Please be patient as the page loads
|
PLXA4_HUMAN
|
||||||
| SwissProt Accessions | Q9HCM2, Q6UWC6, Q6ZW89, Q8N969, Q8ND00, Q8NEN3, Q9NTD4 | Gene names | PLXNA4, KIAA1550, PLXNA4A, PLXNA4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PLXA1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.993958 (rank : 5) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UIW2 | Gene names | PLXNA1, NOV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Semaphorin receptor NOV). | |||||
|
PLXA1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.993491 (rank : 7) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P70206, Q5DTR0 | Gene names | Plxna1, KIAA4053 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Plexin-1) (Plex 1). | |||||
|
PLXA2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.994971 (rank : 3) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O75051, Q6UX61, Q96GN9, Q9BRL1, Q9UIW1 | Gene names | PLXNA2, KIAA0463, OCT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A2 precursor (Semaphorin receptor OCT). | |||||
|
PLXA2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.994506 (rank : 4) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P70207, Q6NVE6, Q6PHN4, Q80TZ7, Q8R1I4 | Gene names | Plxna2, Kiaa0463 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A2 precursor (Plexin-2) (Plex 2). | |||||
|
PLXA3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.993841 (rank : 6) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P51805 | Gene names | PLXNA3, PLXN4, SEX | |||
|
Domain Architecture |

|
|||||
| Description | Plexin-A3 precursor (Plexin-4) (Semaphorin receptor SEX). | |||||
|
PLXA4_HUMAN
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9HCM2, Q6UWC6, Q6ZW89, Q8N969, Q8ND00, Q8NEN3, Q9NTD4 | Gene names | PLXNA4, KIAA1550, PLXNA4A, PLXNA4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
PLXA4_MOUSE
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.999493 (rank : 2) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q80UG2, Q5DTW8, Q8BKK9 | Gene names | Plxna4, Kiaa1550 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
PLXB1_HUMAN
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.941655 (rank : 13) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
PLXB1_MOUSE
|
||||||
| θ value | 0 (rank : 9) | NC score | 0.952214 (rank : 12) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
PLXB2_HUMAN
|
||||||
| θ value | 0 (rank : 10) | NC score | 0.975448 (rank : 8) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O15031, Q7KZU3, Q9BSU7 | Gene names | PLXNB2, KIAA0315 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B2 precursor (MM1). | |||||
|
PLXB3_HUMAN
|
||||||
| θ value | 0 (rank : 11) | NC score | 0.968538 (rank : 10) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9ULL4, Q9HDA4 | Gene names | PLXNB3, KIAA1206 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor. | |||||
|
PLXB3_MOUSE
|
||||||
| θ value | 0 (rank : 12) | NC score | 0.968522 (rank : 11) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9QY40, Q80TH8 | Gene names | Plxnb3, Kiaa1206, Plxn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor (Plexin-6). | |||||
|
PLXD1_HUMAN
|
||||||
| θ value | 0 (rank : 13) | NC score | 0.972516 (rank : 9) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Y4D7, Q6PJS9, Q8IZJ2, Q9BTQ2 | Gene names | PLXND1, KIAA0620 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-D1 precursor. | |||||
|
PLXC1_MOUSE
|
||||||
| θ value | 1.53401e-116 (rank : 14) | NC score | 0.925938 (rank : 14) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9QZC2, Q8CGW1 | Gene names | Plxnc1, Vespr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
|
PLXC1_HUMAN
|
||||||
| θ value | 9.94316e-116 (rank : 15) | NC score | 0.923073 (rank : 15) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O60486, Q59H25 | Gene names | PLXNC1, VESPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
|
MET_MOUSE
|
||||||
| θ value | 5.76516e-39 (rank : 16) | NC score | 0.205747 (rank : 54) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P16056, Q62125 | Gene names | Met | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor receptor precursor (EC 2.7.10.1) (HGF receptor) (Scatter factor receptor) (SF receptor) (HGF/SF receptor) (Met proto-oncogene tyrosine kinase) (c-Met). | |||||
|
MET_HUMAN
|
||||||
| θ value | 4.88048e-38 (rank : 17) | NC score | 0.205605 (rank : 55) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P08581, O60366, Q12875, Q9UDX7, Q9UPL8 | Gene names | MET | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor receptor precursor (EC 2.7.10.1) (HGF receptor) (Scatter factor receptor) (SF receptor) (HGF/SF receptor) (Met proto-oncogene tyrosine kinase) (c-Met). | |||||
|
RON_HUMAN
|
||||||
| θ value | 3.75424e-22 (rank : 18) | NC score | 0.179997 (rank : 56) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q04912 | Gene names | MST1R, RON | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage-stimulating protein receptor precursor (EC 2.7.10.1) (MSP receptor) (p185-Ron) (CDw136 antigen) [Contains: Macrophage- stimulating protein receptor alpha chain; Macrophage-stimulating protein receptor beta chain]. | |||||
|
SEM4B_MOUSE
|
||||||
| θ value | 8.65492e-19 (rank : 19) | NC score | 0.442326 (rank : 16) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62179, Q4PKI6, Q69ZB7 | Gene names | Sema4b, Kiaa1745, Semac, SemC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-4B precursor (Semaphorin C) (Sema C). | |||||
|
SEM4B_HUMAN
|
||||||
| θ value | 2.5182e-18 (rank : 20) | NC score | 0.431620 (rank : 18) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NPR2, Q6UXE3, Q8WVP9, Q96FK5, Q9C0B8, Q9H691, Q9NPM8, Q9NPN0 | Gene names | SEMA4B, KIAA1745 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4B precursor. | |||||
|
RON_MOUSE
|
||||||
| θ value | 3.63628e-17 (rank : 21) | NC score | 0.176774 (rank : 57) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62190, Q62555 | Gene names | Mst1r, Ron, Stk | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage-stimulating protein receptor precursor (EC 2.7.10.1) (MSP receptor) (p185-Ron) (Stem cell-derived tyrosine kinase) (CDw136 antigen) [Contains: Macrophage-stimulating protein receptor alpha chain; Macrophage-stimulating protein receptor beta chain]. | |||||
|
SEM5B_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 22) | NC score | 0.356128 (rank : 30) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9P283, Q6UY12 | Gene names | SEMA5B, KIAA1445 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5B precursor. | |||||
|
SEM5B_MOUSE
|
||||||
| θ value | 2.88119e-14 (rank : 23) | NC score | 0.354943 (rank : 32) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q60519 | Gene names | Sema5b, Semag, SemG | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5B precursor (Semaphorin G) (Sema G). | |||||
|
SEM5A_MOUSE
|
||||||
| θ value | 3.76295e-14 (rank : 24) | NC score | 0.353814 (rank : 33) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q62217 | Gene names | Sema5a, Semaf, SemF | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5A precursor (Semaphorin F) (Sema F). | |||||
|
SEM5A_HUMAN
|
||||||
| θ value | 3.18553e-13 (rank : 25) | NC score | 0.342010 (rank : 42) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13591, O60408, Q1RLL9 | Gene names | SEMA5A, SEMAF | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5A precursor (Semaphorin F) (Sema F). | |||||
|
SEM4A_HUMAN
|
||||||
| θ value | 1.58096e-12 (rank : 26) | NC score | 0.432118 (rank : 17) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9H3S1, Q8WUA9 | Gene names | SEMA4A, SEMB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
SEM6D_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 27) | NC score | 0.378683 (rank : 23) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q76KF0, Q76KF1, Q76KF2, Q76KF3, Q76KF4, Q80TD0 | Gene names | Sema6d, Kiaa1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SEM4A_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 28) | NC score | 0.425640 (rank : 19) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62178 | Gene names | Sema4a, Semab, SemB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
SEM4C_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 29) | NC score | 0.403838 (rank : 20) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9C0C4, Q7Z5X0 | Gene names | SEMA4C, KIAA1739 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4C precursor. | |||||
|
SEM6B_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 30) | NC score | 0.364351 (rank : 27) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H3T3, Q9NRK9 | Gene names | SEMA6B, SEMAZ | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin Z) (Sema Z). | |||||
|
SEM6D_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 31) | NC score | 0.377097 (rank : 24) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8NFY4, Q8NFY3, Q8NFY5, Q8NFY6, Q8NFY7, Q9P249 | Gene names | SEMA6D, KIAA1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SEM3F_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 32) | NC score | 0.348781 (rank : 37) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13275, Q13274, Q13372, Q15704 | Gene names | SEMA3F | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3F precursor (Semaphorin IV) (Sema IV) (Sema III/F). | |||||
|
SEM6B_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 33) | NC score | 0.347124 (rank : 39) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O54951 | Gene names | Sema6b, Seman | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin VIB) (Sema VIB) (Semaphorin N) (Sema N). | |||||
|
SEM4G_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 34) | NC score | 0.390045 (rank : 21) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9WUH7 | Gene names | Sema4g | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
SEM3F_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 35) | NC score | 0.348547 (rank : 38) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O88632, O88633 | Gene names | Sema3f | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3F precursor (Semaphorin IV) (Sema IV). | |||||
|
SEM4C_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 36) | NC score | 0.372398 (rank : 26) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q64151 | Gene names | Sema4c, Semacl1, Semai | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4C precursor (Semaphorin I) (Sema I) (Semaphorin C-like 1) (M-Sema F). | |||||
|
SEM4G_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 37) | NC score | 0.389206 (rank : 22) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NTN9, Q9HCF3 | Gene names | SEMA4G, KIAA1619 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
SEM4F_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 38) | NC score | 0.350141 (rank : 36) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O95754, Q9NS35 | Gene names | SEMA4F, SEMAM, SEMAW | |||
|
Domain Architecture |
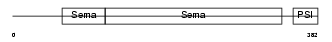
|
|||||
| Description | Semaphorin-4F precursor (Semaphorin W) (Sema W) (Semaphorin M) (Sema M). | |||||
|
SEM4F_MOUSE
|
||||||
| θ value | 4.92598e-06 (rank : 39) | NC score | 0.351353 (rank : 34) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Z123, Q9R1Y1 | Gene names | Sema4f, Semaw | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4F precursor (Semaphorin W) (Sema W). | |||||
|
SEM3B_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 40) | NC score | 0.342037 (rank : 41) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62177 | Gene names | Sema3b, Sema, Semaa | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3B precursor (Semaphorin A) (Sema A). | |||||
|
SEM6A_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 41) | NC score | 0.355347 (rank : 31) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O35464, Q6P5A8, Q6PCN9, Q9EQ71 | Gene names | Sema6a, Semaq | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1) (Semaphorin Q) (Sema Q). | |||||
|
SEM3C_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 42) | NC score | 0.335480 (rank : 45) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62181 | Gene names | Sema3c, Semae, SemE | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3C precursor (Semaphorin E) (Sema E). | |||||
|
SEM7A_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 43) | NC score | 0.374336 (rank : 25) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9QUR8, O88371 | Gene names | Sema7a, Cd108, Semal, Semk1 | |||
|
Domain Architecture |
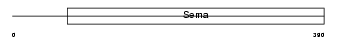
|
|||||
| Description | Semaphorin-7A precursor (Semaphorin L) (Sema L) (Semaphorin K1) (Sema K1) (CD108 antigen) (CDw108). | |||||
|
SEM3C_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 44) | NC score | 0.333929 (rank : 46) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q99985 | Gene names | SEMA3C, SEMAE | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3C precursor (Semaphorin E) (Sema E). | |||||
|
SEM3D_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 45) | NC score | 0.312849 (rank : 50) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O95025, Q6UW77, Q8NCQ1 | Gene names | SEMA3D | |||
|
Domain Architecture |
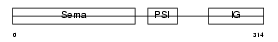
|
|||||
| Description | Semaphorin-3D precursor. | |||||
|
SEM3B_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 46) | NC score | 0.336646 (rank : 44) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13214, Q8TB71, Q8TDV7, Q93018, Q96GX0 | Gene names | SEMA3B, SEMA5 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3B precursor (Semaphorin V) (Sema V) (Sema A(V)). | |||||
|
SEM7A_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 47) | NC score | 0.363071 (rank : 28) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75326 | Gene names | SEMA7A, CD108, SEMAL | |||
|
Domain Architecture |
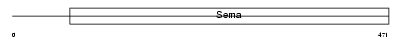
|
|||||
| Description | Semaphorin-7A precursor (Semaphorin L) (Sema L) (Semaphorin K1) (Sema K1) (John-Milton-Hargen human blood group Ag) (JMH blood group antigen) (CD108 antigen) (CDw108). | |||||
|
SEM6A_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 48) | NC score | 0.358185 (rank : 29) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9H2E6, Q9P2H9 | Gene names | SEMA6A, KIAA1368 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1). | |||||
|
SEM3E_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 49) | NC score | 0.315666 (rank : 47) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O15041, Q75M94, Q75M97 | Gene names | SEMA3E, KIAA0331 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-3E precursor. | |||||
|
SEM3E_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 50) | NC score | 0.312063 (rank : 51) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70275, O09078, O09079 | Gene names | Sema3e, Semah, Semh | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3E precursor (Semaphorin H) (Sema H). | |||||
|
SEM3A_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 51) | NC score | 0.314566 (rank : 49) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O08665, Q5BL08, Q62180, Q62215 | Gene names | Sema3a, Semad, SemD | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3A precursor (Semaphorin III) (Sema III) (Semaphorin D) (Sema D). | |||||
|
SEM6C_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 52) | NC score | 0.344615 (rank : 40) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H3T2, Q5JR71, Q8WXT8, Q8WXT9, Q8WXU0, Q96JF8 | Gene names | SEMA6C, KIAA1869, SEMAY | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SEM3D_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 53) | NC score | 0.303465 (rank : 53) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BH34 | Gene names | Sema3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-3D precursor. | |||||
|
SEM3A_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 54) | NC score | 0.310675 (rank : 52) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q14563 | Gene names | SEMA3A | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3A precursor (Semaphorin III) (Sema III). | |||||
|
SEM6C_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 55) | NC score | 0.315412 (rank : 48) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SEM4D_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 56) | NC score | 0.350220 (rank : 35) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O09126 | Gene names | Sema4d, Semacl2, Semaj | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Semaphorin J) (Sema J) (Semaphorin C-like 2) (M-Sema G). | |||||
|
SEM4D_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 57) | NC score | 0.337412 (rank : 43) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q92854, Q7Z5S4 | Gene names | SEMA4D, CD100 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Leukocyte activation antigen CD100) (BB18) (A8) (GR3). | |||||
|
EXOC2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 58) | NC score | 0.120950 (rank : 59) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96KP1, Q5JPC8, Q96AN6, Q9NUZ8, Q9UJM7 | Gene names | EXOC2, SEC5, SEC5L1 | |||
|
Domain Architecture |
|
|||||
| Description | Exocyst complex component 2 (Exocyst complex component Sec5). | |||||
|
EXOC2_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 59) | NC score | 0.116317 (rank : 60) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D4H1 | Gene names | Exoc2, Sec5, Sec5l1 | |||
|
Domain Architecture |
|
|||||
| Description | Exocyst complex component 2 (Exocyst complex component Sec5). | |||||
|
ATRN_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 60) | NC score | 0.072975 (rank : 65) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
RASA3_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 61) | NC score | 0.086168 (rank : 61) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14644 | Gene names | RASA3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein). | |||||
|
ATRN_MOUSE
|
||||||
| θ value | 0.21417 (rank : 62) | NC score | 0.065117 (rank : 66) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WU60, Q9R263, Q9WU77 | Gene names | Atrn, mg, Mgca | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany protein). | |||||
|
SYGP1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 63) | NC score | 0.063887 (rank : 67) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96PV0, Q8TCS2, Q9UGE2 | Gene names | SYNGAP1, KIAA1938 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein SynGAP (Synaptic Ras-GTPase-activating protein 1) (Synaptic Ras-GAP 1) (Neuronal RasGAP). | |||||
|
RASA2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 64) | NC score | 0.034859 (rank : 73) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15283, O00695, Q15284, Q92594, Q99577, Q9UEQ2 | Gene names | RASA2, GAP1M, RASGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
RASA3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 65) | NC score | 0.073560 (rank : 64) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60790 | Gene names | Rasa3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein) (GapIII). | |||||
|
MEGF8_HUMAN
|
||||||
| θ value | 0.813845 (rank : 66) | NC score | 0.055098 (rank : 69) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
ITB8_HUMAN
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.020478 (rank : 80) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P26012 | Gene names | ITGB8 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-8 precursor. | |||||
|
MTMR5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 68) | NC score | 0.020866 (rank : 79) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
ITB1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.023611 (rank : 75) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P05556, P78466, P78467, Q13089, Q13090, Q13091, Q13212, Q14622, Q14647 | Gene names | ITGB1, FNRB | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
ITB1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.023438 (rank : 76) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P09055 | Gene names | Itgb1 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
DDX20_HUMAN
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.004450 (rank : 94) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
ITB6_MOUSE
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.020412 (rank : 81) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z0T9 | Gene names | Itgb6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
ZN226_HUMAN
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | -0.002626 (rank : 105) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NYT6, Q96TE6, Q9NS44 | Gene names | ZNF226 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 226. | |||||
|
ZN567_HUMAN
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | -0.002709 (rank : 106) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N184, Q6N044 | Gene names | ZNF567 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 567. | |||||
|
DAB2P_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.042378 (rank : 72) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5VWQ8, Q8TDL2, Q96SE1, Q9C0C0 | Gene names | DAB2IP, AF9Q34, AIP1, KIAA1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein) (DAB2 interaction protein) (ASK-interacting protein 1). | |||||
|
DAB2P_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.042634 (rank : 71) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3UHC7, Q3TPD5, Q3UH44, Q6JTV1, Q80T97 | Gene names | Dab2ip, Kiaa1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein). | |||||
|
FAN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.005403 (rank : 91) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35242 | Gene names | Nsmaf, Fan | |||
|
Domain Architecture |

|
|||||
| Description | Protein FAN (Factor associated with N-SMase activation) (Factor associated with neutral sphingomyelinase activation). | |||||
|
MEGF8_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.049129 (rank : 70) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P60882, Q80TR3, Q80V41, Q8BMN9, Q8JZW7, Q8K0J3 | Gene names | Megf8, Egfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.010373 (rank : 84) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
ZN507_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | -0.001811 (rank : 104) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TCN5 | Gene names | ZNF507 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 507. | |||||
|
PKHD1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.170182 (rank : 58) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TCZ9 | Gene names | PKHD1, FCYT, TIGM1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney and hepatic disease 1 precursor (Fibrocystin) (Polyductin) (Tigmin). | |||||
|
RASA2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.024093 (rank : 74) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P58069 | Gene names | Rasa2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
RBCC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.001242 (rank : 99) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
EP400_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.007709 (rank : 88) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
PARC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.006802 (rank : 89) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
VIGLN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.011364 (rank : 83) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VDJ3 | Gene names | Hdlbp | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
CADH7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.000307 (rank : 100) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULB5, Q9H157 | Gene names | CDH7, CDH7L1 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-7 precursor. | |||||
|
EP400_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.008462 (rank : 86) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
KLH14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.003630 (rank : 95) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P2G3 | Gene names | KLHL14, KIAA1384 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 14. | |||||
|
PXDC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.086047 (rank : 62) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91ZV7, Q8BM20, Q9CWV5 | Gene names | Plxdc1, Tem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 1 precursor (Tumor endothelial marker 7). | |||||
|
T22D1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.022938 (rank : 77) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62500, Q00992 | Gene names | Tsc22d1, Tgfb1i4, Tsc22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 1 (Transforming growth factor beta-1- induced transcript 4 protein) (Regulatory protein TSC-22) (TGFB- stimulated clone 22 homolog). | |||||
|
ZN687_MOUSE
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | -0.000181 (rank : 101) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
AIM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.008311 (rank : 87) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y4K1, O00296, Q5VWJ2 | Gene names | AIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Absent in melanoma 1 protein. | |||||
|
CREL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.003622 (rank : 96) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91XD7, Q8BGJ8 | Gene names | Creld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.001758 (rank : 98) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
KPCT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.004578 (rank : 93) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 897 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q04759, Q3MJF1, Q64FY5, Q9H508, Q9H549 | Gene names | PRKCQ, PRKCT | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C theta type (EC 2.7.11.13) (nPKC-theta). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | -0.000632 (rank : 103) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
PARC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.006055 (rank : 90) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TT8, Q8BKL9, Q8CGC0, Q8R363 | Gene names | Parc, Kiaa0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein. | |||||
|
PRC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.011835 (rank : 82) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99K43, Q8CE25 | Gene names | Prc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein regulator of cytokinesis 1. | |||||
|
ZN480_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | -0.003400 (rank : 107) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WV37, Q5JPG9, Q6P0Q4, Q8N1M5 | Gene names | ZNF480 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 480. | |||||
|
ACV1C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.009290 (rank : 85) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NER5, Q4ZFZ8, Q86UL1, Q86UL2, Q8TBG2 | Gene names | ACVR1C, ALK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activin receptor type 1C precursor (EC 2.7.11.30) (ACTR-IC) (Activin receptor-like kinase 7) (ALK-7). | |||||
|
GNPAT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.022559 (rank : 78) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P98192, Q9WUT6 | Gene names | Gnpat, Dhapat | |||
|
Domain Architecture |
|
|||||
| Description | Dihydroxyacetone phosphate acyltransferase (EC 2.3.1.42) (DHAP-AT) (DAP-AT) (Glycerone-phosphate O-acyltransferase) (Acyl- CoA:dihydroxyacetonephosphateacyltransferase). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | -0.003672 (rank : 108) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
KCMA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.003172 (rank : 97) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08460, Q64703, Q8VHF1, Q9R196 | Gene names | Kcnma1, Kcnma | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (mSlo) (mSlo1). | |||||
|
MATN2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | -0.000529 (rank : 102) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08746 | Gene names | Matn2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
RPGR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.004641 (rank : 92) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96KN7, Q7Z2W6, Q8IXV5, Q96QA8, Q9HB94, Q9HB95, Q9HBK6, Q9NR40 | Gene names | RPGRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
PXDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.082106 (rank : 63) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IUK5, Q5QCZ7, Q5QCZ8, Q5QCZ9, Q9HCT9 | Gene names | PLXDC1, TEM3, TEM7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 1 precursor (Tumor endothelial marker 7) (Tumor endothelial marker 3). | |||||
|
PXDC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.055230 (rank : 68) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6UX71, Q96E59, Q96PD9, Q96SU9 | Gene names | PLXDC2, TEM7R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 2 precursor (Tumor endothelial marker 7-related protein). | |||||
|
PLXA4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9HCM2, Q6UWC6, Q6ZW89, Q8N969, Q8ND00, Q8NEN3, Q9NTD4 | Gene names | PLXNA4, KIAA1550, PLXNA4A, PLXNA4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
PLXA4_MOUSE
|
||||||
| NC score | 0.999493 (rank : 2) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q80UG2, Q5DTW8, Q8BKK9 | Gene names | Plxna4, Kiaa1550 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
PLXA2_HUMAN
|
||||||
| NC score | 0.994971 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O75051, Q6UX61, Q96GN9, Q9BRL1, Q9UIW1 | Gene names | PLXNA2, KIAA0463, OCT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A2 precursor (Semaphorin receptor OCT). | |||||
|
PLXA2_MOUSE
|
||||||
| NC score | 0.994506 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P70207, Q6NVE6, Q6PHN4, Q80TZ7, Q8R1I4 | Gene names | Plxna2, Kiaa0463 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A2 precursor (Plexin-2) (Plex 2). | |||||
|
PLXA1_HUMAN
|
||||||
| NC score | 0.993958 (rank : 5) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9UIW2 | Gene names | PLXNA1, NOV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Semaphorin receptor NOV). | |||||
|
PLXA3_HUMAN
|
||||||
| NC score | 0.993841 (rank : 6) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P51805 | Gene names | PLXNA3, PLXN4, SEX | |||
|
Domain Architecture |

|
|||||
| Description | Plexin-A3 precursor (Plexin-4) (Semaphorin receptor SEX). | |||||
|
PLXA1_MOUSE
|
||||||
| NC score | 0.993491 (rank : 7) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P70206, Q5DTR0 | Gene names | Plxna1, KIAA4053 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Plexin-1) (Plex 1). | |||||
|
PLXB2_HUMAN
|
||||||
| NC score | 0.975448 (rank : 8) | θ value | 0 (rank : 10) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O15031, Q7KZU3, Q9BSU7 | Gene names | PLXNB2, KIAA0315 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B2 precursor (MM1). | |||||
|
PLXD1_HUMAN
|
||||||
| NC score | 0.972516 (rank : 9) | θ value | 0 (rank : 13) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Y4D7, Q6PJS9, Q8IZJ2, Q9BTQ2 | Gene names | PLXND1, KIAA0620 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-D1 precursor. | |||||
|
PLXB3_HUMAN
|
||||||
| NC score | 0.968538 (rank : 10) | θ value | 0 (rank : 11) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9ULL4, Q9HDA4 | Gene names | PLXNB3, KIAA1206 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor. | |||||
|
PLXB3_MOUSE
|
||||||
| NC score | 0.968522 (rank : 11) | θ value | 0 (rank : 12) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9QY40, Q80TH8 | Gene names | Plxnb3, Kiaa1206, Plxn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor (Plexin-6). | |||||
|
PLXB1_MOUSE
|
||||||
| NC score | 0.952214 (rank : 12) | θ value | 0 (rank : 9) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
PLXB1_HUMAN
|
||||||
| NC score | 0.941655 (rank : 13) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
PLXC1_MOUSE
|
||||||
| NC score | 0.925938 (rank : 14) | θ value | 1.53401e-116 (rank : 14) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9QZC2, Q8CGW1 | Gene names | Plxnc1, Vespr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
|
PLXC1_HUMAN
|
||||||
| NC score | 0.923073 (rank : 15) | θ value | 9.94316e-116 (rank : 15) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O60486, Q59H25 | Gene names | PLXNC1, VESPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
|
SEM4B_MOUSE
|
||||||
| NC score | 0.442326 (rank : 16) | θ value | 8.65492e-19 (rank : 19) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62179, Q4PKI6, Q69ZB7 | Gene names | Sema4b, Kiaa1745, Semac, SemC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-4B precursor (Semaphorin C) (Sema C). | |||||
|
SEM4A_HUMAN
|
||||||
| NC score | 0.432118 (rank : 17) | θ value | 1.58096e-12 (rank : 26) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9H3S1, Q8WUA9 | Gene names | SEMA4A, SEMB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
SEM4B_HUMAN
|
||||||
| NC score | 0.431620 (rank : 18) | θ value | 2.5182e-18 (rank : 20) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NPR2, Q6UXE3, Q8WVP9, Q96FK5, Q9C0B8, Q9H691, Q9NPM8, Q9NPN0 | Gene names | SEMA4B, KIAA1745 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4B precursor. | |||||
|
SEM4A_MOUSE
|
||||||
| NC score | 0.425640 (rank : 19) | θ value | 4.76016e-09 (rank : 28) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62178 | Gene names | Sema4a, Semab, SemB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
SEM4C_HUMAN
|
||||||
| NC score | 0.403838 (rank : 20) | θ value | 4.76016e-09 (rank : 29) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9C0C4, Q7Z5X0 | Gene names | SEMA4C, KIAA1739 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4C precursor. | |||||
|
SEM4G_MOUSE
|
||||||
| NC score | 0.390045 (rank : 21) | θ value | 7.59969e-07 (rank : 34) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9WUH7 | Gene names | Sema4g | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
SEM4G_HUMAN
|
||||||
| NC score | 0.389206 (rank : 22) | θ value | 1.69304e-06 (rank : 37) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NTN9, Q9HCF3 | Gene names | SEMA4G, KIAA1619 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
SEM6D_MOUSE
|
||||||
| NC score | 0.378683 (rank : 23) | θ value | 3.64472e-09 (rank : 27) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q76KF0, Q76KF1, Q76KF2, Q76KF3, Q76KF4, Q80TD0 | Gene names | Sema6d, Kiaa1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SEM6D_HUMAN
|
||||||
| NC score | 0.377097 (rank : 24) | θ value | 4.0297e-08 (rank : 31) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8NFY4, Q8NFY3, Q8NFY5, Q8NFY6, Q8NFY7, Q9P249 | Gene names | SEMA6D, KIAA1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SEM7A_MOUSE
|
||||||
| NC score | 0.374336 (rank : 25) | θ value | 5.44631e-05 (rank : 43) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9QUR8, O88371 | Gene names | Sema7a, Cd108, Semal, Semk1 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-7A precursor (Semaphorin L) (Sema L) (Semaphorin K1) (Sema K1) (CD108 antigen) (CDw108). | |||||
|
SEM4C_MOUSE
|
||||||
| NC score | 0.372398 (rank : 26) | θ value | 9.92553e-07 (rank : 36) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q64151 | Gene names | Sema4c, Semacl1, Semai | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4C precursor (Semaphorin I) (Sema I) (Semaphorin C-like 1) (M-Sema F). | |||||
|
SEM6B_HUMAN
|
||||||
| NC score | 0.364351 (rank : 27) | θ value | 8.11959e-09 (rank : 30) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H3T3, Q9NRK9 | Gene names | SEMA6B, SEMAZ | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin Z) (Sema Z). | |||||
|
SEM7A_HUMAN
|
||||||
| NC score | 0.363071 (rank : 28) | θ value | 0.000158464 (rank : 47) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75326 | Gene names | SEMA7A, CD108, SEMAL | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-7A precursor (Semaphorin L) (Sema L) (Semaphorin K1) (Sema K1) (John-Milton-Hargen human blood group Ag) (JMH blood group antigen) (CD108 antigen) (CDw108). | |||||
|
SEM6A_HUMAN
|
||||||
| NC score | 0.358185 (rank : 29) | θ value | 0.000270298 (rank : 48) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9H2E6, Q9P2H9 | Gene names | SEMA6A, KIAA1368 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1). | |||||
|
SEM5B_HUMAN
|
||||||
| NC score | 0.356128 (rank : 30) | θ value | 2.88119e-14 (rank : 22) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9P283, Q6UY12 | Gene names | SEMA5B, KIAA1445 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5B precursor. | |||||
|
SEM6A_MOUSE
|
||||||
| NC score | 0.355347 (rank : 31) | θ value | 4.1701e-05 (rank : 41) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O35464, Q6P5A8, Q6PCN9, Q9EQ71 | Gene names | Sema6a, Semaq | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1) (Semaphorin Q) (Sema Q). | |||||
|
SEM5B_MOUSE
|
||||||
| NC score | 0.354943 (rank : 32) | θ value | 2.88119e-14 (rank : 23) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q60519 | Gene names | Sema5b, Semag, SemG | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5B precursor (Semaphorin G) (Sema G). | |||||
|
SEM5A_MOUSE
|
||||||
| NC score | 0.353814 (rank : 33) | θ value | 3.76295e-14 (rank : 24) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q62217 | Gene names | Sema5a, Semaf, SemF | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5A precursor (Semaphorin F) (Sema F). | |||||
|
SEM4F_MOUSE
|
||||||
| NC score | 0.351353 (rank : 34) | θ value | 4.92598e-06 (rank : 39) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Z123, Q9R1Y1 | Gene names | Sema4f, Semaw | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4F precursor (Semaphorin W) (Sema W). | |||||
|
SEM4D_MOUSE
|
||||||
| NC score | 0.350220 (rank : 35) | θ value | 0.00665767 (rank : 56) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O09126 | Gene names | Sema4d, Semacl2, Semaj | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Semaphorin J) (Sema J) (Semaphorin C-like 2) (M-Sema G). | |||||
|
SEM4F_HUMAN
|
||||||
| NC score | 0.350141 (rank : 36) | θ value | 2.88788e-06 (rank : 38) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O95754, Q9NS35 | Gene names | SEMA4F, SEMAM, SEMAW | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4F precursor (Semaphorin W) (Sema W) (Semaphorin M) (Sema M). | |||||
|
SEM3F_HUMAN
|
||||||
| NC score | 0.348781 (rank : 37) | θ value | 4.45536e-07 (rank : 32) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13275, Q13274, Q13372, Q15704 | Gene names | SEMA3F | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3F precursor (Semaphorin IV) (Sema IV) (Sema III/F). | |||||
|
SEM3F_MOUSE
|
||||||
| NC score | 0.348547 (rank : 38) | θ value | 9.92553e-07 (rank : 35) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O88632, O88633 | Gene names | Sema3f | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3F precursor (Semaphorin IV) (Sema IV). | |||||
|
SEM6B_MOUSE
|
||||||
| NC score | 0.347124 (rank : 39) | θ value | 5.81887e-07 (rank : 33) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O54951 | Gene names | Sema6b, Seman | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin VIB) (Sema VIB) (Semaphorin N) (Sema N). | |||||
|
SEM6C_HUMAN
|
||||||
| NC score | 0.344615 (rank : 40) | θ value | 0.000461057 (rank : 52) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H3T2, Q5JR71, Q8WXT8, Q8WXT9, Q8WXU0, Q96JF8 | Gene names | SEMA6C, KIAA1869, SEMAY | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SEM3B_MOUSE
|
||||||
| NC score | 0.342037 (rank : 41) | θ value | 6.43352e-06 (rank : 40) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62177 | Gene names | Sema3b, Sema, Semaa | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3B precursor (Semaphorin A) (Sema A). | |||||
|
SEM5A_HUMAN
|
||||||
| NC score | 0.342010 (rank : 42) | θ value | 3.18553e-13 (rank : 25) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13591, O60408, Q1RLL9 | Gene names | SEMA5A, SEMAF | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5A precursor (Semaphorin F) (Sema F). | |||||
|
SEM4D_HUMAN
|
||||||
| NC score | 0.337412 (rank : 43) | θ value | 0.00869519 (rank : 57) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q92854, Q7Z5S4 | Gene names | SEMA4D, CD100 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Leukocyte activation antigen CD100) (BB18) (A8) (GR3). | |||||
|
SEM3B_HUMAN
|
||||||
| NC score | 0.336646 (rank : 44) | θ value | 9.29e-05 (rank : 46) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13214, Q8TB71, Q8TDV7, Q93018, Q96GX0 | Gene names | SEMA3B, SEMA5 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3B precursor (Semaphorin V) (Sema V) (Sema A(V)). | |||||
|
SEM3C_MOUSE
|
||||||
| NC score | 0.335480 (rank : 45) | θ value | 5.44631e-05 (rank : 42) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62181 | Gene names | Sema3c, Semae, SemE | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3C precursor (Semaphorin E) (Sema E). | |||||
|
SEM3C_HUMAN
|
||||||
| NC score | 0.333929 (rank : 46) | θ value | 7.1131e-05 (rank : 44) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q99985 | Gene names | SEMA3C, SEMAE | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3C precursor (Semaphorin E) (Sema E). | |||||
|
SEM3E_HUMAN
|
||||||
| NC score | 0.315666 (rank : 47) | θ value | 0.00035302 (rank : 49) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O15041, Q75M94, Q75M97 | Gene names | SEMA3E, KIAA0331 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-3E precursor. | |||||
|
SEM6C_MOUSE
|
||||||
| NC score | 0.315412 (rank : 48) | θ value | 0.00134147 (rank : 55) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SEM3A_MOUSE
|
||||||
| NC score | 0.314566 (rank : 49) | θ value | 0.000461057 (rank : 51) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O08665, Q5BL08, Q62180, Q62215 | Gene names | Sema3a, Semad, SemD | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3A precursor (Semaphorin III) (Sema III) (Semaphorin D) (Sema D). | |||||
|
SEM3D_HUMAN
|
||||||
| NC score | 0.312849 (rank : 50) | θ value | 7.1131e-05 (rank : 45) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O95025, Q6UW77, Q8NCQ1 | Gene names | SEMA3D | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3D precursor. | |||||
|
SEM3E_MOUSE
|
||||||
| NC score | 0.312063 (rank : 51) | θ value | 0.00035302 (rank : 50) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70275, O09078, O09079 | Gene names | Sema3e, Semah, Semh | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3E precursor (Semaphorin H) (Sema H). | |||||
|
SEM3A_HUMAN
|
||||||
| NC score | 0.310675 (rank : 52) | θ value | 0.00102713 (rank : 54) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q14563 | Gene names | SEMA3A | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3A precursor (Semaphorin III) (Sema III). | |||||
|
SEM3D_MOUSE
|
||||||
| NC score | 0.303465 (rank : 53) | θ value | 0.000602161 (rank : 53) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BH34 | Gene names | Sema3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-3D precursor. | |||||
|
MET_MOUSE
|
||||||
| NC score | 0.205747 (rank : 54) | θ value | 5.76516e-39 (rank : 16) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P16056, Q62125 | Gene names | Met | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor receptor precursor (EC 2.7.10.1) (HGF receptor) (Scatter factor receptor) (SF receptor) (HGF/SF receptor) (Met proto-oncogene tyrosine kinase) (c-Met). | |||||
|
MET_HUMAN
|
||||||
| NC score | 0.205605 (rank : 55) | θ value | 4.88048e-38 (rank : 17) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P08581, O60366, Q12875, Q9UDX7, Q9UPL8 | Gene names | MET | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor receptor precursor (EC 2.7.10.1) (HGF receptor) (Scatter factor receptor) (SF receptor) (HGF/SF receptor) (Met proto-oncogene tyrosine kinase) (c-Met). | |||||
|
RON_HUMAN
|
||||||
| NC score | 0.179997 (rank : 56) | θ value | 3.75424e-22 (rank : 18) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q04912 | Gene names | MST1R, RON | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage-stimulating protein receptor precursor (EC 2.7.10.1) (MSP receptor) (p185-Ron) (CDw136 antigen) [Contains: Macrophage- stimulating protein receptor alpha chain; Macrophage-stimulating protein receptor beta chain]. | |||||
|
RON_MOUSE
|
||||||
| NC score | 0.176774 (rank : 57) | θ value | 3.63628e-17 (rank : 21) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62190, Q62555 | Gene names | Mst1r, Ron, Stk | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage-stimulating protein receptor precursor (EC 2.7.10.1) (MSP receptor) (p185-Ron) (Stem cell-derived tyrosine kinase) (CDw136 antigen) [Contains: Macrophage-stimulating protein receptor alpha chain; Macrophage-stimulating protein receptor beta chain]. | |||||
|
PKHD1_HUMAN
|
||||||
| NC score | 0.170182 (rank : 58) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TCZ9 | Gene names | PKHD1, FCYT, TIGM1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney and hepatic disease 1 precursor (Fibrocystin) (Polyductin) (Tigmin). | |||||
|
EXOC2_HUMAN
|
||||||
| NC score | 0.120950 (rank : 59) | θ value | 0.0113563 (rank : 58) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96KP1, Q5JPC8, Q96AN6, Q9NUZ8, Q9UJM7 | Gene names | EXOC2, SEC5, SEC5L1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 2 (Exocyst complex component Sec5). | |||||
|
EXOC2_MOUSE
|
||||||
| NC score | 0.116317 (rank : 60) | θ value | 0.0113563 (rank : 59) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D4H1 | Gene names | Exoc2, Sec5, Sec5l1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 2 (Exocyst complex component Sec5). | |||||
|
RASA3_HUMAN
|
||||||
| NC score | 0.086168 (rank : 61) | θ value | 0.0736092 (rank : 61) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14644 | Gene names | RASA3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein). | |||||
|
PXDC1_MOUSE
|
||||||
| NC score | 0.086047 (rank : 62) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q91ZV7, Q8BM20, Q9CWV5 | Gene names | Plxdc1, Tem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 1 precursor (Tumor endothelial marker 7). | |||||
|
PXDC1_HUMAN
|
||||||
| NC score | 0.082106 (rank : 63) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IUK5, Q5QCZ7, Q5QCZ8, Q5QCZ9, Q9HCT9 | Gene names | PLXDC1, TEM3, TEM7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 1 precursor (Tumor endothelial marker 7) (Tumor endothelial marker 3). | |||||
|
RASA3_MOUSE
|
||||||
| NC score | 0.073560 (rank : 64) | θ value | 0.62314 (rank : 65) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60790 | Gene names | Rasa3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein) (GapIII). | |||||
|
ATRN_HUMAN
|
||||||
| NC score | 0.072975 (rank : 65) | θ value | 0.0736092 (rank : 60) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
ATRN_MOUSE
|
||||||
| NC score | 0.065117 (rank : 66) | θ value | 0.21417 (rank : 62) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WU60, Q9R263, Q9WU77 | Gene names | Atrn, mg, Mgca | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany protein). | |||||
|
SYGP1_HUMAN
|
||||||
| NC score | 0.063887 (rank : 67) | θ value | 0.21417 (rank : 63) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96PV0, Q8TCS2, Q9UGE2 | Gene names | SYNGAP1, KIAA1938 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein SynGAP (Synaptic Ras-GTPase-activating protein 1) (Synaptic Ras-GAP 1) (Neuronal RasGAP). | |||||
|
PXDC2_HUMAN
|
||||||
| NC score | 0.055230 (rank : 68) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6UX71, Q96E59, Q96PD9, Q96SU9 | Gene names | PLXDC2, TEM7R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 2 precursor (Tumor endothelial marker 7-related protein). | |||||
|
MEGF8_HUMAN
|
||||||
| NC score | 0.055098 (rank : 69) | θ value | 0.813845 (rank : 66) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
MEGF8_MOUSE
|
||||||
| NC score | 0.049129 (rank : 70) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P60882, Q80TR3, Q80V41, Q8BMN9, Q8JZW7, Q8K0J3 | Gene names | Megf8, Egfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
DAB2P_MOUSE
|
||||||
| NC score | 0.042634 (rank : 71) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3UHC7, Q3TPD5, Q3UH44, Q6JTV1, Q80T97 | Gene names | Dab2ip, Kiaa1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein). | |||||
|
DAB2P_HUMAN
|
||||||
| NC score | 0.042378 (rank : 72) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5VWQ8, Q8TDL2, Q96SE1, Q9C0C0 | Gene names | DAB2IP, AF9Q34, AIP1, KIAA1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein) (DAB2 interaction protein) (ASK-interacting protein 1). | |||||
|
RASA2_HUMAN
|
||||||
| NC score | 0.034859 (rank : 73) | θ value | 0.365318 (rank : 64) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15283, O00695, Q15284, Q92594, Q99577, Q9UEQ2 | Gene names | RASA2, GAP1M, RASGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
RASA2_MOUSE
|
||||||
| NC score | 0.024093 (rank : 74) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P58069 | Gene names | Rasa2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
ITB1_HUMAN
|
||||||
| NC score | 0.023611 (rank : 75) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P05556, P78466, P78467, Q13089, Q13090, Q13091, Q13212, Q14622, Q14647 | Gene names | ITGB1, FNRB | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
ITB1_MOUSE
|
||||||
| NC score | 0.023438 (rank : 76) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P09055 | Gene names | Itgb1 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
T22D1_MOUSE
|
||||||
| NC score | 0.022938 (rank : 77) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62500, Q00992 | Gene names | Tsc22d1, Tgfb1i4, Tsc22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 1 (Transforming growth factor beta-1- induced transcript 4 protein) (Regulatory protein TSC-22) (TGFB- stimulated clone 22 homolog). | |||||
|
GNPAT_MOUSE
|
||||||
| NC score | 0.022559 (rank : 78) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P98192, Q9WUT6 | Gene names | Gnpat, Dhapat | |||
|
Domain Architecture |

|
|||||
| Description | Dihydroxyacetone phosphate acyltransferase (EC 2.3.1.42) (DHAP-AT) (DAP-AT) (Glycerone-phosphate O-acyltransferase) (Acyl- CoA:dihydroxyacetonephosphateacyltransferase). | |||||
|
MTMR5_HUMAN
|
||||||
| NC score | 0.020866 (rank : 79) | θ value | 1.06291 (rank : 68) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
ITB8_HUMAN
|
||||||
| NC score | 0.020478 (rank : 80) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P26012 | Gene names | ITGB8 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-8 precursor. | |||||
|
ITB6_MOUSE
|
||||||
| NC score | 0.020412 (rank : 81) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z0T9 | Gene names | Itgb6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
PRC1_MOUSE
|
||||||
| NC score | 0.011835 (rank : 82) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99K43, Q8CE25 | Gene names | Prc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein regulator of cytokinesis 1. | |||||
|
VIGLN_MOUSE
|
||||||
| NC score | 0.011364 (rank : 83) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VDJ3 | Gene names | Hdlbp | |||
|
Domain Architecture |

|
|||||
| Description | Vigilin (High density lipoprotein-binding protein) (HDL-binding protein). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.010373 (rank : 84) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
ACV1C_HUMAN
|
||||||
| NC score | 0.009290 (rank : 85) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NER5, Q4ZFZ8, Q86UL1, Q86UL2, Q8TBG2 | Gene names | ACVR1C, ALK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activin receptor type 1C precursor (EC 2.7.11.30) (ACTR-IC) (Activin receptor-like kinase 7) (ALK-7). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.008462 (rank : 86) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
AIM1_HUMAN
|
||||||
| NC score | 0.008311 (rank : 87) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y4K1, O00296, Q5VWJ2 | Gene names | AIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Absent in melanoma 1 protein. | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.007709 (rank : 88) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
PARC_HUMAN
|
||||||
| NC score | 0.006802 (rank : 89) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
PARC_MOUSE
|
||||||
| NC score | 0.006055 (rank : 90) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TT8, Q8BKL9, Q8CGC0, Q8R363 | Gene names | Parc, Kiaa0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein. | |||||
|
FAN_MOUSE
|
||||||
| NC score | 0.005403 (rank : 91) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35242 | Gene names | Nsmaf, Fan | |||
|
Domain Architecture |

|
|||||
| Description | Protein FAN (Factor associated with N-SMase activation) (Factor associated with neutral sphingomyelinase activation). | |||||
|
RPGR1_HUMAN
|
||||||
| NC score | 0.004641 (rank : 92) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96KN7, Q7Z2W6, Q8IXV5, Q96QA8, Q9HB94, Q9HB95, Q9HBK6, Q9NR40 | Gene names | RPGRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
KPCT_HUMAN
|
||||||
| NC score | 0.004578 (rank : 93) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 897 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q04759, Q3MJF1, Q64FY5, Q9H508, Q9H549 | Gene names | PRKCQ, PRKCT | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C theta type (EC 2.7.11.13) (nPKC-theta). | |||||
|
DDX20_HUMAN
|
||||||
| NC score | 0.004450 (rank : 94) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UHI6, Q96F72, Q9NVM3, Q9UF59, Q9UIY0, Q9Y659 | Gene names | DDX20, DP103, GEMIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ATP-dependent RNA helicase DDX20 (EC 3.6.1.-) (DEAD box protein 20) (DEAD box protein DP 103) (Component of gems 3) (Gemin-3). | |||||
|
KLH14_HUMAN
|
||||||
| NC score | 0.003630 (rank : 95) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P2G3 | Gene names | KLHL14, KIAA1384 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 14. | |||||
|
CREL1_MOUSE
|
||||||
| NC score | 0.003622 (rank : 96) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91XD7, Q8BGJ8 | Gene names | Creld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
KCMA1_MOUSE
|
||||||
| NC score | 0.003172 (rank : 97) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08460, Q64703, Q8VHF1, Q9R196 | Gene names | Kcnma1, Kcnma | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (mSlo) (mSlo1). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.001758 (rank : 98) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
RBCC1_HUMAN
|
||||||
| NC score | 0.001242 (rank : 99) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
CADH7_HUMAN
|
||||||
| NC score | 0.000307 (rank : 100) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULB5, Q9H157 | Gene names | CDH7, CDH7L1 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-7 precursor. | |||||
|
ZN687_MOUSE
|
||||||
| NC score | -0.000181 (rank : 101) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
MATN2_MOUSE
|
||||||
| NC score | -0.000529 (rank : 102) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08746 | Gene names | Matn2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
MYH9_MOUSE
|
||||||
| NC score | -0.000632 (rank : 103) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
ZN507_HUMAN
|
||||||
| NC score | -0.001811 (rank : 104) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TCN5 | Gene names | ZNF507 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 507. | |||||
|
ZN226_HUMAN
|
||||||
| NC score | -0.002626 (rank : 105) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NYT6, Q96TE6, Q9NS44 | Gene names | ZNF226 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 226. | |||||
|
ZN567_HUMAN
|
||||||
| NC score | -0.002709 (rank : 106) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N184, Q6N044 | Gene names | ZNF567 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 567. | |||||
|
ZN480_HUMAN
|
||||||
| NC score | -0.003400 (rank : 107) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WV37, Q5JPG9, Q6P0Q4, Q8N1M5 | Gene names | ZNF480 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 480. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | -0.003672 (rank : 108) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||