Please be patient as the page loads
|
SEM6D_HUMAN
|
||||||
| SwissProt Accessions | Q8NFY4, Q8NFY3, Q8NFY5, Q8NFY6, Q8NFY7, Q9P249 | Gene names | SEMA6D, KIAA1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SEM6A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.987149 (rank : 3) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9H2E6, Q9P2H9 | Gene names | SEMA6A, KIAA1368 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1). | |||||
|
SEM6A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.987146 (rank : 4) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O35464, Q6P5A8, Q6PCN9, Q9EQ71 | Gene names | Sema6a, Semaq | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1) (Semaphorin Q) (Sema Q). | |||||
|
SEM6D_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 169 | |
| SwissProt Accessions | Q8NFY4, Q8NFY3, Q8NFY5, Q8NFY6, Q8NFY7, Q9P249 | Gene names | SEMA6D, KIAA1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SEM6D_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.998164 (rank : 2) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 131 | |
| SwissProt Accessions | Q76KF0, Q76KF1, Q76KF2, Q76KF3, Q76KF4, Q80TD0 | Gene names | Sema6d, Kiaa1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SEM6B_MOUSE
|
||||||
| θ value | 2.93876e-176 (rank : 5) | NC score | 0.977636 (rank : 7) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O54951 | Gene names | Sema6b, Seman | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin VIB) (Sema VIB) (Semaphorin N) (Sema N). | |||||
|
SEM6B_HUMAN
|
||||||
| θ value | 5.54228e-175 (rank : 6) | NC score | 0.980039 (rank : 5) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9H3T3, Q9NRK9 | Gene names | SEMA6B, SEMAZ | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin Z) (Sema Z). | |||||
|
SEM6C_MOUSE
|
||||||
| θ value | 6.56209e-168 (rank : 7) | NC score | 0.965107 (rank : 8) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SEM6C_HUMAN
|
||||||
| θ value | 2.75699e-166 (rank : 8) | NC score | 0.979582 (rank : 6) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9H3T2, Q5JR71, Q8WXT8, Q8WXT9, Q8WXU0, Q96JF8 | Gene names | SEMA6C, KIAA1869, SEMAY | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SEM3D_MOUSE
|
||||||
| θ value | 2.16048e-86 (rank : 9) | NC score | 0.939010 (rank : 28) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BH34 | Gene names | Sema3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-3D precursor. | |||||
|
SEM3D_HUMAN
|
||||||
| θ value | 3.11973e-85 (rank : 10) | NC score | 0.939902 (rank : 25) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O95025, Q6UW77, Q8NCQ1 | Gene names | SEMA3D | |||
|
Domain Architecture |
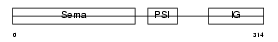
|
|||||
| Description | Semaphorin-3D precursor. | |||||
|
SEM5B_MOUSE
|
||||||
| θ value | 5.32145e-85 (rank : 11) | NC score | 0.799368 (rank : 37) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q60519 | Gene names | Sema5b, Semag, SemG | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5B precursor (Semaphorin G) (Sema G). | |||||
|
SEM5B_HUMAN
|
||||||
| θ value | 4.50487e-84 (rank : 12) | NC score | 0.801896 (rank : 35) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9P283, Q6UY12 | Gene names | SEMA5B, KIAA1445 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5B precursor. | |||||
|
SEM3A_HUMAN
|
||||||
| θ value | 8.49582e-83 (rank : 13) | NC score | 0.940788 (rank : 21) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q14563 | Gene names | SEMA3A | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3A precursor (Semaphorin III) (Sema III). | |||||
|
SEM3A_MOUSE
|
||||||
| θ value | 2.47191e-82 (rank : 14) | NC score | 0.940768 (rank : 22) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O08665, Q5BL08, Q62180, Q62215 | Gene names | Sema3a, Semad, SemD | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3A precursor (Semaphorin III) (Sema III) (Semaphorin D) (Sema D). | |||||
|
SEM3E_HUMAN
|
||||||
| θ value | 7.95186e-81 (rank : 15) | NC score | 0.941127 (rank : 18) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O15041, Q75M94, Q75M97 | Gene names | SEMA3E, KIAA0331 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-3E precursor. | |||||
|
SEM5A_MOUSE
|
||||||
| θ value | 3.94648e-80 (rank : 16) | NC score | 0.801729 (rank : 36) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62217 | Gene names | Sema5a, Semaf, SemF | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5A precursor (Semaphorin F) (Sema F). | |||||
|
SEM3F_MOUSE
|
||||||
| θ value | 6.73165e-80 (rank : 17) | NC score | 0.940972 (rank : 20) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O88632, O88633 | Gene names | Sema3f | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3F precursor (Semaphorin IV) (Sema IV). | |||||
|
SEM5A_HUMAN
|
||||||
| θ value | 8.79181e-80 (rank : 18) | NC score | 0.796423 (rank : 38) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13591, O60408, Q1RLL9 | Gene names | SEMA5A, SEMAF | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5A precursor (Semaphorin F) (Sema F). | |||||
|
SEM3F_HUMAN
|
||||||
| θ value | 1.95861e-79 (rank : 19) | NC score | 0.940005 (rank : 24) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13275, Q13274, Q13372, Q15704 | Gene names | SEMA3F | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3F precursor (Semaphorin IV) (Sema IV) (Sema III/F). | |||||
|
SEM3B_HUMAN
|
||||||
| θ value | 6.30065e-78 (rank : 20) | NC score | 0.944653 (rank : 9) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q13214, Q8TB71, Q8TDV7, Q93018, Q96GX0 | Gene names | SEMA3B, SEMA5 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3B precursor (Semaphorin V) (Sema V) (Sema A(V)). | |||||
|
SEM3C_HUMAN
|
||||||
| θ value | 8.22888e-78 (rank : 21) | NC score | 0.938227 (rank : 30) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q99985 | Gene names | SEMA3C, SEMAE | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3C precursor (Semaphorin E) (Sema E). | |||||
|
SEM3C_MOUSE
|
||||||
| θ value | 2.39424e-77 (rank : 22) | NC score | 0.938388 (rank : 29) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62181 | Gene names | Sema3c, Semae, SemE | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3C precursor (Semaphorin E) (Sema E). | |||||
|
SEM3E_MOUSE
|
||||||
| θ value | 2.39424e-77 (rank : 23) | NC score | 0.939282 (rank : 26) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70275, O09078, O09079 | Gene names | Sema3e, Semah, Semh | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3E precursor (Semaphorin H) (Sema H). | |||||
|
SEM4D_MOUSE
|
||||||
| θ value | 5.8972e-76 (rank : 24) | NC score | 0.942048 (rank : 17) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O09126 | Gene names | Sema4d, Semacl2, Semaj | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Semaphorin J) (Sema J) (Semaphorin C-like 2) (M-Sema G). | |||||
|
SEM4D_HUMAN
|
||||||
| θ value | 1.71583e-75 (rank : 25) | NC score | 0.940590 (rank : 23) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q92854, Q7Z5S4 | Gene names | SEMA4D, CD100 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Leukocyte activation antigen CD100) (BB18) (A8) (GR3). | |||||
|
SEM3B_MOUSE
|
||||||
| θ value | 2.73936e-73 (rank : 26) | NC score | 0.943864 (rank : 10) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62177 | Gene names | Sema3b, Sema, Semaa | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3B precursor (Semaphorin A) (Sema A). | |||||
|
SEM4C_HUMAN
|
||||||
| θ value | 1.15091e-71 (rank : 27) | NC score | 0.943265 (rank : 13) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9C0C4, Q7Z5X0 | Gene names | SEMA4C, KIAA1739 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4C precursor. | |||||
|
SEM4C_MOUSE
|
||||||
| θ value | 1.50314e-71 (rank : 28) | NC score | 0.943679 (rank : 11) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q64151 | Gene names | Sema4c, Semacl1, Semai | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4C precursor (Semaphorin I) (Sema I) (Semaphorin C-like 1) (M-Sema F). | |||||
|
SEM4B_HUMAN
|
||||||
| θ value | 1.27248e-70 (rank : 29) | NC score | 0.943173 (rank : 14) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NPR2, Q6UXE3, Q8WVP9, Q96FK5, Q9C0B8, Q9H691, Q9NPM8, Q9NPN0 | Gene names | SEMA4B, KIAA1745 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4B precursor. | |||||
|
SEM4B_MOUSE
|
||||||
| θ value | 2.3998e-69 (rank : 30) | NC score | 0.942386 (rank : 15) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62179, Q4PKI6, Q69ZB7 | Gene names | Sema4b, Kiaa1745, Semac, SemC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-4B precursor (Semaphorin C) (Sema C). | |||||
|
SEM4A_HUMAN
|
||||||
| θ value | 2.40537e-61 (rank : 31) | NC score | 0.943663 (rank : 12) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H3S1, Q8WUA9 | Gene names | SEMA4A, SEMB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
SEM4F_MOUSE
|
||||||
| θ value | 3.14151e-61 (rank : 32) | NC score | 0.941101 (rank : 19) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Z123, Q9R1Y1 | Gene names | Sema4f, Semaw | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4F precursor (Semaphorin W) (Sema W). | |||||
|
SEM4A_MOUSE
|
||||||
| θ value | 1.01059e-59 (rank : 33) | NC score | 0.942294 (rank : 16) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62178 | Gene names | Sema4a, Semab, SemB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
SEM4G_MOUSE
|
||||||
| θ value | 2.25136e-59 (rank : 34) | NC score | 0.934338 (rank : 32) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9WUH7 | Gene names | Sema4g | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
SEM4F_HUMAN
|
||||||
| θ value | 2.48917e-58 (rank : 35) | NC score | 0.939247 (rank : 27) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O95754, Q9NS35 | Gene names | SEMA4F, SEMAM, SEMAW | |||
|
Domain Architecture |
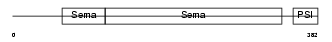
|
|||||
| Description | Semaphorin-4F precursor (Semaphorin W) (Sema W) (Semaphorin M) (Sema M). | |||||
|
SEM4G_HUMAN
|
||||||
| θ value | 2.48917e-58 (rank : 36) | NC score | 0.935683 (rank : 31) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9NTN9, Q9HCF3 | Gene names | SEMA4G, KIAA1619 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
SEM7A_HUMAN
|
||||||
| θ value | 2.59387e-31 (rank : 37) | NC score | 0.908821 (rank : 33) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75326 | Gene names | SEMA7A, CD108, SEMAL | |||
|
Domain Architecture |
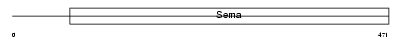
|
|||||
| Description | Semaphorin-7A precursor (Semaphorin L) (Sema L) (Semaphorin K1) (Sema K1) (John-Milton-Hargen human blood group Ag) (JMH blood group antigen) (CD108 antigen) (CDw108). | |||||
|
SEM7A_MOUSE
|
||||||
| θ value | 1.28732e-30 (rank : 38) | NC score | 0.907032 (rank : 34) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9QUR8, O88371 | Gene names | Sema7a, Cd108, Semal, Semk1 | |||
|
Domain Architecture |
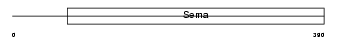
|
|||||
| Description | Semaphorin-7A precursor (Semaphorin L) (Sema L) (Semaphorin K1) (Sema K1) (CD108 antigen) (CDw108). | |||||
|
PLXB2_HUMAN
|
||||||
| θ value | 5.08577e-11 (rank : 39) | NC score | 0.412288 (rank : 39) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O15031, Q7KZU3, Q9BSU7 | Gene names | PLXNB2, KIAA0315 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B2 precursor (MM1). | |||||
|
PLXB1_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 40) | NC score | 0.354664 (rank : 47) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
PLXA2_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 41) | NC score | 0.362598 (rank : 43) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75051, Q6UX61, Q96GN9, Q9BRL1, Q9UIW1 | Gene names | PLXNA2, KIAA0463, OCT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A2 precursor (Semaphorin receptor OCT). | |||||
|
PLXA4_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 42) | NC score | 0.376109 (rank : 41) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q80UG2, Q5DTW8, Q8BKK9 | Gene names | Plxna4, Kiaa1550 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
PLXA1_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 43) | NC score | 0.348858 (rank : 49) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UIW2 | Gene names | PLXNA1, NOV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Semaphorin receptor NOV). | |||||
|
PLXB1_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 44) | NC score | 0.355708 (rank : 46) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
PLXA4_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 45) | NC score | 0.377097 (rank : 40) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9HCM2, Q6UWC6, Q6ZW89, Q8N969, Q8ND00, Q8NEN3, Q9NTD4 | Gene names | PLXNA4, KIAA1550, PLXNA4A, PLXNA4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
PLXA1_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 46) | NC score | 0.354475 (rank : 48) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70206, Q5DTR0 | Gene names | Plxna1, KIAA4053 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Plexin-1) (Plex 1). | |||||
|
PLXA2_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 47) | NC score | 0.359589 (rank : 45) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70207, Q6NVE6, Q6PHN4, Q80TZ7, Q8R1I4 | Gene names | Plxna2, Kiaa0463 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A2 precursor (Plexin-2) (Plex 2). | |||||
|
PLXA3_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 48) | NC score | 0.343729 (rank : 50) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P51805 | Gene names | PLXNA3, PLXN4, SEX | |||
|
Domain Architecture |

|
|||||
| Description | Plexin-A3 precursor (Plexin-4) (Semaphorin receptor SEX). | |||||
|
PLXB3_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 49) | NC score | 0.321348 (rank : 52) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9ULL4, Q9HDA4 | Gene names | PLXNB3, KIAA1206 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor. | |||||
|
PLXD1_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 50) | NC score | 0.307362 (rank : 53) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y4D7, Q6PJS9, Q8IZJ2, Q9BTQ2 | Gene names | PLXND1, KIAA0620 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-D1 precursor. | |||||
|
PLXC1_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 51) | NC score | 0.372664 (rank : 42) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9QZC2, Q8CGW1 | Gene names | Plxnc1, Vespr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
|
PHAR2_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 52) | NC score | 0.054308 (rank : 68) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75167, Q68DM2 | Gene names | PHACTR2, C6orf56, KIAA0680 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 2. | |||||
|
PLXB3_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 53) | NC score | 0.322309 (rank : 51) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9QY40, Q80TH8 | Gene names | Plxnb3, Kiaa1206, Plxn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor (Plexin-6). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 54) | NC score | 0.047126 (rank : 79) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 55) | NC score | 0.042215 (rank : 82) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
CT032_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 56) | NC score | 0.036987 (rank : 84) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NQ75, Q96K09, Q9BYL5 | Gene names | HEFL, C20orf32 | |||
|
Domain Architecture |
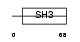
|
|||||
| Description | HEF-like protein. | |||||
|
PLXC1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 57) | NC score | 0.362428 (rank : 44) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O60486, Q59H25 | Gene names | PLXNC1, VESPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 58) | NC score | 0.040061 (rank : 83) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
DDEF1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 59) | NC score | 0.017476 (rank : 119) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
WIRE_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 60) | NC score | 0.054203 (rank : 69) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
TLE1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 61) | NC score | 0.021725 (rank : 110) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62440 | Gene names | Tle1, Grg1 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 1 (Groucho-related protein 1) (Grg- 1). | |||||
|
MBB1A_MOUSE
|
||||||
| θ value | 0.163984 (rank : 62) | NC score | 0.031744 (rank : 90) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 0.163984 (rank : 63) | NC score | 0.033056 (rank : 87) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.163984 (rank : 64) | NC score | 0.025984 (rank : 100) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | 0.163984 (rank : 65) | NC score | 0.045787 (rank : 80) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
ABI1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 66) | NC score | 0.015155 (rank : 130) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CBW3, Q60747, Q91ZM5, Q923I9, Q99KH4 | Gene names | Abi1, Ssh3bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 1 (Abelson interactor 1) (Abi-1) (Spectrin SH3 domain- binding protein 1) (Eps8 SH3 domain-binding protein) (Eps8-binding protein) (e3B1) (Ablphilin-1). | |||||
|
RPB1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 67) | NC score | 0.024880 (rank : 102) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 68) | NC score | 0.024907 (rank : 101) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
PKN1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 69) | NC score | -0.003580 (rank : 189) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P70268, Q7TST2 | Gene names | Pkn1, Pkn, Prk1, Prkcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N1 (EC 2.7.11.13) (Protein kinase C- like 1) (Protein-kinase C-related kinase 1) (Protein kinase C-like PKN) (Serine-threonine protein kinase N). | |||||
|
SOX30_MOUSE
|
||||||
| θ value | 0.279714 (rank : 70) | NC score | 0.022125 (rank : 109) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
TLE1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 71) | NC score | 0.020666 (rank : 111) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q04724, Q5T3G4, Q969V9 | Gene names | TLE1 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 1 (ESG1) (E(Sp1) homolog). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 72) | NC score | 0.018682 (rank : 116) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
ZN473_MOUSE
|
||||||
| θ value | 0.279714 (rank : 73) | NC score | -0.004598 (rank : 191) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 824 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BI67, Q8BI98, Q8BIB7 | Gene names | Znf473, Zfp100, Zfp473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 473 homolog (Zinc finger protein 100) (Zfp-100). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 0.365318 (rank : 74) | NC score | 0.015554 (rank : 128) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 75) | NC score | 0.018199 (rank : 118) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 76) | NC score | 0.026393 (rank : 99) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
TLE4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 77) | NC score | 0.018771 (rank : 115) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62441, Q9JKQ9 | Gene names | Tle4, Grg4 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 4 (Groucho-related protein 4) (Grg- 4). | |||||
|
ZDH17_HUMAN
|
||||||
| θ value | 0.47712 (rank : 78) | NC score | 0.010826 (rank : 144) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IUH5, O75407, Q7Z2I0, Q86W89, Q86YK0, Q9P088, Q9UPZ8 | Gene names | ZDHHC17, HIP14, HIP3, KIAA0946 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC17 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 17) (DHHC-17) (Huntingtin-interacting protein 14) (Huntingtin-interacting protein 3) (Huntingtin-interacting protein HYPH) (Putative NF-kappa-B-activating protein 205) (Putative MAPK- activating protein PM11). | |||||
|
ZDH17_MOUSE
|
||||||
| θ value | 0.47712 (rank : 79) | NC score | 0.011033 (rank : 143) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80TN5, Q8BJ08, Q8BYQ2, Q8BYQ6 | Gene names | Zdhhc17, Hip14, Kiaa0946 | |||
|
Domain Architecture |
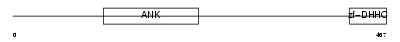
|
|||||
| Description | Palmitoyltransferase ZDHHC17 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 17) (DHHC-17) (Huntingtin-interacting protein 14). | |||||
|
FGD1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 80) | NC score | 0.020453 (rank : 112) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 0.62314 (rank : 81) | NC score | 0.043563 (rank : 81) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
SOS1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 82) | NC score | 0.015828 (rank : 126) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
SOS1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 83) | NC score | 0.016541 (rank : 124) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
WASF3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 84) | NC score | 0.023294 (rank : 105) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UPY6, O94974 | Gene names | WASF3, KIAA0900, SCAR3, WAVE3 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein family member 3 (WASP-family protein member 3) (Protein WAVE-3) (Verprolin homology domain-containing protein 3). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 85) | NC score | 0.026887 (rank : 98) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 86) | NC score | 0.024477 (rank : 103) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CIC_MOUSE
|
||||||
| θ value | 0.813845 (rank : 87) | NC score | 0.023449 (rank : 104) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 88) | NC score | 0.031294 (rank : 91) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
NFRKB_MOUSE
|
||||||
| θ value | 0.813845 (rank : 89) | NC score | 0.031922 (rank : 89) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
PAPOA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 90) | NC score | 0.014346 (rank : 133) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P51003, Q86SX4, Q86TV0, Q8IYF5, Q9BVU2 | Gene names | PAPOLA, PAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase alpha). | |||||
|
RIN3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 91) | NC score | 0.016546 (rank : 123) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P59729 | Gene names | Rin3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
BRCA2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 92) | NC score | 0.011459 (rank : 139) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51587, O00183, O15008, Q13879 | Gene names | BRCA2, FANCD1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein (Fanconi anemia group D1 protein). | |||||
|
GLIS1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 93) | NC score | -0.003912 (rank : 190) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NBF1, Q8N9V9 | Gene names | GLIS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS1 (GLI-similar 1). | |||||
|
NFRKB_HUMAN
|
||||||
| θ value | 1.06291 (rank : 94) | NC score | 0.030056 (rank : 94) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6P4R8, Q12869, Q15312, Q9H048 | Gene names | NFRKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 95) | NC score | 0.033267 (rank : 86) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
AHTF1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 96) | NC score | 0.016509 (rank : 125) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WYP5, Q7Z4E3, Q8IZA4, Q96EH9, Q9Y4Q6 | Gene names | AHCTF1, ELYS, TMBS62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 97) | NC score | 0.002895 (rank : 173) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
DAB2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 98) | NC score | 0.022748 (rank : 106) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
FGD1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 99) | NC score | 0.016830 (rank : 121) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
GGN_MOUSE
|
||||||
| θ value | 1.38821 (rank : 100) | NC score | 0.036664 (rank : 85) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.38821 (rank : 101) | NC score | 0.022198 (rank : 108) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
ABI1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 102) | NC score | 0.011683 (rank : 137) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IZP0, O15147, O76049, O95060, Q5W070, Q5W072, Q8TB63, Q96S81, Q9NXZ9, Q9NYB8 | Gene names | ABI1, SSH3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 1 (Abelson interactor 1) (Abi-1) (Spectrin SH3 domain- binding protein 1) (Eps8 SH3 domain-binding protein) (Eps8-binding protein) (e3B1) (Nap1-binding protein) (Nap1BP) (Abl-binding protein 4) (AblBP4). | |||||
|
EDC3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 103) | NC score | 0.026940 (rank : 97) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96F86, Q9H797 | Gene names | EDC3, YJDC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of mRNA decapping protein 3 (YjeF domain-containing protein 1). | |||||
|
EDC3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 104) | NC score | 0.027032 (rank : 96) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K2D3, Q3THV7 | Gene names | Edc3, Yjdc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of mRNA decapping protein 3 (YjeF domain-containing protein 1). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 1.81305 (rank : 105) | NC score | 0.014181 (rank : 134) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 106) | NC score | 0.011074 (rank : 142) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
RBMS2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 107) | NC score | 0.009110 (rank : 149) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15434 | Gene names | RBMS2, SCR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 2 (Suppressor of CDC2 with RNA-binding motif 3). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 1.81305 (rank : 108) | NC score | 0.000419 (rank : 182) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TRAM2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 109) | NC score | 0.014120 (rank : 135) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15035 | Gene names | TRAM2, KIAA0057 | |||
|
Domain Architecture |

|
|||||
| Description | Translocation-associated membrane protein 2. | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 110) | NC score | 0.032253 (rank : 88) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
DDEF2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 111) | NC score | 0.007875 (rank : 157) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
DMD_HUMAN
|
||||||
| θ value | 2.36792 (rank : 112) | NC score | 0.000422 (rank : 181) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DMD_MOUSE
|
||||||
| θ value | 2.36792 (rank : 113) | NC score | -0.000100 (rank : 183) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 2.36792 (rank : 114) | NC score | 0.015595 (rank : 127) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 115) | NC score | 0.030153 (rank : 93) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 116) | NC score | 0.030672 (rank : 92) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
TISB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 117) | NC score | 0.015062 (rank : 131) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q07352, Q13851 | Gene names | ZFP36L1, BERG36, BRF1, ERF1, TIS11B | |||
|
Domain Architecture |
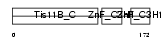
|
|||||
| Description | Butyrate response factor 1 (TIS11B protein) (EGF-response factor 1) (ERF-1). | |||||
|
TLE4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 118) | NC score | 0.016990 (rank : 120) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q04727, Q5T1Y2, Q9BZ07, Q9BZ08, Q9BZ09, Q9NSL3, Q9ULF9 | Gene names | TLE4, KIAA1261 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 4. | |||||
|
TSC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 119) | NC score | 0.003976 (rank : 169) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92574 | Gene names | TSC1, KIAA0243, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Hamartin (Tuberous sclerosis 1 protein). | |||||
|
HXA11_HUMAN
|
||||||
| θ value | 3.0926 (rank : 120) | NC score | 0.000913 (rank : 177) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P31270 | Gene names | HOXA11, HOX1I | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A11 (Hox-1I). | |||||
|
HXA11_MOUSE
|
||||||
| θ value | 3.0926 (rank : 121) | NC score | 0.000941 (rank : 176) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P31311, Q3V026 | Gene names | Hoxa11, Hox-1.9, Hoxa-11 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A11 (Hox-1.9). | |||||
|
MAST2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 122) | NC score | -0.003045 (rank : 188) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 3.0926 (rank : 123) | NC score | 0.014576 (rank : 132) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 3.0926 (rank : 124) | NC score | 0.028876 (rank : 95) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 125) | NC score | 0.009374 (rank : 148) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
DAB2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 126) | NC score | 0.018247 (rank : 117) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P98078, Q3U3K1, Q91W56, Q923E1 | Gene names | Dab2, Doc2 | |||
|
Domain Architecture |
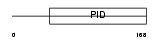
|
|||||
| Description | Disabled homolog 2 (DOC-2) (Mitogen-responsive phosphoprotein). | |||||
|
EYA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 127) | NC score | 0.007689 (rank : 159) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99502 | Gene names | EYA1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
LAMC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 128) | NC score | 0.000687 (rank : 180) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P11047 | Gene names | LAMC1, LAMB2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
MN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 129) | NC score | 0.015513 (rank : 129) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q10571 | Gene names | MN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tumor suppressor protein MN1. | |||||
|
RBM12_HUMAN
|
||||||
| θ value | 4.03905 (rank : 130) | NC score | 0.008891 (rank : 151) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NTZ6, O94865, Q8N3B1, Q9H196 | Gene names | RBM12, KIAA0765 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
RTN1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 131) | NC score | 0.006987 (rank : 164) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K0T0, Q8K4S4 | Gene names | Rtn1, Nsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 4.03905 (rank : 132) | NC score | 0.022707 (rank : 107) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 4.03905 (rank : 133) | NC score | 0.016556 (rank : 122) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 4.03905 (rank : 134) | NC score | 0.007181 (rank : 162) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 135) | NC score | 0.011447 (rank : 140) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | 5.27518 (rank : 136) | NC score | 0.001111 (rank : 175) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CM018_HUMAN
|
||||||
| θ value | 5.27518 (rank : 137) | NC score | 0.011529 (rank : 138) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H714, Q5W051, Q6PJ74, Q6PK94, Q86XH7, Q8N5J6 | Gene names | C13orf18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf18. | |||||
|
K0423_HUMAN
|
||||||
| θ value | 5.27518 (rank : 138) | NC score | 0.013443 (rank : 136) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y4F4, Q68D66, Q6PG27 | Gene names | KIAA0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
MIRO2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 139) | NC score | 0.005942 (rank : 165) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8JZN7 | Gene names | Rhot2, Arht2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial Rho GTPase 2 (EC 3.6.5.-) (MIRO-2) (Ras homolog gene family member T2). | |||||
|
POLS_HUMAN
|
||||||
| θ value | 5.27518 (rank : 140) | NC score | 0.008761 (rank : 152) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5XG87, O43289, Q9Y6C1 | Gene names | POLS, TRF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase sigma (EC 2.7.7.7) (Topoisomerase-related function protein 4-1) (TRF4-1) (LAK-1) (DNA polymerase kappa). | |||||
|
RBM16_HUMAN
|
||||||
| θ value | 5.27518 (rank : 141) | NC score | 0.007727 (rank : 158) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
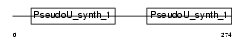
RBMS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 142) | NC score | 0.008045 (rank : 154) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29558, Q15433, Q8WV20 | Gene names | RBMS1, MSSP, MSSP1, SCR2 | |||
|
Domain Architecture |
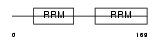
|
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1) (Suppressor of CDC2 with RNA- binding motif 2). | |||||
|
SYP2L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 143) | NC score | 0.004785 (rank : 167) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H987 | Gene names | SYNPO2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
CN032_HUMAN
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.020453 (rank : 113) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | -0.000608 (rank : 185) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
DACT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.011117 (rank : 141) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NYF0, Q86TY0 | Gene names | DACT1, DPR1, HNG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (hDPR1) (Heptacellular carcinoma novel gene 3 protein). | |||||
|
FOXO4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.007196 (rank : 161) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WVH3 | Gene names | Mllt7, Afx, Afx1, Fkhr3, Foxo4 | |||
|
Domain Architecture |
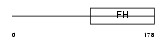
|
|||||
| Description | Fork head domain transcription factor AFX1 (Afxh) (Forkhead box protein O4). | |||||
|
GP112_HUMAN
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.009022 (rank : 150) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
LAMB3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.000840 (rank : 179) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13751, O14947, Q14733, Q9UJK4, Q9UJL1 | Gene names | LAMB3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Laminin B1k chain) (Kalinin B1 chain). | |||||
|
MBD5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.010605 (rank : 145) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P267, Q9NUV6 | Gene names | MBD5, KIAA1461 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 5 (Methyl-CpG-binding protein MBD5). | |||||
|
NHS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.008038 (rank : 155) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6T4R5, Q5J7Q0, Q5J7Q1, Q68DR5 | Gene names | NHS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nance-Horan syndrome protein (Congenital cataracts and dental anomalies protein). | |||||
|
PRKDC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.003219 (rank : 172) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | -0.000275 (rank : 184) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
TRUA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | 0.009859 (rank : 146) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y606, Q8WYT2, Q9BU44 | Gene names | PUS1 | |||
|
Domain Architecture |
|
|||||
| Description | tRNA pseudouridine synthase A (EC 5.4.99.12) (tRNA-uridine isomerase I) (tRNA pseudouridylate synthase I). | |||||
|
YETS2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 155) | NC score | 0.007426 (rank : 160) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
C3AR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | -0.002144 (rank : 186) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O09047, O35951 | Gene names | C3ar1, C3r1 | |||
|
Domain Architecture |

|
|||||
| Description | C3a anaphylatoxin chemotactic receptor (C3a-R) (C3AR) (Complement component 3a receptor 1). | |||||
|
CD2L7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | -0.005083 (rank : 192) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.019740 (rank : 114) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CRTC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.007885 (rank : 156) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
FOXO4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.003615 (rank : 170) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P98177, O43821, Q13720, Q3KPF1, Q8TDK9 | Gene names | MLLT7, AFX, AFX1, FOXO4 | |||
|
Domain Architecture |

|
|||||
| Description | Fork head domain transcription factor AFX1 (Forkhead box protein O4). | |||||
|
HPS4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.009508 (rank : 147) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99KG7 | Gene names | Hps4, Le | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 4 protein homolog (Light-ear protein) (Le protein). | |||||
|
ITCH_MOUSE
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.000895 (rank : 178) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
K0460_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.008334 (rank : 153) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5VT52, O75048, Q5VT53, Q6MZL4, Q86XD2 | Gene names | KIAA0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
LRRC7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | -0.002226 (rank : 187) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TE7 | Gene names | Lrrc7, Kiaa1365, Lap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 7 (Protein LAP1) (Densin-180). | |||||
|
MIRO2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.004154 (rank : 168) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IXI1, Q8NF53, Q96C13, Q96S17, Q9BT60, Q9H7M8 | Gene names | RHOT2, ARHT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial Rho GTPase 2 (EC 3.6.5.-) (MIRO-2) (hMiro-2) (Ras homolog gene family member T2). | |||||
|
PYGO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.005375 (rank : 166) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y3Y4 | Gene names | PYGO1 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 1. | |||||
|
STIM2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.003372 (rank : 171) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P246, Q96BF1, Q9BQH2, Q9H8R1 | Gene names | STIM2, KIAA1482 | |||
|
Domain Architecture |
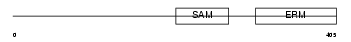
|
|||||
| Description | Stromal interaction molecule 2 precursor. | |||||
|
TISB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.007101 (rank : 163) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23950 | Gene names | Zfp36l1, Brf1, Tis11b | |||
|
Domain Architecture |
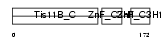
|
|||||
| Description | Butyrate response factor 1 (TIS11B protein). | |||||
|
UBP34_MOUSE
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.001570 (rank : 174) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
BAI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.057380 (rank : 63) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14514 | Gene names | BAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
BAI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.057101 (rank : 65) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3UHD1, Q3UH36, Q8CGM0 | Gene names | Bai1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
BAI2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.053399 (rank : 71) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60241, Q5T6K0, Q8NGW8 | Gene names | BAI2 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 2 precursor. | |||||
|
BAI2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.053350 (rank : 72) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CGM1, Q3TYC8, Q3UN11, Q3UNE2, Q6PGN0 | Gene names | Bai2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 2 precursor. | |||||
|
BAI3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.051652 (rank : 75) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60242, O60297 | Gene names | BAI3, KIAA0550 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 3 precursor. | |||||
|
BAI3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.053620 (rank : 70) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80ZF8 | Gene names | Bai3 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 3 precursor. | |||||
|
PROP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.098705 (rank : 54) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P27918, O15134, O15135, O15136, O75826 | Gene names | CFP, PFC | |||
|
Domain Architecture |
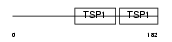
|
|||||
| Description | Properdin precursor (Factor P). | |||||
|
PROP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.096185 (rank : 55) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11680, Q3TB98, Q3U779 | Gene names | Cfp, Pfc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Properdin precursor (Factor P). | |||||
|
SPON1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.071376 (rank : 57) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCB6, O94862, Q8NCD7, Q8WUR5 | Gene names | SPON1, KIAA0762, VSGP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spondin-1 precursor (F-spondin) (Vascular smooth muscle cell growth- promoting factor). | |||||
|
SPON1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.073044 (rank : 56) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VCC9, Q8K2Q8 | Gene names | Spon1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spondin-1 precursor (F-spondin). | |||||
|
SSPO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.055578 (rank : 67) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
THSD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.052498 (rank : 74) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NS62, Q6P3U1, Q6UXZ2 | Gene names | THSD1, TMTSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thrombospondin type-1 domain-containing protein 1 precursor (Transmembrane molecule with thrombospondin module). | |||||
|
TSP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.050417 (rank : 78) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P07996, Q15667 | Gene names | THBS1, TSP, TSP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-1 precursor. | |||||
|
TSP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.050748 (rank : 77) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35441 | Gene names | Thbs1, Tsp1 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-1 precursor. | |||||
|
TSP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.051560 (rank : 76) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35442 | Gene names | THBS2, TSP2 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-2 precursor. | |||||
|
TSP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.052857 (rank : 73) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q03350 | Gene names | Thbs2, Tsp2 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-2 precursor. | |||||
|
UNC5A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.058657 (rank : 62) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K1S4, Q6PEF7, Q80T71 | Gene names | Unc5a, Kiaa1976, Unc5h1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5A precursor (Unc-5 homolog A) (Unc-5 homolog 1). | |||||
|
UNC5B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.061273 (rank : 60) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IZJ1, Q86SN3, Q8N1Y2, Q9H9F3 | Gene names | UNC5B, P53RDL1, UNC5H2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5B precursor (Unc-5 homolog B) (Unc-5 homolog 2) (p53-regulated receptor for death and life protein 1). | |||||
|
UNC5B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.060318 (rank : 61) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K1S3, Q3U4F2, Q6PFH0, Q80Y85, Q9D398 | Gene names | Unc5b, Unc5h2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5B precursor (Unc-5 homolog B) (Unc-5 homolog 2). | |||||
|
UNC5C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.056563 (rank : 66) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95185, Q8IUT0 | Gene names | UNC5C, UNC5H3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5C precursor (Unc-5 homolog C) (Unc-5 homolog 3). | |||||
|
UNC5C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.057373 (rank : 64) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08747, Q8CD16 | Gene names | Unc5c, Rcm, Unc5h3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5C precursor (Unc-5 homolog C) (Unc-5 homolog 3) (Rostral cerebellar malformation protein). | |||||
|
UNC5D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.068188 (rank : 58) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6UXZ4, Q8WYP7 | Gene names | UNC5D, KIAA1777, UNC5H4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5D precursor (Unc-5 homolog D) (Unc-5 homolog 4). | |||||
|
UNC5D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.068064 (rank : 59) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K1S2 | Gene names | Unc5d, Unc5h4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5D precursor (Unc-5 homolog D) (Unc-5 homolog 4). | |||||
|
SEM6D_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 169 | |
| SwissProt Accessions | Q8NFY4, Q8NFY3, Q8NFY5, Q8NFY6, Q8NFY7, Q9P249 | Gene names | SEMA6D, KIAA1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SEM6D_MOUSE
|
||||||
| NC score | 0.998164 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 131 | |
| SwissProt Accessions | Q76KF0, Q76KF1, Q76KF2, Q76KF3, Q76KF4, Q80TD0 | Gene names | Sema6d, Kiaa1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SEM6A_HUMAN
|
||||||
| NC score | 0.987149 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9H2E6, Q9P2H9 | Gene names | SEMA6A, KIAA1368 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1). | |||||
|
SEM6A_MOUSE
|
||||||
| NC score | 0.987146 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O35464, Q6P5A8, Q6PCN9, Q9EQ71 | Gene names | Sema6a, Semaq | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1) (Semaphorin Q) (Sema Q). | |||||
|
SEM6B_HUMAN
|
||||||
| NC score | 0.980039 (rank : 5) | θ value | 5.54228e-175 (rank : 6) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9H3T3, Q9NRK9 | Gene names | SEMA6B, SEMAZ | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin Z) (Sema Z). | |||||
|
SEM6C_HUMAN
|
||||||
| NC score | 0.979582 (rank : 6) | θ value | 2.75699e-166 (rank : 8) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9H3T2, Q5JR71, Q8WXT8, Q8WXT9, Q8WXU0, Q96JF8 | Gene names | SEMA6C, KIAA1869, SEMAY | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SEM6B_MOUSE
|
||||||
| NC score | 0.977636 (rank : 7) | θ value | 2.93876e-176 (rank : 5) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O54951 | Gene names | Sema6b, Seman | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin VIB) (Sema VIB) (Semaphorin N) (Sema N). | |||||
|
SEM6C_MOUSE
|
||||||
| NC score | 0.965107 (rank : 8) | θ value | 6.56209e-168 (rank : 7) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SEM3B_HUMAN
|
||||||
| NC score | 0.944653 (rank : 9) | θ value | 6.30065e-78 (rank : 20) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q13214, Q8TB71, Q8TDV7, Q93018, Q96GX0 | Gene names | SEMA3B, SEMA5 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3B precursor (Semaphorin V) (Sema V) (Sema A(V)). | |||||
|
SEM3B_MOUSE
|
||||||
| NC score | 0.943864 (rank : 10) | θ value | 2.73936e-73 (rank : 26) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62177 | Gene names | Sema3b, Sema, Semaa | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3B precursor (Semaphorin A) (Sema A). | |||||
|
SEM4C_MOUSE
|
||||||
| NC score | 0.943679 (rank : 11) | θ value | 1.50314e-71 (rank : 28) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q64151 | Gene names | Sema4c, Semacl1, Semai | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4C precursor (Semaphorin I) (Sema I) (Semaphorin C-like 1) (M-Sema F). | |||||
|
SEM4A_HUMAN
|
||||||
| NC score | 0.943663 (rank : 12) | θ value | 2.40537e-61 (rank : 31) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H3S1, Q8WUA9 | Gene names | SEMA4A, SEMB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
SEM4C_HUMAN
|
||||||
| NC score | 0.943265 (rank : 13) | θ value | 1.15091e-71 (rank : 27) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9C0C4, Q7Z5X0 | Gene names | SEMA4C, KIAA1739 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4C precursor. | |||||
|
SEM4B_HUMAN
|
||||||
| NC score | 0.943173 (rank : 14) | θ value | 1.27248e-70 (rank : 29) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NPR2, Q6UXE3, Q8WVP9, Q96FK5, Q9C0B8, Q9H691, Q9NPM8, Q9NPN0 | Gene names | SEMA4B, KIAA1745 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4B precursor. | |||||
|
SEM4B_MOUSE
|
||||||
| NC score | 0.942386 (rank : 15) | θ value | 2.3998e-69 (rank : 30) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62179, Q4PKI6, Q69ZB7 | Gene names | Sema4b, Kiaa1745, Semac, SemC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-4B precursor (Semaphorin C) (Sema C). | |||||
|
SEM4A_MOUSE
|
||||||
| NC score | 0.942294 (rank : 16) | θ value | 1.01059e-59 (rank : 33) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62178 | Gene names | Sema4a, Semab, SemB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
SEM4D_MOUSE
|
||||||
| NC score | 0.942048 (rank : 17) | θ value | 5.8972e-76 (rank : 24) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O09126 | Gene names | Sema4d, Semacl2, Semaj | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Semaphorin J) (Sema J) (Semaphorin C-like 2) (M-Sema G). | |||||
|
SEM3E_HUMAN
|
||||||
| NC score | 0.941127 (rank : 18) | θ value | 7.95186e-81 (rank : 15) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O15041, Q75M94, Q75M97 | Gene names | SEMA3E, KIAA0331 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-3E precursor. | |||||
|
SEM4F_MOUSE
|
||||||
| NC score | 0.941101 (rank : 19) | θ value | 3.14151e-61 (rank : 32) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9Z123, Q9R1Y1 | Gene names | Sema4f, Semaw | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4F precursor (Semaphorin W) (Sema W). | |||||
|
SEM3F_MOUSE
|
||||||
| NC score | 0.940972 (rank : 20) | θ value | 6.73165e-80 (rank : 17) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O88632, O88633 | Gene names | Sema3f | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3F precursor (Semaphorin IV) (Sema IV). | |||||
|
SEM3A_HUMAN
|
||||||
| NC score | 0.940788 (rank : 21) | θ value | 8.49582e-83 (rank : 13) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q14563 | Gene names | SEMA3A | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3A precursor (Semaphorin III) (Sema III). | |||||
|
SEM3A_MOUSE
|
||||||
| NC score | 0.940768 (rank : 22) | θ value | 2.47191e-82 (rank : 14) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O08665, Q5BL08, Q62180, Q62215 | Gene names | Sema3a, Semad, SemD | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3A precursor (Semaphorin III) (Sema III) (Semaphorin D) (Sema D). | |||||
|
SEM4D_HUMAN
|
||||||
| NC score | 0.940590 (rank : 23) | θ value | 1.71583e-75 (rank : 25) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q92854, Q7Z5S4 | Gene names | SEMA4D, CD100 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Leukocyte activation antigen CD100) (BB18) (A8) (GR3). | |||||
|
SEM3F_HUMAN
|
||||||
| NC score | 0.940005 (rank : 24) | θ value | 1.95861e-79 (rank : 19) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13275, Q13274, Q13372, Q15704 | Gene names | SEMA3F | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3F precursor (Semaphorin IV) (Sema IV) (Sema III/F). | |||||
|
SEM3D_HUMAN
|
||||||
| NC score | 0.939902 (rank : 25) | θ value | 3.11973e-85 (rank : 10) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O95025, Q6UW77, Q8NCQ1 | Gene names | SEMA3D | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3D precursor. | |||||
|
SEM3E_MOUSE
|
||||||
| NC score | 0.939282 (rank : 26) | θ value | 2.39424e-77 (rank : 23) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70275, O09078, O09079 | Gene names | Sema3e, Semah, Semh | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3E precursor (Semaphorin H) (Sema H). | |||||
|
SEM4F_HUMAN
|
||||||
| NC score | 0.939247 (rank : 27) | θ value | 2.48917e-58 (rank : 35) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O95754, Q9NS35 | Gene names | SEMA4F, SEMAM, SEMAW | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4F precursor (Semaphorin W) (Sema W) (Semaphorin M) (Sema M). | |||||
|
SEM3D_MOUSE
|
||||||
| NC score | 0.939010 (rank : 28) | θ value | 2.16048e-86 (rank : 9) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BH34 | Gene names | Sema3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-3D precursor. | |||||
|
SEM3C_MOUSE
|
||||||
| NC score | 0.938388 (rank : 29) | θ value | 2.39424e-77 (rank : 22) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62181 | Gene names | Sema3c, Semae, SemE | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3C precursor (Semaphorin E) (Sema E). | |||||
|
SEM3C_HUMAN
|
||||||
| NC score | 0.938227 (rank : 30) | θ value | 8.22888e-78 (rank : 21) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q99985 | Gene names | SEMA3C, SEMAE | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3C precursor (Semaphorin E) (Sema E). | |||||
|
SEM4G_HUMAN
|
||||||
| NC score | 0.935683 (rank : 31) | θ value | 2.48917e-58 (rank : 36) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9NTN9, Q9HCF3 | Gene names | SEMA4G, KIAA1619 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
SEM4G_MOUSE
|
||||||
| NC score | 0.934338 (rank : 32) | θ value | 2.25136e-59 (rank : 34) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9WUH7 | Gene names | Sema4g | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
SEM7A_HUMAN
|
||||||
| NC score | 0.908821 (rank : 33) | θ value | 2.59387e-31 (rank : 37) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75326 | Gene names | SEMA7A, CD108, SEMAL | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-7A precursor (Semaphorin L) (Sema L) (Semaphorin K1) (Sema K1) (John-Milton-Hargen human blood group Ag) (JMH blood group antigen) (CD108 antigen) (CDw108). | |||||
|
SEM7A_MOUSE
|
||||||
| NC score | 0.907032 (rank : 34) | θ value | 1.28732e-30 (rank : 38) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9QUR8, O88371 | Gene names | Sema7a, Cd108, Semal, Semk1 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-7A precursor (Semaphorin L) (Sema L) (Semaphorin K1) (Sema K1) (CD108 antigen) (CDw108). | |||||
|
SEM5B_HUMAN
|
||||||
| NC score | 0.801896 (rank : 35) | θ value | 4.50487e-84 (rank : 12) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9P283, Q6UY12 | Gene names | SEMA5B, KIAA1445 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5B precursor. | |||||
|
SEM5A_MOUSE
|
||||||
| NC score | 0.801729 (rank : 36) | θ value | 3.94648e-80 (rank : 16) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62217 | Gene names | Sema5a, Semaf, SemF | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5A precursor (Semaphorin F) (Sema F). | |||||
|
SEM5B_MOUSE
|
||||||
| NC score | 0.799368 (rank : 37) | θ value | 5.32145e-85 (rank : 11) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q60519 | Gene names | Sema5b, Semag, SemG | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5B precursor (Semaphorin G) (Sema G). | |||||
|
SEM5A_HUMAN
|
||||||
| NC score | 0.796423 (rank : 38) | θ value | 8.79181e-80 (rank : 18) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13591, O60408, Q1RLL9 | Gene names | SEMA5A, SEMAF | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5A precursor (Semaphorin F) (Sema F). | |||||
|
PLXB2_HUMAN
|
||||||
| NC score | 0.412288 (rank : 39) | θ value | 5.08577e-11 (rank : 39) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O15031, Q7KZU3, Q9BSU7 | Gene names | PLXNB2, KIAA0315 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B2 precursor (MM1). | |||||
|
PLXA4_HUMAN
|
||||||
| NC score | 0.377097 (rank : 40) | θ value | 4.0297e-08 (rank : 45) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9HCM2, Q6UWC6, Q6ZW89, Q8N969, Q8ND00, Q8NEN3, Q9NTD4 | Gene names | PLXNA4, KIAA1550, PLXNA4A, PLXNA4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
PLXA4_MOUSE
|
||||||
| NC score | 0.376109 (rank : 41) | θ value | 1.80886e-08 (rank : 42) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q80UG2, Q5DTW8, Q8BKK9 | Gene names | Plxna4, Kiaa1550 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
PLXC1_MOUSE
|
||||||
| NC score | 0.372664 (rank : 42) | θ value | 0.00102713 (rank : 51) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9QZC2, Q8CGW1 | Gene names | Plxnc1, Vespr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
|
PLXA2_HUMAN
|
||||||
| NC score | 0.362598 (rank : 43) | θ value | 1.06045e-08 (rank : 41) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75051, Q6UX61, Q96GN9, Q9BRL1, Q9UIW1 | Gene names | PLXNA2, KIAA0463, OCT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A2 precursor (Semaphorin receptor OCT). | |||||
|
PLXC1_HUMAN
|
||||||
| NC score | 0.362428 (rank : 44) | θ value | 0.0113563 (rank : 57) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O60486, Q59H25 | Gene names | PLXNC1, VESPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
|
PLXA2_MOUSE
|
||||||
| NC score | 0.359589 (rank : 45) | θ value | 9.92553e-07 (rank : 47) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70207, Q6NVE6, Q6PHN4, Q80TZ7, Q8R1I4 | Gene names | Plxna2, Kiaa0463 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A2 precursor (Plexin-2) (Plex 2). | |||||
|
PLXB1_MOUSE
|
||||||
| NC score | 0.355708 (rank : 46) | θ value | 3.08544e-08 (rank : 44) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
PLXB1_HUMAN
|
||||||
| NC score | 0.354664 (rank : 47) | θ value | 1.63604e-09 (rank : 40) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
PLXA1_MOUSE
|
||||||
| NC score | 0.354475 (rank : 48) | θ value | 1.53129e-07 (rank : 46) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70206, Q5DTR0 | Gene names | Plxna1, KIAA4053 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Plexin-1) (Plex 1). | |||||
|
PLXA1_HUMAN
|
||||||
| NC score | 0.348858 (rank : 49) | θ value | 3.08544e-08 (rank : 43) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UIW2 | Gene names | PLXNA1, NOV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Semaphorin receptor NOV). | |||||
|
PLXA3_HUMAN
|
||||||
| NC score | 0.343729 (rank : 50) | θ value | 1.69304e-06 (rank : 48) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P51805 | Gene names | PLXNA3, PLXN4, SEX | |||
|
Domain Architecture |

|
|||||
| Description | Plexin-A3 precursor (Plexin-4) (Semaphorin receptor SEX). | |||||
|
PLXB3_MOUSE
|
||||||
| NC score | 0.322309 (rank : 51) | θ value | 0.00175202 (rank : 53) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9QY40, Q80TH8 | Gene names | Plxnb3, Kiaa1206, Plxn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor (Plexin-6). | |||||
|
PLXB3_HUMAN
|
||||||
| NC score | 0.321348 (rank : 52) | θ value | 0.000270298 (rank : 49) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9ULL4, Q9HDA4 | Gene names | PLXNB3, KIAA1206 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor. | |||||
|
PLXD1_HUMAN
|
||||||
| NC score | 0.307362 (rank : 53) | θ value | 0.000602161 (rank : 50) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y4D7, Q6PJS9, Q8IZJ2, Q9BTQ2 | Gene names | PLXND1, KIAA0620 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-D1 precursor. | |||||
|
PROP_HUMAN
|
||||||
| NC score | 0.098705 (rank : 54) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P27918, O15134, O15135, O15136, O75826 | Gene names | CFP, PFC | |||
|
Domain Architecture |

|
|||||
| Description | Properdin precursor (Factor P). | |||||
|
PROP_MOUSE
|
||||||
| NC score | 0.096185 (rank : 55) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P11680, Q3TB98, Q3U779 | Gene names | Cfp, Pfc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Properdin precursor (Factor P). | |||||
|
SPON1_MOUSE
|
||||||
| NC score | 0.073044 (rank : 56) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VCC9, Q8K2Q8 | Gene names | Spon1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spondin-1 precursor (F-spondin). | |||||
|
SPON1_HUMAN
|
||||||
| NC score | 0.071376 (rank : 57) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCB6, O94862, Q8NCD7, Q8WUR5 | Gene names | SPON1, KIAA0762, VSGP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spondin-1 precursor (F-spondin) (Vascular smooth muscle cell growth- promoting factor). | |||||
|
UNC5D_HUMAN
|
||||||
| NC score | 0.068188 (rank : 58) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6UXZ4, Q8WYP7 | Gene names | UNC5D, KIAA1777, UNC5H4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5D precursor (Unc-5 homolog D) (Unc-5 homolog 4). | |||||
|
UNC5D_MOUSE
|
||||||
| NC score | 0.068064 (rank : 59) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K1S2 | Gene names | Unc5d, Unc5h4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5D precursor (Unc-5 homolog D) (Unc-5 homolog 4). | |||||
|
UNC5B_HUMAN
|
||||||
| NC score | 0.061273 (rank : 60) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IZJ1, Q86SN3, Q8N1Y2, Q9H9F3 | Gene names | UNC5B, P53RDL1, UNC5H2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5B precursor (Unc-5 homolog B) (Unc-5 homolog 2) (p53-regulated receptor for death and life protein 1). | |||||
|
UNC5B_MOUSE
|
||||||
| NC score | 0.060318 (rank : 61) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K1S3, Q3U4F2, Q6PFH0, Q80Y85, Q9D398 | Gene names | Unc5b, Unc5h2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5B precursor (Unc-5 homolog B) (Unc-5 homolog 2). | |||||
|
UNC5A_MOUSE
|
||||||
| NC score | 0.058657 (rank : 62) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K1S4, Q6PEF7, Q80T71 | Gene names | Unc5a, Kiaa1976, Unc5h1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5A precursor (Unc-5 homolog A) (Unc-5 homolog 1). | |||||
|
BAI1_HUMAN
|
||||||
| NC score | 0.057380 (rank : 63) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O14514 | Gene names | BAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
UNC5C_MOUSE
|
||||||
| NC score | 0.057373 (rank : 64) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08747, Q8CD16 | Gene names | Unc5c, Rcm, Unc5h3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5C precursor (Unc-5 homolog C) (Unc-5 homolog 3) (Rostral cerebellar malformation protein). | |||||
|
BAI1_MOUSE
|
||||||
| NC score | 0.057101 (rank : 65) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3UHD1, Q3UH36, Q8CGM0 | Gene names | Bai1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
UNC5C_HUMAN
|
||||||
| NC score | 0.056563 (rank : 66) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95185, Q8IUT0 | Gene names | UNC5C, UNC5H3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin receptor UNC5C precursor (Unc-5 homolog C) (Unc-5 homolog 3). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.055578 (rank : 67) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
PHAR2_HUMAN
|
||||||
| NC score | 0.054308 (rank : 68) | θ value | 0.00175202 (rank : 52) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75167, Q68DM2 | Gene names | PHACTR2, C6orf56, KIAA0680 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatase and actin regulator 2. | |||||
|
WIRE_MOUSE
|
||||||
| NC score | 0.054203 (rank : 69) | θ value | 0.0736092 (rank : 60) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
BAI3_MOUSE
|
||||||
| NC score | 0.053620 (rank : 70) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80ZF8 | Gene names | Bai3 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 3 precursor. | |||||
|
BAI2_HUMAN
|
||||||
| NC score | 0.053399 (rank : 71) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60241, Q5T6K0, Q8NGW8 | Gene names | BAI2 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 2 precursor. | |||||
|
BAI2_MOUSE
|
||||||
| NC score | 0.053350 (rank : 72) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CGM1, Q3TYC8, Q3UN11, Q3UNE2, Q6PGN0 | Gene names | Bai2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 2 precursor. | |||||
|
TSP2_MOUSE
|
||||||
| NC score | 0.052857 (rank : 73) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q03350 | Gene names | Thbs2, Tsp2 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-2 precursor. | |||||
|
THSD1_HUMAN
|
||||||
| NC score | 0.052498 (rank : 74) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NS62, Q6P3U1, Q6UXZ2 | Gene names | THSD1, TMTSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thrombospondin type-1 domain-containing protein 1 precursor (Transmembrane molecule with thrombospondin module). | |||||
|
BAI3_HUMAN
|
||||||
| NC score | 0.051652 (rank : 75) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60242, O60297 | Gene names | BAI3, KIAA0550 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 3 precursor. | |||||
|
TSP2_HUMAN
|
||||||
| NC score | 0.051560 (rank : 76) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35442 | Gene names | THBS2, TSP2 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-2 precursor. | |||||
|
TSP1_MOUSE
|
||||||
| NC score | 0.050748 (rank : 77) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35441 | Gene names | Thbs1, Tsp1 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-1 precursor. | |||||
|
TSP1_HUMAN
|
||||||
| NC score | 0.050417 (rank : 78) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P07996, Q15667 | Gene names | THBS1, TSP, TSP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-1 precursor. | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.047126 (rank : 79) | θ value | 0.00509761 (rank : 54) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.045787 (rank : 80) | θ value | 0.163984 (rank : 65) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.043563 (rank : 81) | θ value | 0.62314 (rank : 81) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.042215 (rank : 82) | θ value | 0.00665767 (rank : 55) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.040061 (rank : 83) | θ value | 0.0193708 (rank : 58) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
CT032_HUMAN
|
||||||
| NC score | 0.036987 (rank : 84) | θ value | 0.00869519 (rank : 56) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NQ75, Q96K09, Q9BYL5 | Gene names | HEFL, C20orf32 | |||
|
Domain Architecture |

|
|||||
| Description | HEF-like protein. | |||||
|
GGN_MOUSE
|
||||||
| NC score | 0.036664 (rank : 85) | θ value | 1.38821 (rank : 100) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.033267 (rank : 86) | θ value | 1.06291 (rank : 95) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.033056 (rank : 87) | θ value | 0.163984 (rank : 63) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.032253 (rank : 88) | θ value | 1.81305 (rank : 110) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
NFRKB_MOUSE
|
||||||
| NC score | 0.031922 (rank : 89) | θ value | 0.813845 (rank : 89) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
MBB1A_MOUSE
|
||||||
| NC score | 0.031744 (rank : 90) | θ value | 0.163984 (rank : 62) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.031294 (rank : 91) | θ value | 0.813845 (rank : 88) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.030672 (rank : 92) | θ value | 2.36792 (rank : 116) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.030153 (rank : 93) | θ value | 2.36792 (rank : 115) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
NFRKB_HUMAN
|
||||||
| NC score | 0.030056 (rank : 94) | θ value | 1.06291 (rank : 94) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6P4R8, Q12869, Q15312, Q9H048 | Gene names | NFRKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.028876 (rank : 95) | θ value | 3.0926 (rank : 124) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
EDC3_MOUSE
|
||||||
| NC score | 0.027032 (rank : 96) | θ value | 1.81305 (rank : 104) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K2D3, Q3THV7 | Gene names | Edc3, Yjdc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of mRNA decapping protein 3 (YjeF domain-containing protein 1). | |||||
|
EDC3_HUMAN
|
||||||
| NC score | 0.026940 (rank : 97) | θ value | 1.81305 (rank : 103) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96F86, Q9H797 | Gene names | EDC3, YJDC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of mRNA decapping protein 3 (YjeF domain-containing protein 1). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.026887 (rank : 98) | θ value | 0.62314 (rank : 85) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.026393 (rank : 99) | θ value | 0.47712 (rank : 76) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.025984 (rank : 100) | θ value | 0.163984 (rank : 64) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
RPB1_MOUSE
|
||||||
| NC score | 0.024907 (rank : 101) | θ value | 0.21417 (rank : 68) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_HUMAN
|
||||||
| NC score | 0.024880 (rank : 102) | θ value | 0.21417 (rank : 67) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.024477 (rank : 103) | θ value | 0.813845 (rank : 86) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.023449 (rank : 104) | θ value | 0.813845 (rank : 87) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
WASF3_HUMAN
|
||||||
| NC score | 0.023294 (rank : 105) | θ value | 0.62314 (rank : 84) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9UPY6, O94974 | Gene names | WASF3, KIAA0900, SCAR3, WAVE3 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein family member 3 (WASP-family protein member 3) (Protein WAVE-3) (Verprolin homology domain-containing protein 3). | |||||
|
DAB2_HUMAN
|
||||||
| NC score | 0.022748 (rank : 106) | θ value | 1.38821 (rank : 98) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P98082, Q13598, Q9BTY0, Q9UK04 | Gene names | DAB2, DOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (Differentially expressed protein 2) (DOC-2). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.022707 (rank : 107) | θ value | 4.03905 (rank : 132) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.022198 (rank : 108) | θ value | 1.38821 (rank : 101) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SOX30_MOUSE
|
||||||
| NC score | 0.022125 (rank : 109) | θ value | 0.279714 (rank : 70) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
TLE1_MOUSE
|
||||||
| NC score | 0.021725 (rank : 110) | θ value | 0.125558 (rank : 61) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62440 | Gene names | Tle1, Grg1 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 1 (Groucho-related protein 1) (Grg- 1). | |||||
|
TLE1_HUMAN
|
||||||
| NC score | 0.020666 (rank : 111) | θ value | 0.279714 (rank : 71) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q04724, Q5T3G4, Q969V9 | Gene names | TLE1 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 1 (ESG1) (E(Sp1) homolog). | |||||
|
FGD1_MOUSE
|
||||||
| NC score | 0.020453 (rank : 112) | θ value | 0.62314 (rank : 80) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P52734 | Gene names | Fgd1 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein homolog) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.020453 (rank : 113) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.019740 (rank : 114) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
TLE4_MOUSE
|
||||||
| NC score | 0.018771 (rank : 115) | θ value | 0.47712 (rank : 77) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62441, Q9JKQ9 | Gene names | Tle4, Grg4 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 4 (Groucho-related protein 4) (Grg- 4). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.018682 (rank : 116) | θ value | 0.279714 (rank : 72) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
DAB2_MOUSE
|
||||||
| NC score | 0.018247 (rank : 117) | θ value | 4.03905 (rank : 126) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P98078, Q3U3K1, Q91W56, Q923E1 | Gene names | Dab2, Doc2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (DOC-2) (Mitogen-responsive phosphoprotein). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.018199 (rank : 118) | θ value | 0.365318 (rank : 75) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
DDEF1_MOUSE
|
||||||
| NC score | 0.017476 (rank : 119) | θ value | 0.0736092 (rank : 59) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QWY8, O08612, Q80TG8, Q80UV6, Q99LV8, Q9Z2B6 | Gene names | Ddef1, Asap1, Kiaa1249, Shag1 | |||
|
Domain Architecture |

|
|||||
| Description | 130 kDa phosphatidylinositol 4,5-biphosphate-dependent ARF1 GTPase- activating protein (PIP2-dependent ARF1 GAP) (ADP-ribosylation factor- directed GTPase-activating protein 1) (ARF GTPase-activating protein 1) (Development and differentiation-enhancing factor 1) (Differentiation-enhancing factor 1) (DEF-1). | |||||
|
TLE4_HUMAN
|
||||||
| NC score | 0.016990 (rank : 120) | θ value | 2.36792 (rank : 118) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q04727, Q5T1Y2, Q9BZ07, Q9BZ08, Q9BZ09, Q9NSL3, Q9ULF9 | Gene names | TLE4, KIAA1261 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 4. | |||||
|
FGD1_HUMAN
|
||||||
| NC score | 0.016830 (rank : 121) | θ value | 1.38821 (rank : 99) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.016556 (rank : 122) | θ value | 4.03905 (rank : 133) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
RIN3_MOUSE
|
||||||
| NC score | 0.016546 (rank : 123) | θ value | 0.813845 (rank : 91) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P59729 | Gene names | Rin3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
SOS1_MOUSE
|
||||||
| NC score | 0.016541 (rank : 124) | θ value | 0.62314 (rank : 83) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62245, Q62244 | Gene names | Sos1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1) (mSOS-1). | |||||
|
AHTF1_HUMAN
|
||||||
| NC score | 0.016509 (rank : 125) | θ value | 1.38821 (rank : 96) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WYP5, Q7Z4E3, Q8IZA4, Q96EH9, Q9Y4Q6 | Gene names | AHCTF1, ELYS, TMBS62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
SOS1_HUMAN
|
||||||
| NC score | 0.015828 (rank : 126) | θ value | 0.62314 (rank : 82) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q07889 | Gene names | SOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Son of sevenless homolog 1 (SOS-1). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.015595 (rank : 127) | θ value | 2.36792 (rank : 114) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.015554 (rank : 128) | θ value | 0.365318 (rank : 74) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
MN1_HUMAN
|
||||||
| NC score | 0.015513 (rank : 129) | θ value | 4.03905 (rank : 129) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q10571 | Gene names | MN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tumor suppressor protein MN1. | |||||
|
ABI1_MOUSE
|
||||||
| NC score | 0.015155 (rank : 130) | θ value | 0.21417 (rank : 66) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CBW3, Q60747, Q91ZM5, Q923I9, Q99KH4 | Gene names | Abi1, Ssh3bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 1 (Abelson interactor 1) (Abi-1) (Spectrin SH3 domain- binding protein 1) (Eps8 SH3 domain-binding protein) (Eps8-binding protein) (e3B1) (Ablphilin-1). | |||||
|
TISB_HUMAN
|
||||||
| NC score | 0.015062 (rank : 131) | θ value | 2.36792 (rank : 117) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q07352, Q13851 | Gene names | ZFP36L1, BERG36, BRF1, ERF1, TIS11B | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 1 (TIS11B protein) (EGF-response factor 1) (ERF-1). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.014576 (rank : 132) | θ value | 3.0926 (rank : 123) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PAPOA_HUMAN
|
||||||
| NC score | 0.014346 (rank : 133) | θ value | 0.813845 (rank : 90) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P51003, Q86SX4, Q86TV0, Q8IYF5, Q9BVU2 | Gene names | PAPOLA, PAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase alpha). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.014181 (rank : 134) | θ value | 1.81305 (rank : 105) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
TRAM2_HUMAN
|
||||||
| NC score | 0.014120 (rank : 135) | θ value | 1.81305 (rank : 109) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15035 | Gene names | TRAM2, KIAA0057 | |||
|
Domain Architecture |

|
|||||
| Description | Translocation-associated membrane protein 2. | |||||
|
K0423_HUMAN
|
||||||
| NC score | 0.013443 (rank : 136) | θ value | 5.27518 (rank : 138) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y4F4, Q68D66, Q6PG27 | Gene names | KIAA0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
ABI1_HUMAN
|
||||||
| NC score | 0.011683 (rank : 137) | θ value | 1.81305 (rank : 102) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IZP0, O15147, O76049, O95060, Q5W070, Q5W072, Q8TB63, Q96S81, Q9NXZ9, Q9NYB8 | Gene names | ABI1, SSH3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Abl interactor 1 (Abelson interactor 1) (Abi-1) (Spectrin SH3 domain- binding protein 1) (Eps8 SH3 domain-binding protein) (Eps8-binding protein) (e3B1) (Nap1-binding protein) (Nap1BP) (Abl-binding protein 4) (AblBP4). | |||||
|
CM018_HUMAN
|
||||||
| NC score | 0.011529 (rank : 138) | θ value | 5.27518 (rank : 137) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H714, Q5W051, Q6PJ74, Q6PK94, Q86XH7, Q8N5J6 | Gene names | C13orf18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf18. | |||||
|
BRCA2_HUMAN
|
||||||
| NC score | 0.011459 (rank : 139) | θ value | 1.06291 (rank : 92) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51587, O00183, O15008, Q13879 | Gene names | BRCA2, FANCD1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein (Fanconi anemia group D1 protein). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.011447 (rank : 140) | θ value | 5.27518 (rank : 135) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
DACT1_HUMAN
|
||||||
| NC score | 0.011117 (rank : 141) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NYF0, Q86TY0 | Gene names | DACT1, DPR1, HNG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dapper homolog 1 (hDPR1) (Heptacellular carcinoma novel gene 3 protein). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.011074 (rank : 142) | θ value | 1.81305 (rank : 106) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
ZDH17_MOUSE
|
||||||
| NC score | 0.011033 (rank : 143) | θ value | 0.47712 (rank : 79) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80TN5, Q8BJ08, Q8BYQ2, Q8BYQ6 | Gene names | Zdhhc17, Hip14, Kiaa0946 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC17 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 17) (DHHC-17) (Huntingtin-interacting protein 14). | |||||
|
ZDH17_HUMAN
|
||||||
| NC score | 0.010826 (rank : 144) | θ value | 0.47712 (rank : 78) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IUH5, O75407, Q7Z2I0, Q86W89, Q86YK0, Q9P088, Q9UPZ8 | Gene names | ZDHHC17, HIP14, HIP3, KIAA0946 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC17 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 17) (DHHC-17) (Huntingtin-interacting protein 14) (Huntingtin-interacting protein 3) (Huntingtin-interacting protein HYPH) (Putative NF-kappa-B-activating protein 205) (Putative MAPK- activating protein PM11). | |||||
|
MBD5_HUMAN
|
||||||
| NC score | 0.010605 (rank : 145) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P267, Q9NUV6 | Gene names | MBD5, KIAA1461 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 5 (Methyl-CpG-binding protein MBD5). | |||||
|
TRUA_HUMAN
|
||||||
| NC score | 0.009859 (rank : 146) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y606, Q8WYT2, Q9BU44 | Gene names | PUS1 | |||
|
Domain Architecture |

|
|||||
| Description | tRNA pseudouridine synthase A (EC 5.4.99.12) (tRNA-uridine isomerase I) (tRNA pseudouridylate synthase I). | |||||
|
HPS4_MOUSE
|
||||||
| NC score | 0.009508 (rank : 147) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99KG7 | Gene names | Hps4, Le | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 4 protein homolog (Light-ear protein) (Le protein). | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.009374 (rank : 148) | θ value | 4.03905 (rank : 125) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
RBMS2_HUMAN
|
||||||
| NC score | 0.009110 (rank : 149) | θ value | 1.81305 (rank : 107) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15434 | Gene names | RBMS2, SCR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 2 (Suppressor of CDC2 with RNA-binding motif 3). | |||||
|
GP112_HUMAN
|
||||||
| NC score | 0.009022 (rank : 150) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
RBM12_HUMAN
|
||||||
| NC score | 0.008891 (rank : 151) | θ value | 4.03905 (rank : 130) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NTZ6, O94865, Q8N3B1, Q9H196 | Gene names | RBM12, KIAA0765 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
POLS_HUMAN
|
||||||
| NC score | 0.008761 (rank : 152) | θ value | 5.27518 (rank : 140) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5XG87, O43289, Q9Y6C1 | Gene names | POLS, TRF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase sigma (EC 2.7.7.7) (Topoisomerase-related function protein 4-1) (TRF4-1) (LAK-1) (DNA polymerase kappa). | |||||
|
K0460_HUMAN
|
||||||
| NC score | 0.008334 (rank : 153) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5VT52, O75048, Q5VT53, Q6MZL4, Q86XD2 | Gene names | KIAA0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
RBMS1_HUMAN
|
||||||
| NC score | 0.008045 (rank : 154) | θ value | 5.27518 (rank : 142) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29558, Q15433, Q8WV20 | Gene names | RBMS1, MSSP, MSSP1, SCR2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif, single-stranded-interacting protein 1 (Single- stranded DNA-binding protein MSSP-1) (Suppressor of CDC2 with RNA- binding motif 2). | |||||
|
NHS_HUMAN
|
||||||
| NC score | 0.008038 (rank : 155) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6T4R5, Q5J7Q0, Q5J7Q1, Q68DR5 | Gene names | NHS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nance-Horan syndrome protein (Congenital cataracts and dental anomalies protein). | |||||
|
CRTC1_MOUSE
|
||||||
| NC score | 0.007885 (rank : 156) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
DDEF2_MOUSE
|
||||||
| NC score | 0.007875 (rank : 157) | θ value | 2.36792 (rank : 111) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7SIG6, Q501K1, Q66JN2 | Gene names | Ddef2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Development and differentiation-enhancing factor 2 (Pyk2 C-terminus- associated protein) (PAP) (Paxillin-associated protein with ARFGAP activity 3) (PAG3). | |||||
|
RBM16_HUMAN
|
||||||
| NC score | 0.007727 (rank : 158) | θ value | 5.27518 (rank : 141) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
EYA1_HUMAN
|
||||||
| NC score | 0.007689 (rank : 159) | θ value | 4.03905 (rank : 127) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99502 | Gene names | EYA1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
YETS2_HUMAN
|
||||||
| NC score | 0.007426 (rank : 160) | θ value | 6.88961 (rank : 155) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
FOXO4_MOUSE
|
||||||
| NC score | 0.007196 (rank : 161) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WVH3 | Gene names | Mllt7, Afx, Afx1, Fkhr3, Foxo4 | |||
|
Domain Architecture |

|
|||||
| Description | Fork head domain transcription factor AFX1 (Afxh) (Forkhead box protein O4). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.007181 (rank : 162) | θ value | 4.03905 (rank : 134) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
TISB_MOUSE
|
||||||
| NC score | 0.007101 (rank : 163) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23950 | Gene names | Zfp36l1, Brf1, Tis11b | |||
|
Domain Architecture |

|
|||||
| Description | Butyrate response factor 1 (TIS11B protein). | |||||
|
RTN1_MOUSE
|
||||||
| NC score | 0.006987 (rank : 164) | θ value | 4.03905 (rank : 131) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K0T0, Q8K4S4 | Gene names | Rtn1, Nsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
MIRO2_MOUSE
|
||||||
| NC score | 0.005942 (rank : 165) | θ value | 5.27518 (rank : 139) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8JZN7 | Gene names | Rhot2, Arht2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial Rho GTPase 2 (EC 3.6.5.-) (MIRO-2) (Ras homolog gene family member T2). | |||||
|
PYGO1_HUMAN
|
||||||
| NC score | 0.005375 (rank : 166) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y3Y4 | Gene names | PYGO1 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 1. | |||||
|
SYP2L_HUMAN
|
||||||
| NC score | 0.004785 (rank : 167) | θ value | 5.27518 (rank : 143) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H987 | Gene names | SYNPO2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
MIRO2_HUMAN
|
||||||
| NC score | 0.004154 (rank : 168) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IXI1, Q8NF53, Q96C13, Q96S17, Q9BT60, Q9H7M8 | Gene names | RHOT2, ARHT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial Rho GTPase 2 (EC 3.6.5.-) (MIRO-2) (hMiro-2) (Ras homolog gene family member T2). | |||||
|
TSC1_HUMAN
|
||||||
| NC score | 0.003976 (rank : 169) | θ value | 2.36792 (rank : 119) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92574 | Gene names | TSC1, KIAA0243, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Hamartin (Tuberous sclerosis 1 protein). | |||||
|
FOXO4_HUMAN
|
||||||
| NC score | 0.003615 (rank : 170) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P98177, O43821, Q13720, Q3KPF1, Q8TDK9 | Gene names | MLLT7, AFX, AFX1, FOXO4 | |||
|
Domain Architecture |

|
|||||
| Description | Fork head domain transcription factor AFX1 (Forkhead box protein O4). | |||||
|
STIM2_HUMAN
|
||||||
| NC score | 0.003372 (rank : 171) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P246, Q96BF1, Q9BQH2, Q9H8R1 | Gene names | STIM2, KIAA1482 | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 2 precursor. | |||||
|
PRKDC_HUMAN
|
||||||
| NC score | 0.003219 (rank : 172) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.002895 (rank : 173) | θ value | 1.38821 (rank : 97) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
UBP34_MOUSE
|
||||||
| NC score | 0.001570 (rank : 174) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZQ93, Q3UPN0, Q6P563, Q7TMJ6, Q8CCH0 | Gene names | Usp34, Kiaa0570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.001111 (rank : 175) | θ value | 5.27518 (rank : 136) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
HXA11_MOUSE
|
||||||
| NC score | 0.000941 (rank : 176) | θ value | 3.0926 (rank : 121) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P31311, Q3V026 | Gene names | Hoxa11, Hox-1.9, Hoxa-11 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A11 (Hox-1.9). | |||||
|
HXA11_HUMAN
|
||||||
| NC score | 0.000913 (rank : 177) | θ value | 3.0926 (rank : 120) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P31270 | Gene names | HOXA11, HOX1I | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A11 (Hox-1I). | |||||
|
ITCH_MOUSE
|
||||||
| NC score | 0.000895 (rank : 178) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
LAMB3_HUMAN
|
||||||
| NC score | 0.000840 (rank : 179) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13751, O14947, Q14733, Q9UJK4, Q9UJL1 | Gene names | LAMB3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Laminin B1k chain) (Kalinin B1 chain). | |||||
|
LAMC1_HUMAN
|
||||||
| NC score | 0.000687 (rank : 180) | θ value | 4.03905 (rank : 128) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P11047 | Gene names | LAMC1, LAMB2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.000422 (rank : 181) | θ value | 2.36792 (rank : 112) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.000419 (rank : 182) | θ value | 1.81305 (rank : 108) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
DMD_MOUSE
|
||||||
| NC score | -0.000100 (rank : 183) | θ value | 2.36792 (rank : 113) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
SPEG_MOUSE
|
||||||
| NC score | -0.000275 (rank : 184) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | -0.000608 (rank : 185) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
C3AR_MOUSE
|
||||||
| NC score | -0.002144 (rank : 186) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O09047, O35951 | Gene names | C3ar1, C3r1 | |||
|
Domain Architecture |

|
|||||
| Description | C3a anaphylatoxin chemotactic receptor (C3a-R) (C3AR) (Complement component 3a receptor 1). | |||||
|
LRRC7_MOUSE
|
||||||
| NC score | -0.002226 (rank : 187) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TE7 | Gene names | Lrrc7, Kiaa1365, Lap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 7 (Protein LAP1) (Densin-180). | |||||
|
MAST2_HUMAN
|
||||||
| NC score | -0.003045 (rank : 188) | θ value | 3.0926 (rank : 122) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
PKN1_MOUSE
|
||||||
| NC score | -0.003580 (rank : 189) | θ value | 0.279714 (rank : 69) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P70268, Q7TST2 | Gene names | Pkn1, Pkn, Prk1, Prkcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N1 (EC 2.7.11.13) (Protein kinase C- like 1) (Protein-kinase C-related kinase 1) (Protein kinase C-like PKN) (Serine-threonine protein kinase N). | |||||
|
GLIS1_HUMAN
|
||||||
| NC score | -0.003912 (rank : 190) | θ value | 1.06291 (rank : 93) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NBF1, Q8N9V9 | Gene names | GLIS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS1 (GLI-similar 1). | |||||
|
ZN473_MOUSE
|
||||||
| NC score | -0.004598 (rank : 191) | θ value | 0.279714 (rank : 73) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 824 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BI67, Q8BI98, Q8BIB7 | Gene names | Znf473, Zfp100, Zfp473 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 473 homolog (Zinc finger protein 100) (Zfp-100). | |||||
|
CD2L7_HUMAN
|
||||||
| NC score | -0.005083 (rank : 192) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 169 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||