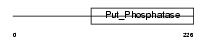
|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
EYA3_HUMAN
|
||||||
| θ value | 5.07123e-136 (rank : 7) | NC score | 0.965248 (rank : 8) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99504, O95463, Q99813 | Gene names | EYA3 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 3 (EC 3.1.3.48). | |||||
|
EYA3_MOUSE
|
||||||
| θ value | 1.87085e-122 (rank : 8) | NC score | 0.965523 (rank : 7) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97480, P97768 | Gene names | Eya3 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 3 (EC 3.1.3.48). | |||||
|
EWS_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 9) | NC score | 0.164335 (rank : 10) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q01844, Q92635 | Gene names | EWSR1, EWS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS (EWS oncogene) (Ewing sarcoma breakpoint region 1 protein). | |||||
|
EWS_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 10) | NC score | 0.161887 (rank : 11) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61545 | Gene names | Ewsr1, Ews, Ewsh | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS. | |||||
|
FUS_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 11) | NC score | 0.166955 (rank : 9) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P56959 | Gene names | Fus | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Pigpen protein). | |||||
|
FUS_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 12) | NC score | 0.157121 (rank : 12) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 13) | NC score | 0.084972 (rank : 13) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
RBM14_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 14) | NC score | 0.066134 (rank : 16) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C2Q3, Q3TJB6, Q91Z21, Q9DBI6 | Gene names | Rbm14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14). | |||||
|
RBM14_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 15) | NC score | 0.054032 (rank : 23) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96PK6, O75932, Q53GV1, Q68DQ9, Q96PK5 | Gene names | RBM14, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14) (RRM-containing coactivator activator/modulator) (Synaptotagmin-interacting protein) (SYT-interacting protein). | |||||
|
SOX8_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 16) | NC score | 0.031818 (rank : 38) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q04886 | Gene names | Sox8, Sox-8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
TRIM2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 17) | NC score | 0.039048 (rank : 32) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9C040, O60272, Q9BSI9, Q9UFZ1 | Gene names | TRIM2, KIAA0517, RNF86 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 2 (RING finger protein 86). | |||||
|
TRIM2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 18) | NC score | 0.039157 (rank : 31) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ESN6, Q3UHP5, Q8C981 | Gene names | Trim2, Narf | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 2 (Neural activity-related RING finger protein). | |||||
|
NR4A1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 19) | NC score | 0.013038 (rank : 76) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P22736 | Gene names | NR4A1, GFRP1, HMR, NAK1 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A1 (Orphan nuclear receptor HMR) (Early response protein NAK1) (TR3 orphan receptor) (ST-59). | |||||
|
DDX17_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 20) | NC score | 0.030329 (rank : 40) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92841, Q69YT1, Q6ICD6 | Gene names | DDX17 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 21) | NC score | 0.045718 (rank : 27) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
RNF12_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 22) | NC score | 0.037540 (rank : 36) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NVW2, Q9Y598 | Gene names | RNF12, RLIM | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM) (NY-REN-43 antigen). | |||||
|
GPS2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.059901 (rank : 21) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13227 | Gene names | GPS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein pathway suppressor 2 (Protein GPS2). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.017180 (rank : 66) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
DDX17_MOUSE
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.025190 (rank : 49) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q501J6, Q6P5G1, Q8BIN2 | Gene names | Ddx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17). | |||||
|
HNRL1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 26) | NC score | 0.072044 (rank : 14) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VDM6, Q3U201, Q3UPB0, Q6AZA7, Q8BY45, Q8K365 | Gene names | Hnrpul1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1. | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 0.21417 (rank : 27) | NC score | 0.037756 (rank : 35) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 0.21417 (rank : 28) | NC score | 0.039842 (rank : 30) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
GATA4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 29) | NC score | 0.027807 (rank : 44) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43694 | Gene names | GATA4 | |||
|
Domain Architecture |
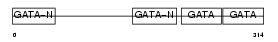
|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
HNRL1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 30) | NC score | 0.070414 (rank : 15) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BUJ2, O76022, Q6ZSZ0, Q7L8P4, Q8N6Z4, Q96G37, Q9HAL3, Q9UG75 | Gene names | HNRPUL1, E1BAP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1 (Adenovirus early region 1B-associated protein 5) (E1B-55 kDa-associated protein 5) (E1B-AP5). | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.040043 (rank : 29) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.038949 (rank : 33) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.022568 (rank : 52) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.030304 (rank : 41) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.013427 (rank : 74) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
RBM10_HUMAN
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.025320 (rank : 48) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P98175, Q14136 | Gene names | RBM10, DXS8237E, KIAA0122 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 10 (RNA-binding motif protein 10). | |||||
|
BAG4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.046936 (rank : 25) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95429, O95818 | Gene names | BAG4, SODD | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
BAG4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.055932 (rank : 22) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CI61, Q3TRL9, Q91VT5, Q9CWG2 | Gene names | Bag4, Sodd | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
SOX30_MOUSE
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.024505 (rank : 50) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
PO121_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.027508 (rank : 45) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.021686 (rank : 55) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
ANX11_MOUSE
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.015048 (rank : 70) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97384 | Gene names | Anxa11, Anx11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.008755 (rank : 83) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
PDC6I_HUMAN
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.024079 (rank : 51) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WUM4, Q9BX86, Q9NUN0, Q9P2H2, Q9UKL5 | Gene names | PDCD6IP, AIP1, ALIX, KIAA1375 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95). | |||||
|
PUM2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.038874 (rank : 34) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TB72, O00234, Q53TV7, Q8WY43, Q9HAN2 | Gene names | PUM2, KIAA0235, PUMH2 | |||
|
Domain Architecture |

|
|||||
| Description | Pumilio homolog 2 (Pumilio-2). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.025461 (rank : 47) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
DDX3Y_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.019086 (rank : 61) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
HRBL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.027826 (rank : 43) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
RPB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.062931 (rank : 17) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.062864 (rank : 18) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
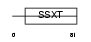
SSXT_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.029378 (rank : 42) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15532, Q16404, Q9BXC6 | Gene names | SS18, SSXT, SYT | |||
|
Domain Architecture |
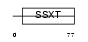
|
|||||
| Description | SSXT protein (Synovial sarcoma, translocated to X chromosome) (SYT protein). | |||||
|
ZN198_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.012890 (rank : 77) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UBW7, O43212, O43434, O60898, Q5W0Q4, Q63HP0, Q8NE39, Q9H538, Q9UEU2 | Gene names | ZNF198, FIM, RAMP, ZMYM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 198 (Zinc finger MYM-type protein 2) (Fused in myeloproliferative disorders protein) (Rearranged in atypical myeloproliferative disorder protein). | |||||
|
ZN560_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | -0.000629 (rank : 92) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96MR9, Q495S9, Q495T1 | Gene names | ZNF560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 560. | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.046311 (rank : 26) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
FOXE2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.006312 (rank : 86) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99526 | Gene names | FOXE2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E2 (HNF-3/fork head-like protein 5) (HFKL5) (HFKH4). | |||||
|
PLK4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | -0.001378 (rank : 95) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00444, Q8IYF0, Q96Q95 | Gene names | PLK4, SAK, STK18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PLK4 (EC 2.7.11.21) (Polo-like kinase 4) (PLK-4) (Serine/threonine-protein kinase Sak) (Serine/threonine- protein kinase 18). | |||||
|
SMAD4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.011577 (rank : 80) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
K0999_MOUSE
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | -0.001157 (rank : 94) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6P4S6, Q641L5, Q66JZ5, Q6ZQ09, Q8K075, Q9CYD5 | Gene names | Kiaa0999 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized serine/threonine-protein kinase KIAA0999 (EC 2.7.11.1). | |||||
|
MCM3A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.052743 (rank : 24) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WUU9 | Gene names | Mcm3ap, Ganp, Map80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
MN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.020139 (rank : 58) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q10571 | Gene names | MN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tumor suppressor protein MN1. | |||||
|
PUM2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.035370 (rank : 37) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80U58, Q80UZ9, Q91YW4, Q925A0, Q9ERC7 | Gene names | Pum2, Kiaa0235 | |||
|
Domain Architecture |

|
|||||
| Description | Pumilio homolog 2. | |||||
|
RFX1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.013667 (rank : 73) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P22670 | Gene names | RFX1 | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II regulatory factor RFX1 (RFX) (Enhancer factor C) (EF-C). | |||||
|
SEM6D_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.007689 (rank : 84) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NFY4, Q8NFY3, Q8NFY5, Q8NFY6, Q8NFY7, Q9P249 | Gene names | SEMA6D, KIAA1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
CBP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.020789 (rank : 56) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CING_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.000829 (rank : 91) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CNOT4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.014603 (rank : 72) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95628, O95339, O95627, Q8IYM7, Q8NCL0, Q9NPQ1, Q9NZN6 | Gene names | CNOT4, NOT4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
COE1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.010283 (rank : 81) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UH73, Q8IW11 | Gene names | EBF1, COE1, EBF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE1 (OE-1) (O/E-1) (Early B-cell factor). | |||||
|
COE1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.010283 (rank : 82) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07802 | Gene names | Ebf1, Coe1, Ebf | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE1 (OE-1) (O/E-1) (Early B-cell factor). | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.001083 (rank : 90) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
PARM1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.022279 (rank : 54) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
SNPC4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.016384 (rank : 68) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
ZCH14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.011994 (rank : 79) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WYQ9, O60324, Q9UFP0 | Gene names | ZCCHC14, KIAA0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
ZFAN5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.014871 (rank : 71) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O76080 | Gene names | ZFAND5, ZA20D2, ZNF216 | |||
|
Domain Architecture |

|
|||||
| Description | AN1-type zinc finger protein 5 (Zinc finger A20 domain-containing protein 2) (Zinc finger protein 216). | |||||
|
CDSN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.030562 (rank : 39) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15517, O43509, Q5SQ85, Q5STD2, Q7LA70, Q7LA71, Q86Z04, Q8IZU4, Q8IZU5, Q8IZU6, Q8N5P3, Q95IF9, Q9NP52, Q9NPE0, Q9NPG5, Q9NRH4, Q9NRH5, Q9NRH6, Q9NRH7, Q9NRH8, Q9UBH8, Q9UIN6, Q9UIN7, Q9UIN8, Q9UIN9, Q9UIP0 | Gene names | CDSN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor (S protein). | |||||
|
CDSN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.019255 (rank : 60) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TPC1 | Gene names | Cdsn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor. | |||||
|
DDX3X_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.017753 (rank : 64) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
ILF3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.018290 (rank : 63) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12906, O43409, Q6P1X1, Q86XY7, Q99544, Q99545, Q9BZH4, Q9BZH5, Q9NQ95, Q9NQ96, Q9NQ97, Q9NQ98, Q9NQ99, Q9NQA0, Q9NQA1, Q9NQA2, Q9NRN2, Q9NRN3, Q9NRN4, Q9UMZ9, Q9UN00, Q9UN84, Q9UNA2 | Gene names | ILF3, DRBF, MPHOSPH4, NF90 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3 (Nuclear factor of activated T- cells 90 kDa) (NF-AT-90) (Double-stranded RNA-binding protein 76) (DRBP76) (Translational control protein 80) (TCP80) (Nuclear factor associated with dsRNA) (NFAR) (M-phase phosphoprotein 4) (MPP4). | |||||
|
NFRKB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.018668 (rank : 62) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
PL10_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.017504 (rank : 65) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
S12A5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.004047 (rank : 88) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H2X9, Q5VZ41, Q9H4Z0, Q9ULP4 | Gene names | SLC12A5, KCC2, KIAA1176 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (Erythroid K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (hKCC2). | |||||
|
SEM6D_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.007207 (rank : 85) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q76KF0, Q76KF1, Q76KF2, Q76KF3, Q76KF4, Q80TD0 | Gene names | Sema6d, Kiaa1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SSXT_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.040579 (rank : 28) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62280 | Gene names | Ss18, Ssxt, Syt | |||
|
Domain Architecture |
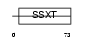
|
|||||
| Description | SSXT protein (SYT protein) (Synovial sarcoma-associated Ss18-alpha). | |||||
|
SYNPO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.019942 (rank : 59) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CC35, Q99JI0 | Gene names | Synpo, Kiaa1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
ZIC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | -0.000951 (rank : 93) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P46684 | Gene names | Zic1, Zic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZIC 1 (Zinc finger protein of the cerebellum 1). | |||||
|
ANXA7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.013211 (rank : 75) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P20073 | Gene names | ANXA7, ANX7, SNX | |||
|
Domain Architecture |
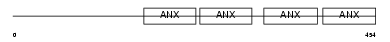
|
|||||
| Description | Annexin A7 (Annexin VII) (Synexin). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.022315 (rank : 53) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
CUZD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.005983 (rank : 87) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70412, Q9CTZ7 | Gene names | Cuzd1, Itmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Integral membrane-associated protein 1). | |||||
|
HRBL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.016771 (rank : 67) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
HXA5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.002762 (rank : 89) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20719, O43367, Q96CY6 | Gene names | HOXA5, HOX1C | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A5 (Hox-1C). | |||||
|
NUP62_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.020373 (rank : 57) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q63850, Q99JN7 | Gene names | Nup62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
S18L1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.026979 (rank : 46) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75177, Q5JXJ3, Q8NE69, Q9BR55, Q9H4K6 | Gene names | SS18L1, KIAA0693 | |||
|
Domain Architecture |
|
|||||
| Description | SS18-like protein 1 (SYT homolog 1). | |||||
|
SFR15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.016175 (rank : 69) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
ZFAN5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.012680 (rank : 78) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88878 | Gene names | Zfand5, Za20d2, Zfp216, Znf216 | |||
|
Domain Architecture |

|
|||||
| Description | AN1-type zinc finger protein 5 (Zinc finger A20 domain-containing protein 2) (Zinc finger protein 216). | |||||
|
RBP56_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.062756 (rank : 19) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92804, Q92751 | Gene names | TAF15, RBP56, TAF2N | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 2N (RNA-binding protein 56) (TAFII68) (TAF(II)68). | |||||
|
TFG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.060524 (rank : 20) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92734, Q15656 | Gene names | TFG | |||
|
Domain Architecture |

|
|||||
| Description | Protein TFG (TRK-fused gene protein). | |||||
|
EYA1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q99502 | Gene names | EYA1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
EYA1_MOUSE
|
||||||
| NC score | 0.989351 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P97767, O08818 | Gene names | Eya1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
EYA4_MOUSE
|
||||||
| NC score | 0.975708 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z191 | Gene names | Eya4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
EYA2_MOUSE
|
||||||
| NC score | 0.975222 (rank : 4) | θ value | 1.71888e-184 (rank : 5) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O08575, P97925 | Gene names | Eya2, Eab1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
EYA2_HUMAN
|
||||||
| NC score | 0.974414 (rank : 5) | θ value | 1.40939e-178 (rank : 6) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00167, Q5JSW8, Q96CV6, Q96H97, Q99503, Q99812, Q9BWF6, Q9H4S3, Q9H4S9, Q9NPZ4, Q9UIX7 | Gene names | EYA2, EAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
EYA4_HUMAN
|
||||||
| NC score | 0.970248 (rank : 6) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95677, O95464, O95679, Q8IW39, Q9NTR7 | Gene names | EYA4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
EYA3_MOUSE
|
||||||
| NC score | 0.965523 (rank : 7) | θ value | 1.87085e-122 (rank : 8) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97480, P97768 | Gene names | Eya3 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 3 (EC 3.1.3.48). | |||||
|
EYA3_HUMAN
|
||||||
| NC score | 0.965248 (rank : 8) | θ value | 5.07123e-136 (rank : 7) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99504, O95463, Q99813 | Gene names | EYA3 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 3 (EC 3.1.3.48). | |||||
|
FUS_MOUSE
|
||||||
| NC score | 0.166955 (rank : 9) | θ value | 0.000602161 (rank : 11) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P56959 | Gene names | Fus | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Pigpen protein). | |||||
|
EWS_HUMAN
|
||||||
| NC score | 0.164335 (rank : 10) | θ value | 0.000158464 (rank : 9) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q01844, Q92635 | Gene names | EWSR1, EWS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS (EWS oncogene) (Ewing sarcoma breakpoint region 1 protein). | |||||
|
EWS_MOUSE
|
||||||
| NC score | 0.161887 (rank : 11) | θ value | 0.00020696 (rank : 10) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61545 | Gene names | Ewsr1, Ews, Ewsh | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS. | |||||
|
FUS_HUMAN
|
||||||
| NC score | 0.157121 (rank : 12) | θ value | 0.000786445 (rank : 12) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.084972 (rank : 13) | θ value | 0.000786445 (rank : 13) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
HNRL1_MOUSE
|
||||||
| NC score | 0.072044 (rank : 14) | θ value | 0.21417 (rank : 26) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VDM6, Q3U201, Q3UPB0, Q6AZA7, Q8BY45, Q8K365 | Gene names | Hnrpul1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1. | |||||
|
HNRL1_HUMAN
|
||||||
| NC score | 0.070414 (rank : 15) | θ value | 0.279714 (rank : 30) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BUJ2, O76022, Q6ZSZ0, Q7L8P4, Q8N6Z4, Q96G37, Q9HAL3, Q9UG75 | Gene names | HNRPUL1, E1BAP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1 (Adenovirus early region 1B-associated protein 5) (E1B-55 kDa-associated protein 5) (E1B-AP5). | |||||
|
RBM14_MOUSE
|
||||||
| NC score | 0.066134 (rank : 16) | θ value | 0.00102713 (rank : 14) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8C2Q3, Q3TJB6, Q91Z21, Q9DBI6 | Gene names | Rbm14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14). | |||||
|
RPB1_HUMAN
|
||||||
| NC score | 0.062931 (rank : 17) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| NC score | 0.062864 (rank : 18) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RBP56_HUMAN
|
||||||
| NC score | 0.062756 (rank : 19) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92804, Q92751 | Gene names | TAF15, RBP56, TAF2N | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 2N (RNA-binding protein 56) (TAFII68) (TAF(II)68). | |||||
|
TFG_HUMAN
|
||||||
| NC score | 0.060524 (rank : 20) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92734, Q15656 | Gene names | TFG | |||
|
Domain Architecture |

|
|||||
| Description | Protein TFG (TRK-fused gene protein). | |||||
|
GPS2_HUMAN
|
||||||
| NC score | 0.059901 (rank : 21) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13227 | Gene names | GPS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein pathway suppressor 2 (Protein GPS2). | |||||
|
BAG4_MOUSE
|
||||||
| NC score | 0.055932 (rank : 22) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CI61, Q3TRL9, Q91VT5, Q9CWG2 | Gene names | Bag4, Sodd | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
RBM14_HUMAN
|
||||||
| NC score | 0.054032 (rank : 23) | θ value | 0.00175202 (rank : 15) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96PK6, O75932, Q53GV1, Q68DQ9, Q96PK5 | Gene names | RBM14, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14) (RRM-containing coactivator activator/modulator) (Synaptotagmin-interacting protein) (SYT-interacting protein). | |||||
|
MCM3A_MOUSE
|
||||||
| NC score | 0.052743 (rank : 24) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WUU9 | Gene names | Mcm3ap, Ganp, Map80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
BAG4_HUMAN
|
||||||
| NC score | 0.046936 (rank : 25) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95429, O95818 | Gene names | BAG4, SODD | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 4 (BCL2-associated athanogene 4) (BAG-4) (Silencer of death domains). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.046311 (rank : 26) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.045718 (rank : 27) | θ value | 0.0961366 (rank : 21) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
SSXT_MOUSE
|
||||||
| NC score | 0.040579 (rank : 28) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62280 | Gene names | Ss18, Ssxt, Syt | |||
|
Domain Architecture |

|
|||||
| Description | SSXT protein (SYT protein) (Synovial sarcoma-associated Ss18-alpha). | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.040043 (rank : 29) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.039842 (rank : 30) | θ value | 0.21417 (rank : 28) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
TRIM2_MOUSE
|
||||||
| NC score | 0.039157 (rank : 31) | θ value | 0.0252991 (rank : 18) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ESN6, Q3UHP5, Q8C981 | Gene names | Trim2, Narf | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 2 (Neural activity-related RING finger protein). | |||||
|
TRIM2_HUMAN
|
||||||
| NC score | 0.039048 (rank : 32) | θ value | 0.0252991 (rank : 17) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9C040, O60272, Q9BSI9, Q9UFZ1 | Gene names | TRIM2, KIAA0517, RNF86 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 2 (RING finger protein 86). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.038949 (rank : 33) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
PUM2_HUMAN
|
||||||
| NC score | 0.038874 (rank : 34) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TB72, O00234, Q53TV7, Q8WY43, Q9HAN2 | Gene names | PUM2, KIAA0235, PUMH2 | |||
|
Domain Architecture |

|
|||||
| Description | Pumilio homolog 2 (Pumilio-2). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.037756 (rank : 35) | θ value | 0.21417 (rank : 27) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
RNF12_HUMAN
|
||||||
| NC score | 0.037540 (rank : 36) | θ value | 0.0961366 (rank : 22) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NVW2, Q9Y598 | Gene names | RNF12, RLIM | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM) (NY-REN-43 antigen). | |||||
|
PUM2_MOUSE
|
||||||
| NC score | 0.035370 (rank : 37) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80U58, Q80UZ9, Q91YW4, Q925A0, Q9ERC7 | Gene names | Pum2, Kiaa0235 | |||
|
Domain Architecture |

|
|||||
| Description | Pumilio homolog 2. | |||||
|
SOX8_MOUSE
|
||||||
| NC score | 0.031818 (rank : 38) | θ value | 0.0148317 (rank : 16) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q04886 | Gene names | Sox8, Sox-8 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-8. | |||||
|
CDSN_HUMAN
|
||||||
| NC score | 0.030562 (rank : 39) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15517, O43509, Q5SQ85, Q5STD2, Q7LA70, Q7LA71, Q86Z04, Q8IZU4, Q8IZU5, Q8IZU6, Q8N5P3, Q95IF9, Q9NP52, Q9NPE0, Q9NPG5, Q9NRH4, Q9NRH5, Q9NRH6, Q9NRH7, Q9NRH8, Q9UBH8, Q9UIN6, Q9UIN7, Q9UIN8, Q9UIN9, Q9UIP0 | Gene names | CDSN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor (S protein). | |||||
|
DDX17_HUMAN
|
||||||
| NC score | 0.030329 (rank : 40) | θ value | 0.0563607 (rank : 20) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92841, Q69YT1, Q6ICD6 | Gene names | DDX17 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17) (RNA-dependent helicase p72) (DEAD box protein p72). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.030304 (rank : 41) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
SSXT_HUMAN
|
||||||
| NC score | 0.029378 (rank : 42) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15532, Q16404, Q9BXC6 | Gene names | SS18, SSXT, SYT | |||
|
Domain Architecture |

|
|||||
| Description | SSXT protein (Synovial sarcoma, translocated to X chromosome) (SYT protein). | |||||
|
HRBL_HUMAN
|
||||||
| NC score | 0.027826 (rank : 43) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95081, O75429, Q96AB9, Q96GL4 | Gene names | HRBL, RABR | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
GATA4_HUMAN
|
||||||
| NC score | 0.027807 (rank : 44) | θ value | 0.279714 (rank : 29) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43694 | Gene names | GATA4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.027508 (rank : 45) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
S18L1_HUMAN
|
||||||
| NC score | 0.026979 (rank : 46) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75177, Q5JXJ3, Q8NE69, Q9BR55, Q9H4K6 | Gene names | SS18L1, KIAA0693 | |||
|
Domain Architecture |

|
|||||
| Description | SS18-like protein 1 (SYT homolog 1). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.025461 (rank : 47) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
RBM10_HUMAN
|
||||||
| NC score | 0.025320 (rank : 48) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P98175, Q14136 | Gene names | RBM10, DXS8237E, KIAA0122 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 10 (RNA-binding motif protein 10). | |||||
|
DDX17_MOUSE
|
||||||
| NC score | 0.025190 (rank : 49) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q501J6, Q6P5G1, Q8BIN2 | Gene names | Ddx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX17 (EC 3.6.1.-) (DEAD box protein 17). | |||||
|
SOX30_MOUSE
|
||||||
| NC score | 0.024505 (rank : 50) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
PDC6I_HUMAN
|
||||||
| NC score | 0.024079 (rank : 51) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WUM4, Q9BX86, Q9NUN0, Q9P2H2, Q9UKL5 | Gene names | PDCD6IP, AIP1, ALIX, KIAA1375 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.022568 (rank : 52) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.022315 (rank : 53) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
PARM1_MOUSE
|
||||||
| NC score | 0.022279 (rank : 54) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.021686 (rank : 55) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.020789 (rank : 56) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
NUP62_MOUSE
|
||||||
| NC score | 0.020373 (rank : 57) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q63850, Q99JN7 | Gene names | Nup62 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore glycoprotein p62 (62 kDa nucleoporin). | |||||
|
MN1_HUMAN
|
||||||
| NC score | 0.020139 (rank : 58) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q10571 | Gene names | MN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tumor suppressor protein MN1. | |||||
|
SYNPO_MOUSE
|
||||||
| NC score | 0.019942 (rank : 59) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CC35, Q99JI0 | Gene names | Synpo, Kiaa1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
CDSN_MOUSE
|
||||||
| NC score | 0.019255 (rank : 60) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TPC1 | Gene names | Cdsn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor. | |||||
|
DDX3Y_HUMAN
|
||||||
| NC score | 0.019086 (rank : 61) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
NFRKB_MOUSE
|
||||||
| NC score | 0.018668 (rank : 62) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
ILF3_HUMAN
|
||||||
| NC score | 0.018290 (rank : 63) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12906, O43409, Q6P1X1, Q86XY7, Q99544, Q99545, Q9BZH4, Q9BZH5, Q9NQ95, Q9NQ96, Q9NQ97, Q9NQ98, Q9NQ99, Q9NQA0, Q9NQA1, Q9NQA2, Q9NRN2, Q9NRN3, Q9NRN4, Q9UMZ9, Q9UN00, Q9UN84, Q9UNA2 | Gene names | ILF3, DRBF, MPHOSPH4, NF90 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3 (Nuclear factor of activated T- cells 90 kDa) (NF-AT-90) (Double-stranded RNA-binding protein 76) (DRBP76) (Translational control protein 80) (TCP80) (Nuclear factor associated with dsRNA) (NFAR) (M-phase phosphoprotein 4) (MPP4). | |||||
|
DDX3X_HUMAN
|
||||||
| NC score | 0.017753 (rank : 64) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
PL10_MOUSE
|
||||||
| NC score | 0.017504 (rank : 65) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.017180 (rank : 66) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
HRBL_MOUSE
|
||||||
| NC score | 0.016771 (rank : 67) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
SNPC4_HUMAN
|
||||||
| NC score | 0.016384 (rank : 68) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5SXM2, Q9Y6P7 | Gene names | SNAPC4, SNAP190 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA- activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor subunit alpha) (PSE-binding factor subunit alpha) (PTF subunit alpha). | |||||
|
SFR15_HUMAN
|
||||||
| NC score | 0.016175 (rank : 69) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
ANX11_MOUSE
|
||||||
| NC score | 0.015048 (rank : 70) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97384 | Gene names | Anxa11, Anx11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50). | |||||
|
ZFAN5_HUMAN
|
||||||
| NC score | 0.014871 (rank : 71) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O76080 | Gene names | ZFAND5, ZA20D2, ZNF216 | |||
|
Domain Architecture |

|
|||||
| Description | AN1-type zinc finger protein 5 (Zinc finger A20 domain-containing protein 2) (Zinc finger protein 216). | |||||
|
CNOT4_HUMAN
|
||||||
| NC score | 0.014603 (rank : 72) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95628, O95339, O95627, Q8IYM7, Q8NCL0, Q9NPQ1, Q9NZN6 | Gene names | CNOT4, NOT4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
RFX1_HUMAN
|
||||||
| NC score | 0.013667 (rank : 73) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P22670 | Gene names | RFX1 | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II regulatory factor RFX1 (RFX) (Enhancer factor C) (EF-C). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.013427 (rank : 74) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
ANXA7_HUMAN
|
||||||
| NC score | 0.013211 (rank : 75) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P20073 | Gene names | ANXA7, ANX7, SNX | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A7 (Annexin VII) (Synexin). | |||||
|
NR4A1_HUMAN
|
||||||
| NC score | 0.013038 (rank : 76) | θ value | 0.0330416 (rank : 19) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P22736 | Gene names | NR4A1, GFRP1, HMR, NAK1 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A1 (Orphan nuclear receptor HMR) (Early response protein NAK1) (TR3 orphan receptor) (ST-59). | |||||
|
ZN198_HUMAN
|
||||||
| NC score | 0.012890 (rank : 77) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UBW7, O43212, O43434, O60898, Q5W0Q4, Q63HP0, Q8NE39, Q9H538, Q9UEU2 | Gene names | ZNF198, FIM, RAMP, ZMYM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 198 (Zinc finger MYM-type protein 2) (Fused in myeloproliferative disorders protein) (Rearranged in atypical myeloproliferative disorder protein). | |||||
|
ZFAN5_MOUSE
|
||||||
| NC score | 0.012680 (rank : 78) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88878 | Gene names | Zfand5, Za20d2, Zfp216, Znf216 | |||
|
Domain Architecture |

|
|||||
| Description | AN1-type zinc finger protein 5 (Zinc finger A20 domain-containing protein 2) (Zinc finger protein 216). | |||||
|
ZCH14_HUMAN
|
||||||
| NC score | 0.011994 (rank : 79) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WYQ9, O60324, Q9UFP0 | Gene names | ZCCHC14, KIAA0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
SMAD4_MOUSE
|
||||||
| NC score | 0.011577 (rank : 80) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
COE1_HUMAN
|
||||||
| NC score | 0.010283 (rank : 81) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UH73, Q8IW11 | Gene names | EBF1, COE1, EBF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE1 (OE-1) (O/E-1) (Early B-cell factor). | |||||
|
COE1_MOUSE
|
||||||
| NC score | 0.010283 (rank : 82) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07802 | Gene names | Ebf1, Coe1, Ebf | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE1 (OE-1) (O/E-1) (Early B-cell factor). | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.008755 (rank : 83) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
SEM6D_HUMAN
|
||||||
| NC score | 0.007689 (rank : 84) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NFY4, Q8NFY3, Q8NFY5, Q8NFY6, Q8NFY7, Q9P249 | Gene names | SEMA6D, KIAA1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SEM6D_MOUSE
|
||||||
| NC score | 0.007207 (rank : 85) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q76KF0, Q76KF1, Q76KF2, Q76KF3, Q76KF4, Q80TD0 | Gene names | Sema6d, Kiaa1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
FOXE2_HUMAN
|
||||||
| NC score | 0.006312 (rank : 86) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99526 | Gene names | FOXE2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein E2 (HNF-3/fork head-like protein 5) (HFKL5) (HFKH4). | |||||
|
CUZD1_MOUSE
|
||||||
| NC score | 0.005983 (rank : 87) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70412, Q9CTZ7 | Gene names | Cuzd1, Itmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Integral membrane-associated protein 1). | |||||
|
S12A5_HUMAN
|
||||||
| NC score | 0.004047 (rank : 88) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H2X9, Q5VZ41, Q9H4Z0, Q9ULP4 | Gene names | SLC12A5, KCC2, KIAA1176 | |||
|
Domain Architecture |

|
|||||
| Description | Solute carrier family 12 member 5 (Electroneutral potassium-chloride cotransporter 2) (Erythroid K-Cl cotransporter 2) (Neuronal K-Cl cotransporter) (hKCC2). | |||||
|
HXA5_HUMAN
|
||||||
| NC score | 0.002762 (rank : 89) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20719, O43367, Q96CY6 | Gene names | HOXA5, HOX1C | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A5 (Hox-1C). | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.001083 (rank : 90) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.000829 (rank : 91) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
ZN560_HUMAN
|
||||||
| NC score | -0.000629 (rank : 92) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96MR9, Q495S9, Q495T1 | Gene names | ZNF560 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 560. | |||||
|
ZIC1_MOUSE
|
||||||
| NC score | -0.000951 (rank : 93) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P46684 | Gene names | Zic1, Zic | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZIC 1 (Zinc finger protein of the cerebellum 1). | |||||
|
K0999_MOUSE
|
||||||
| NC score | -0.001157 (rank : 94) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6P4S6, Q641L5, Q66JZ5, Q6ZQ09, Q8K075, Q9CYD5 | Gene names | Kiaa0999 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized serine/threonine-protein kinase KIAA0999 (EC 2.7.11.1). | |||||
|
PLK4_HUMAN
|
||||||
| NC score | -0.001378 (rank : 95) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00444, Q8IYF0, Q96Q95 | Gene names | PLK4, SAK, STK18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase PLK4 (EC 2.7.11.21) (Polo-like kinase 4) (PLK-4) (Serine/threonine-protein kinase Sak) (Serine/threonine- protein kinase 18). | |||||