Please be patient as the page loads
|
PLXC1_HUMAN
|
||||||
| SwissProt Accessions | O60486, Q59H25 | Gene names | PLXNC1, VESPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PLXC1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | O60486, Q59H25 | Gene names | PLXNC1, VESPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
|
PLXC1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.997922 (rank : 2) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9QZC2, Q8CGW1 | Gene names | Plxnc1, Vespr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
|
PLXD1_HUMAN
|
||||||
| θ value | 8.0589e-150 (rank : 3) | NC score | 0.943883 (rank : 3) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Y4D7, Q6PJS9, Q8IZJ2, Q9BTQ2 | Gene names | PLXND1, KIAA0620 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-D1 precursor. | |||||
|
PLXA4_MOUSE
|
||||||
| θ value | 2.00348e-116 (rank : 4) | NC score | 0.923254 (rank : 7) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q80UG2, Q5DTW8, Q8BKK9 | Gene names | Plxna4, Kiaa1550 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
PLXA4_HUMAN
|
||||||
| θ value | 9.94316e-116 (rank : 5) | NC score | 0.923073 (rank : 8) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9HCM2, Q6UWC6, Q6ZW89, Q8N969, Q8ND00, Q8NEN3, Q9NTD4 | Gene names | PLXNA4, KIAA1550, PLXNA4A, PLXNA4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
PLXA1_HUMAN
|
||||||
| θ value | 1.29861e-115 (rank : 6) | NC score | 0.922490 (rank : 11) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UIW2 | Gene names | PLXNA1, NOV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Semaphorin receptor NOV). | |||||
|
PLXA2_HUMAN
|
||||||
| θ value | 1.09934e-114 (rank : 7) | NC score | 0.923636 (rank : 6) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O75051, Q6UX61, Q96GN9, Q9BRL1, Q9UIW1 | Gene names | PLXNA2, KIAA0463, OCT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A2 precursor (Semaphorin receptor OCT). | |||||
|
PLXA1_MOUSE
|
||||||
| θ value | 7.12574e-114 (rank : 8) | NC score | 0.922021 (rank : 13) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P70206, Q5DTR0 | Gene names | Plxna1, KIAA4053 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Plexin-1) (Plex 1). | |||||
|
PLXA2_MOUSE
|
||||||
| θ value | 1.48581e-111 (rank : 9) | NC score | 0.923040 (rank : 9) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P70207, Q6NVE6, Q6PHN4, Q80TZ7, Q8R1I4 | Gene names | Plxna2, Kiaa0463 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A2 precursor (Plexin-2) (Plex 2). | |||||
|
PLXA3_HUMAN
|
||||||
| θ value | 2.5344e-111 (rank : 10) | NC score | 0.925531 (rank : 5) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P51805 | Gene names | PLXNA3, PLXN4, SEX | |||
|
Domain Architecture |

|
|||||
| Description | Plexin-A3 precursor (Plexin-4) (Semaphorin receptor SEX). | |||||
|
PLXB1_MOUSE
|
||||||
| θ value | 2.5344e-111 (rank : 11) | NC score | 0.918552 (rank : 14) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
PLXB1_HUMAN
|
||||||
| θ value | 4.32305e-111 (rank : 12) | NC score | 0.908261 (rank : 15) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
PLXB2_HUMAN
|
||||||
| θ value | 3.42535e-108 (rank : 13) | NC score | 0.927886 (rank : 4) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O15031, Q7KZU3, Q9BSU7 | Gene names | PLXNB2, KIAA0315 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B2 precursor (MM1). | |||||
|
PLXB3_HUMAN
|
||||||
| θ value | 9.035e-101 (rank : 14) | NC score | 0.922825 (rank : 10) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9ULL4, Q9HDA4 | Gene names | PLXNB3, KIAA1206 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor. | |||||
|
PLXB3_MOUSE
|
||||||
| θ value | 1.18001e-100 (rank : 15) | NC score | 0.922145 (rank : 12) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9QY40, Q80TH8 | Gene names | Plxnb3, Kiaa1206, Plxn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor (Plexin-6). | |||||
|
SEM4G_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 16) | NC score | 0.403769 (rank : 21) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9WUH7 | Gene names | Sema4g | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
SEM4B_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 17) | NC score | 0.428653 (rank : 18) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NPR2, Q6UXE3, Q8WVP9, Q96FK5, Q9C0B8, Q9H691, Q9NPM8, Q9NPN0 | Gene names | SEMA4B, KIAA1745 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4B precursor. | |||||
|
SEM4C_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 18) | NC score | 0.412041 (rank : 20) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9C0C4, Q7Z5X0 | Gene names | SEMA4C, KIAA1739 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4C precursor. | |||||
|
SEM4F_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 19) | NC score | 0.365707 (rank : 26) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O95754, Q9NS35 | Gene names | SEMA4F, SEMAM, SEMAW | |||
|
Domain Architecture |
|
|||||
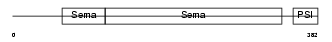
| Description | Semaphorin-4F precursor (Semaphorin W) (Sema W) (Semaphorin M) (Sema M). | |||||
|
SEM4B_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 20) | NC score | 0.438247 (rank : 16) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62179, Q4PKI6, Q69ZB7 | Gene names | Sema4b, Kiaa1745, Semac, SemC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-4B precursor (Semaphorin C) (Sema C). | |||||
|
SEM4A_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 21) | NC score | 0.430106 (rank : 17) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9H3S1, Q8WUA9 | Gene names | SEMA4A, SEMB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
SEM4F_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 22) | NC score | 0.366312 (rank : 25) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Z123, Q9R1Y1 | Gene names | Sema4f, Semaw | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4F precursor (Semaphorin W) (Sema W). | |||||
|
SEM4G_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 23) | NC score | 0.401082 (rank : 22) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NTN9, Q9HCF3 | Gene names | SEMA4G, KIAA1619 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
SEM4C_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 24) | NC score | 0.381580 (rank : 23) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q64151 | Gene names | Sema4c, Semacl1, Semai | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4C precursor (Semaphorin I) (Sema I) (Semaphorin C-like 1) (M-Sema F). | |||||
|
SEM4D_MOUSE
|
||||||
| θ value | 1.29631e-06 (rank : 25) | NC score | 0.362207 (rank : 29) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O09126 | Gene names | Sema4d, Semacl2, Semaj | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Semaphorin J) (Sema J) (Semaphorin C-like 2) (M-Sema G). | |||||
|
MET_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 26) | NC score | 0.163321 (rank : 55) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P08581, O60366, Q12875, Q9UDX7, Q9UPL8 | Gene names | MET | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor receptor precursor (EC 2.7.10.1) (HGF receptor) (Scatter factor receptor) (SF receptor) (HGF/SF receptor) (Met proto-oncogene tyrosine kinase) (c-Met). | |||||
|
SEM4A_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 27) | NC score | 0.424197 (rank : 19) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62178 | Gene names | Sema4a, Semab, SemB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
SEM3E_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 28) | NC score | 0.323024 (rank : 47) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O15041, Q75M94, Q75M97 | Gene names | SEMA3E, KIAA0331 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-3E precursor. | |||||
|
MET_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 29) | NC score | 0.164738 (rank : 54) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P16056, Q62125 | Gene names | Met | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor receptor precursor (EC 2.7.10.1) (HGF receptor) (Scatter factor receptor) (SF receptor) (HGF/SF receptor) (Met proto-oncogene tyrosine kinase) (c-Met). | |||||
|
SEM4D_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 30) | NC score | 0.350933 (rank : 31) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q92854, Q7Z5S4 | Gene names | SEMA4D, CD100 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Leukocyte activation antigen CD100) (BB18) (A8) (GR3). | |||||
|
SEM3E_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 31) | NC score | 0.317969 (rank : 49) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70275, O09078, O09079 | Gene names | Sema3e, Semah, Semh | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3E precursor (Semaphorin H) (Sema H). | |||||
|
SEM3A_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 32) | NC score | 0.321588 (rank : 48) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O08665, Q5BL08, Q62180, Q62215 | Gene names | Sema3a, Semad, SemD | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3A precursor (Semaphorin III) (Sema III) (Semaphorin D) (Sema D). | |||||
|
SEM3A_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 33) | NC score | 0.317906 (rank : 50) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q14563 | Gene names | SEMA3A | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3A precursor (Semaphorin III) (Sema III). | |||||
|
SEM3C_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 34) | NC score | 0.338136 (rank : 40) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q62181 | Gene names | Sema3c, Semae, SemE | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3C precursor (Semaphorin E) (Sema E). | |||||
|
SEM5A_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 35) | NC score | 0.333489 (rank : 44) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q62217 | Gene names | Sema5a, Semaf, SemF | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5A precursor (Semaphorin F) (Sema F). | |||||
|
SEM3F_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 36) | NC score | 0.348584 (rank : 33) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O88632, O88633 | Gene names | Sema3f | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3F precursor (Semaphorin IV) (Sema IV). | |||||
|
SEM5A_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 37) | NC score | 0.324890 (rank : 46) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13591, O60408, Q1RLL9 | Gene names | SEMA5A, SEMAF | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5A precursor (Semaphorin F) (Sema F). | |||||
|
SEM6C_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 38) | NC score | 0.317295 (rank : 51) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SEM5B_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 39) | NC score | 0.338224 (rank : 39) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9P283, Q6UY12 | Gene names | SEMA5B, KIAA1445 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5B precursor. | |||||
|
SEM3C_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 40) | NC score | 0.335042 (rank : 42) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q99985 | Gene names | SEMA3C, SEMAE | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3C precursor (Semaphorin E) (Sema E). | |||||
|
SEM3F_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 41) | NC score | 0.347553 (rank : 34) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13275, Q13274, Q13372, Q15704 | Gene names | SEMA3F | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3F precursor (Semaphorin IV) (Sema IV) (Sema III/F). | |||||
|
SEM5B_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 42) | NC score | 0.334774 (rank : 43) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q60519 | Gene names | Sema5b, Semag, SemG | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5B precursor (Semaphorin G) (Sema G). | |||||
|
SEM6C_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 43) | NC score | 0.340909 (rank : 37) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H3T2, Q5JR71, Q8WXT8, Q8WXT9, Q8WXU0, Q96JF8 | Gene names | SEMA6C, KIAA1869, SEMAY | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SEM3D_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 44) | NC score | 0.308199 (rank : 53) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BH34 | Gene names | Sema3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-3D precursor. | |||||
|
SEM3D_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 45) | NC score | 0.315267 (rank : 52) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O95025, Q6UW77, Q8NCQ1 | Gene names | SEMA3D | |||
|
Domain Architecture |
|
|||||
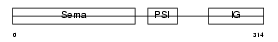
| Description | Semaphorin-3D precursor. | |||||
|
SEM6D_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 46) | NC score | 0.364071 (rank : 27) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q76KF0, Q76KF1, Q76KF2, Q76KF3, Q76KF4, Q80TD0 | Gene names | Sema6d, Kiaa1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
PXDC1_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 47) | NC score | 0.134440 (rank : 60) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IUK5, Q5QCZ7, Q5QCZ8, Q5QCZ9, Q9HCT9 | Gene names | PLXDC1, TEM3, TEM7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 1 precursor (Tumor endothelial marker 7) (Tumor endothelial marker 3). | |||||
|
SEM7A_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 48) | NC score | 0.368128 (rank : 24) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9QUR8, O88371 | Gene names | Sema7a, Cd108, Semal, Semk1 | |||
|
Domain Architecture |
|
|||||
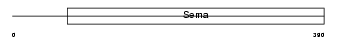
| Description | Semaphorin-7A precursor (Semaphorin L) (Sema L) (Semaphorin K1) (Sema K1) (CD108 antigen) (CDw108). | |||||
|
RON_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 49) | NC score | 0.139099 (rank : 57) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q04912 | Gene names | MST1R, RON | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage-stimulating protein receptor precursor (EC 2.7.10.1) (MSP receptor) (p185-Ron) (CDw136 antigen) [Contains: Macrophage- stimulating protein receptor alpha chain; Macrophage-stimulating protein receptor beta chain]. | |||||
|
SEM3B_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 50) | NC score | 0.335256 (rank : 41) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13214, Q8TB71, Q8TDV7, Q93018, Q96GX0 | Gene names | SEMA3B, SEMA5 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3B precursor (Semaphorin V) (Sema V) (Sema A(V)). | |||||
|
SEM6A_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 51) | NC score | 0.348627 (rank : 32) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H2E6, Q9P2H9 | Gene names | SEMA6A, KIAA1368 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1). | |||||
|
SEM6A_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 52) | NC score | 0.345773 (rank : 36) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O35464, Q6P5A8, Q6PCN9, Q9EQ71 | Gene names | Sema6a, Semaq | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1) (Semaphorin Q) (Sema Q). | |||||
|
SEM6D_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 53) | NC score | 0.362428 (rank : 28) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8NFY4, Q8NFY3, Q8NFY5, Q8NFY6, Q8NFY7, Q9P249 | Gene names | SEMA6D, KIAA1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
PXDC1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 54) | NC score | 0.135355 (rank : 58) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91ZV7, Q8BM20, Q9CWV5 | Gene names | Plxdc1, Tem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 1 precursor (Tumor endothelial marker 7). | |||||
|
SEM3B_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 55) | NC score | 0.338266 (rank : 38) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62177 | Gene names | Sema3b, Sema, Semaa | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3B precursor (Semaphorin A) (Sema A). | |||||
|
CSMD3_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 56) | NC score | 0.015503 (rank : 81) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z407, Q96PZ3 | Gene names | CSMD3, KIAA1894 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 3 precursor (CUB and sushi multiple domains protein 3). | |||||
|
SEM6B_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 57) | NC score | 0.347372 (rank : 35) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H3T3, Q9NRK9 | Gene names | SEMA6B, SEMAZ | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin Z) (Sema Z). | |||||
|
RBP2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 58) | NC score | 0.010954 (rank : 83) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49792, Q13074, Q15280, Q53TE2, Q59FH7 | Gene names | RANBP2, NUP358 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-binding protein 2 (RanBP2) (Nuclear pore complex protein Nup358) (Nucleoporin Nup358) (358 kDa nucleoporin) (P270). | |||||
|
PXDC2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 59) | NC score | 0.092308 (rank : 62) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9DC11, Q6PET5, Q91ZV6 | Gene names | Plxdc2, Tem7r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 2 precursor (Tumor endothelial marker 7-related protein). | |||||
|
PXDC2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 60) | NC score | 0.097240 (rank : 61) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6UX71, Q96E59, Q96PD9, Q96SU9 | Gene names | PLXDC2, TEM7R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 2 precursor (Tumor endothelial marker 7-related protein). | |||||
|
WAPL_HUMAN
|
||||||
| θ value | 0.47712 (rank : 61) | NC score | 0.045158 (rank : 69) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z5K2, Q5VSK5, Q8IX10, Q92549 | Gene names | WAPAL, FOE, KIAA0261, WAPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wings apart-like protein homolog (Friend of EBNA2 protein). | |||||
|
WAPL_MOUSE
|
||||||
| θ value | 0.47712 (rank : 62) | NC score | 0.044776 (rank : 71) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q65Z40, Q6ZQE9 | Gene names | Wapal, Kiaa0261, Wapl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wings apart-like protein homolog (Dioxin-inducible factor 2) (DIF-2). | |||||
|
CASP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 63) | NC score | 0.008622 (rank : 86) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 64) | NC score | 0.010080 (rank : 84) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
MEGF8_HUMAN
|
||||||
| θ value | 0.813845 (rank : 65) | NC score | 0.052085 (rank : 67) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
RON_MOUSE
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.134935 (rank : 59) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q62190, Q62555 | Gene names | Mst1r, Ron, Stk | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage-stimulating protein receptor precursor (EC 2.7.10.1) (MSP receptor) (p185-Ron) (Stem cell-derived tyrosine kinase) (CDw136 antigen) [Contains: Macrophage-stimulating protein receptor alpha chain; Macrophage-stimulating protein receptor beta chain]. | |||||
|
MYCT_HUMAN
|
||||||
| θ value | 1.38821 (rank : 67) | NC score | 0.015597 (rank : 80) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96QE2 | Gene names | SLC2A13 | |||
|
Domain Architecture |

|
|||||
| Description | Proton myo-inositol cotransporter (H(+)-myo-inositol cotransporter) (Hmit). | |||||
|
SEM7A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 68) | NC score | 0.352032 (rank : 30) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75326 | Gene names | SEMA7A, CD108, SEMAL | |||
|
Domain Architecture |
|
|||||
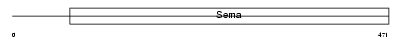
| Description | Semaphorin-7A precursor (Semaphorin L) (Sema L) (Semaphorin K1) (Sema K1) (John-Milton-Hargen human blood group Ag) (JMH blood group antigen) (CD108 antigen) (CDw108). | |||||
|
SEM6B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.328942 (rank : 45) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O54951 | Gene names | Sema6b, Seman | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin VIB) (Sema VIB) (Semaphorin N) (Sema N). | |||||
|
POTE1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.015603 (rank : 79) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NUX5, O95018, Q9H662, Q9NW19, Q9UG95 | Gene names | POT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protection of telomeres 1 (hPot1) (POT1-like telomere end-binding protein). | |||||
|
ZDH15_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.009031 (rank : 85) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96MV8, Q3SY30, Q6UWH3 | Gene names | ZDHHC15 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC15 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 15) (DHHC-15). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.004718 (rank : 94) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CO020_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.023467 (rank : 72) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H611 | Gene names | C15orf20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf20. | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.004741 (rank : 93) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.003736 (rank : 98) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
MEGF8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.044835 (rank : 70) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P60882, Q80TR3, Q80V41, Q8BMN9, Q8JZW7, Q8K0J3 | Gene names | Megf8, Egfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
AIM2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.019938 (rank : 73) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14862 | Gene names | AIM2 | |||
|
Domain Architecture |
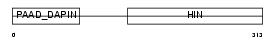
|
|||||
| Description | Interferon-inducible protein AIM2 (Absent in melanoma 2). | |||||
|
CASP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.007196 (rank : 89) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CUBN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.006628 (rank : 90) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60494, Q5VTA6, Q96RU9 | Gene names | CUBN, IFCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor) (Intrinsic factor-vitamin B12 receptor) (460 kDa receptor) (Intestinal intrinsic factor receptor). | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.006620 (rank : 91) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
ITB6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.015921 (rank : 77) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z0T9 | Gene names | Itgb6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.002648 (rank : 99) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
SNX17_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.015761 (rank : 78) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15036, Q53HN7, Q6IAS3 | Gene names | SNX17, KIAA0064 | |||
|
Domain Architecture |
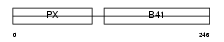
|
|||||
| Description | Sorting nexin-17. | |||||
|
ITB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.019270 (rank : 74) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P05556, P78466, P78467, Q13089, Q13090, Q13091, Q13212, Q14622, Q14647 | Gene names | ITGB1, FNRB | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
ITB1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.019184 (rank : 75) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P09055 | Gene names | Itgb1 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
SNX17_MOUSE
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.014766 (rank : 82) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVL3, Q8R0N8 | Gene names | Snx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-17. | |||||
|
TPSNR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.005257 (rank : 92) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BX59, Q9NWB8 | Gene names | TAPBPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tapasin-related protein precursor (TAPASIN-R) (Tapasin-like) (TAP- binding protein-related protein) (TAPBP-R) (TAP-binding protein-like). | |||||
|
TRS85_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.018771 (rank : 76) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2L5, Q9H0L2 | Gene names | KIAA1012 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein TRS85 homolog. | |||||
|
LIMA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.004301 (rank : 95) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ERG0, Q9ERG1 | Gene names | Lima1, D15Ertd366e, Eplin | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm) (mEPLIN). | |||||
|
WDR24_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.004295 (rank : 96) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96S15, Q96GC7, Q9H0B7 | Gene names | WDR24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 24. | |||||
|
ZDH15_MOUSE
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.007441 (rank : 88) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BGJ0, Q3TV63, Q8BMB7 | Gene names | Zdhhc15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Palmitoyltransferase ZDHHC15 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 15) (DHHC-15). | |||||
|
DMBT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.004140 (rank : 97) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60997, Q80YC6, Q9JMJ9 | Gene names | Dmbt1, Crpd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (CRP-ductin) (Vomeroglandin). | |||||
|
HERC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.001645 (rank : 100) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15034 | Gene names | HERC3, KIAA0032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain protein 3. | |||||
|
TINF2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.008182 (rank : 87) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QXG9 | Gene names | Tinf2, Tin2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TERF1-interacting nuclear factor 2 (TRF1-interacting nuclear protein 2). | |||||
|
ATRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.058477 (rank : 65) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
ATRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.051354 (rank : 68) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WU60, Q9R263, Q9WU77 | Gene names | Atrn, mg, Mgca | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany protein). | |||||
|
EXOC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.081196 (rank : 63) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96KP1, Q5JPC8, Q96AN6, Q9NUZ8, Q9UJM7 | Gene names | EXOC2, SEC5, SEC5L1 | |||
|
Domain Architecture |
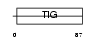
|
|||||
| Description | Exocyst complex component 2 (Exocyst complex component Sec5). | |||||
|
EXOC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.076370 (rank : 64) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D4H1 | Gene names | Exoc2, Sec5, Sec5l1 | |||
|
Domain Architecture |
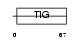
|
|||||
| Description | Exocyst complex component 2 (Exocyst complex component Sec5). | |||||
|
PKHD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.139580 (rank : 56) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TCZ9 | Gene names | PKHD1, FCYT, TIGM1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney and hepatic disease 1 precursor (Fibrocystin) (Polyductin) (Tigmin). | |||||
|
RASA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.053989 (rank : 66) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14644 | Gene names | RASA3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein). | |||||
|
PLXC1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | O60486, Q59H25 | Gene names | PLXNC1, VESPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
|
PLXC1_MOUSE
|
||||||
| NC score | 0.997922 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9QZC2, Q8CGW1 | Gene names | Plxnc1, Vespr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
|
PLXD1_HUMAN
|
||||||
| NC score | 0.943883 (rank : 3) | θ value | 8.0589e-150 (rank : 3) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Y4D7, Q6PJS9, Q8IZJ2, Q9BTQ2 | Gene names | PLXND1, KIAA0620 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-D1 precursor. | |||||
|
PLXB2_HUMAN
|
||||||
| NC score | 0.927886 (rank : 4) | θ value | 3.42535e-108 (rank : 13) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O15031, Q7KZU3, Q9BSU7 | Gene names | PLXNB2, KIAA0315 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B2 precursor (MM1). | |||||
|
PLXA3_HUMAN
|
||||||
| NC score | 0.925531 (rank : 5) | θ value | 2.5344e-111 (rank : 10) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P51805 | Gene names | PLXNA3, PLXN4, SEX | |||
|
Domain Architecture |

|
|||||
| Description | Plexin-A3 precursor (Plexin-4) (Semaphorin receptor SEX). | |||||
|
PLXA2_HUMAN
|
||||||
| NC score | 0.923636 (rank : 6) | θ value | 1.09934e-114 (rank : 7) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O75051, Q6UX61, Q96GN9, Q9BRL1, Q9UIW1 | Gene names | PLXNA2, KIAA0463, OCT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A2 precursor (Semaphorin receptor OCT). | |||||
|
PLXA4_MOUSE
|
||||||
| NC score | 0.923254 (rank : 7) | θ value | 2.00348e-116 (rank : 4) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q80UG2, Q5DTW8, Q8BKK9 | Gene names | Plxna4, Kiaa1550 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
PLXA4_HUMAN
|
||||||
| NC score | 0.923073 (rank : 8) | θ value | 9.94316e-116 (rank : 5) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9HCM2, Q6UWC6, Q6ZW89, Q8N969, Q8ND00, Q8NEN3, Q9NTD4 | Gene names | PLXNA4, KIAA1550, PLXNA4A, PLXNA4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A4 precursor. | |||||
|
PLXA2_MOUSE
|
||||||
| NC score | 0.923040 (rank : 9) | θ value | 1.48581e-111 (rank : 9) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P70207, Q6NVE6, Q6PHN4, Q80TZ7, Q8R1I4 | Gene names | Plxna2, Kiaa0463 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A2 precursor (Plexin-2) (Plex 2). | |||||
|
PLXB3_HUMAN
|
||||||
| NC score | 0.922825 (rank : 10) | θ value | 9.035e-101 (rank : 14) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9ULL4, Q9HDA4 | Gene names | PLXNB3, KIAA1206 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor. | |||||
|
PLXA1_HUMAN
|
||||||
| NC score | 0.922490 (rank : 11) | θ value | 1.29861e-115 (rank : 6) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UIW2 | Gene names | PLXNA1, NOV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Semaphorin receptor NOV). | |||||
|
PLXB3_MOUSE
|
||||||
| NC score | 0.922145 (rank : 12) | θ value | 1.18001e-100 (rank : 15) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9QY40, Q80TH8 | Gene names | Plxnb3, Kiaa1206, Plxn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor (Plexin-6). | |||||
|
PLXA1_MOUSE
|
||||||
| NC score | 0.922021 (rank : 13) | θ value | 7.12574e-114 (rank : 8) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P70206, Q5DTR0 | Gene names | Plxna1, KIAA4053 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-A1 precursor (Plexin-1) (Plex 1). | |||||
|
PLXB1_MOUSE
|
||||||
| NC score | 0.918552 (rank : 14) | θ value | 2.5344e-111 (rank : 11) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
PLXB1_HUMAN
|
||||||
| NC score | 0.908261 (rank : 15) | θ value | 4.32305e-111 (rank : 12) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
SEM4B_MOUSE
|
||||||
| NC score | 0.438247 (rank : 16) | θ value | 1.17247e-07 (rank : 20) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62179, Q4PKI6, Q69ZB7 | Gene names | Sema4b, Kiaa1745, Semac, SemC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-4B precursor (Semaphorin C) (Sema C). | |||||
|
SEM4A_HUMAN
|
||||||
| NC score | 0.430106 (rank : 17) | θ value | 1.53129e-07 (rank : 21) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9H3S1, Q8WUA9 | Gene names | SEMA4A, SEMB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
SEM4B_HUMAN
|
||||||
| NC score | 0.428653 (rank : 18) | θ value | 2.36244e-08 (rank : 17) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9NPR2, Q6UXE3, Q8WVP9, Q96FK5, Q9C0B8, Q9H691, Q9NPM8, Q9NPN0 | Gene names | SEMA4B, KIAA1745 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4B precursor. | |||||
|
SEM4A_MOUSE
|
||||||
| NC score | 0.424197 (rank : 19) | θ value | 6.43352e-06 (rank : 27) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62178 | Gene names | Sema4a, Semab, SemB | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4A precursor (Semaphorin B) (Sema B). | |||||
|
SEM4C_HUMAN
|
||||||
| NC score | 0.412041 (rank : 20) | θ value | 3.08544e-08 (rank : 18) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9C0C4, Q7Z5X0 | Gene names | SEMA4C, KIAA1739 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4C precursor. | |||||
|
SEM4G_MOUSE
|
||||||
| NC score | 0.403769 (rank : 21) | θ value | 1.80886e-08 (rank : 16) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9WUH7 | Gene names | Sema4g | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
SEM4G_HUMAN
|
||||||
| NC score | 0.401082 (rank : 22) | θ value | 2.61198e-07 (rank : 23) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NTN9, Q9HCF3 | Gene names | SEMA4G, KIAA1619 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4G precursor. | |||||
|
SEM4C_MOUSE
|
||||||
| NC score | 0.381580 (rank : 23) | θ value | 4.45536e-07 (rank : 24) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q64151 | Gene names | Sema4c, Semacl1, Semai | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4C precursor (Semaphorin I) (Sema I) (Semaphorin C-like 1) (M-Sema F). | |||||
|
SEM7A_MOUSE
|
||||||
| NC score | 0.368128 (rank : 24) | θ value | 0.00509761 (rank : 48) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9QUR8, O88371 | Gene names | Sema7a, Cd108, Semal, Semk1 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-7A precursor (Semaphorin L) (Sema L) (Semaphorin K1) (Sema K1) (CD108 antigen) (CDw108). | |||||
|
SEM4F_MOUSE
|
||||||
| NC score | 0.366312 (rank : 25) | θ value | 1.53129e-07 (rank : 22) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Z123, Q9R1Y1 | Gene names | Sema4f, Semaw | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4F precursor (Semaphorin W) (Sema W). | |||||
|
SEM4F_HUMAN
|
||||||
| NC score | 0.365707 (rank : 26) | θ value | 3.08544e-08 (rank : 19) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O95754, Q9NS35 | Gene names | SEMA4F, SEMAM, SEMAW | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4F precursor (Semaphorin W) (Sema W) (Semaphorin M) (Sema M). | |||||
|
SEM6D_MOUSE
|
||||||
| NC score | 0.364071 (rank : 27) | θ value | 0.00298849 (rank : 46) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q76KF0, Q76KF1, Q76KF2, Q76KF3, Q76KF4, Q80TD0 | Gene names | Sema6d, Kiaa1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SEM6D_HUMAN
|
||||||
| NC score | 0.362428 (rank : 28) | θ value | 0.0113563 (rank : 53) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8NFY4, Q8NFY3, Q8NFY5, Q8NFY6, Q8NFY7, Q9P249 | Gene names | SEMA6D, KIAA1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SEM4D_MOUSE
|
||||||
| NC score | 0.362207 (rank : 29) | θ value | 1.29631e-06 (rank : 25) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O09126 | Gene names | Sema4d, Semacl2, Semaj | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Semaphorin J) (Sema J) (Semaphorin C-like 2) (M-Sema G). | |||||
|
SEM7A_HUMAN
|
||||||
| NC score | 0.352032 (rank : 30) | θ value | 1.38821 (rank : 68) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O75326 | Gene names | SEMA7A, CD108, SEMAL | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-7A precursor (Semaphorin L) (Sema L) (Semaphorin K1) (Sema K1) (John-Milton-Hargen human blood group Ag) (JMH blood group antigen) (CD108 antigen) (CDw108). | |||||
|
SEM4D_HUMAN
|
||||||
| NC score | 0.350933 (rank : 31) | θ value | 3.19293e-05 (rank : 30) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q92854, Q7Z5S4 | Gene names | SEMA4D, CD100 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-4D precursor (Leukocyte activation antigen CD100) (BB18) (A8) (GR3). | |||||
|
SEM6A_HUMAN
|
||||||
| NC score | 0.348627 (rank : 32) | θ value | 0.00869519 (rank : 51) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H2E6, Q9P2H9 | Gene names | SEMA6A, KIAA1368 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1). | |||||
|
SEM3F_MOUSE
|
||||||
| NC score | 0.348584 (rank : 33) | θ value | 0.000270298 (rank : 36) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O88632, O88633 | Gene names | Sema3f | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3F precursor (Semaphorin IV) (Sema IV). | |||||
|
SEM3F_HUMAN
|
||||||
| NC score | 0.347553 (rank : 34) | θ value | 0.00102713 (rank : 41) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13275, Q13274, Q13372, Q15704 | Gene names | SEMA3F | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3F precursor (Semaphorin IV) (Sema IV) (Sema III/F). | |||||
|
SEM6B_HUMAN
|
||||||
| NC score | 0.347372 (rank : 35) | θ value | 0.0736092 (rank : 57) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H3T3, Q9NRK9 | Gene names | SEMA6B, SEMAZ | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin Z) (Sema Z). | |||||
|
SEM6A_MOUSE
|
||||||
| NC score | 0.345773 (rank : 36) | θ value | 0.0113563 (rank : 52) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O35464, Q6P5A8, Q6PCN9, Q9EQ71 | Gene names | Sema6a, Semaq | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6A precursor (Semaphorin VIA) (Sema VIA) (Semaphorin 6A-1) (SEMA6A-1) (Semaphorin Q) (Sema Q). | |||||
|
SEM6C_HUMAN
|
||||||
| NC score | 0.340909 (rank : 37) | θ value | 0.00134147 (rank : 43) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H3T2, Q5JR71, Q8WXT8, Q8WXT9, Q8WXU0, Q96JF8 | Gene names | SEMA6C, KIAA1869, SEMAY | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SEM3B_MOUSE
|
||||||
| NC score | 0.338266 (rank : 38) | θ value | 0.0431538 (rank : 55) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62177 | Gene names | Sema3b, Sema, Semaa | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3B precursor (Semaphorin A) (Sema A). | |||||
|
SEM5B_HUMAN
|
||||||
| NC score | 0.338224 (rank : 39) | θ value | 0.00035302 (rank : 39) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9P283, Q6UY12 | Gene names | SEMA5B, KIAA1445 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5B precursor. | |||||
|
SEM3C_MOUSE
|
||||||
| NC score | 0.338136 (rank : 40) | θ value | 0.000121331 (rank : 34) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q62181 | Gene names | Sema3c, Semae, SemE | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3C precursor (Semaphorin E) (Sema E). | |||||
|
SEM3B_HUMAN
|
||||||
| NC score | 0.335256 (rank : 41) | θ value | 0.00869519 (rank : 50) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13214, Q8TB71, Q8TDV7, Q93018, Q96GX0 | Gene names | SEMA3B, SEMA5 | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3B precursor (Semaphorin V) (Sema V) (Sema A(V)). | |||||
|
SEM3C_HUMAN
|
||||||
| NC score | 0.335042 (rank : 42) | θ value | 0.000602161 (rank : 40) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q99985 | Gene names | SEMA3C, SEMAE | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3C precursor (Semaphorin E) (Sema E). | |||||
|
SEM5B_MOUSE
|
||||||
| NC score | 0.334774 (rank : 43) | θ value | 0.00102713 (rank : 42) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q60519 | Gene names | Sema5b, Semag, SemG | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5B precursor (Semaphorin G) (Sema G). | |||||
|
SEM5A_MOUSE
|
||||||
| NC score | 0.333489 (rank : 44) | θ value | 0.000121331 (rank : 35) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q62217 | Gene names | Sema5a, Semaf, SemF | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5A precursor (Semaphorin F) (Sema F). | |||||
|
SEM6B_MOUSE
|
||||||
| NC score | 0.328942 (rank : 45) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O54951 | Gene names | Sema6b, Seman | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin VIB) (Sema VIB) (Semaphorin N) (Sema N). | |||||
|
SEM5A_HUMAN
|
||||||
| NC score | 0.324890 (rank : 46) | θ value | 0.000270298 (rank : 37) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13591, O60408, Q1RLL9 | Gene names | SEMA5A, SEMAF | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-5A precursor (Semaphorin F) (Sema F). | |||||
|
SEM3E_HUMAN
|
||||||
| NC score | 0.323024 (rank : 47) | θ value | 1.09739e-05 (rank : 28) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O15041, Q75M94, Q75M97 | Gene names | SEMA3E, KIAA0331 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-3E precursor. | |||||
|
SEM3A_MOUSE
|
||||||
| NC score | 0.321588 (rank : 48) | θ value | 9.29e-05 (rank : 32) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O08665, Q5BL08, Q62180, Q62215 | Gene names | Sema3a, Semad, SemD | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3A precursor (Semaphorin III) (Sema III) (Semaphorin D) (Sema D). | |||||
|
SEM3E_MOUSE
|
||||||
| NC score | 0.317969 (rank : 49) | θ value | 5.44631e-05 (rank : 31) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70275, O09078, O09079 | Gene names | Sema3e, Semah, Semh | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3E precursor (Semaphorin H) (Sema H). | |||||
|
SEM3A_HUMAN
|
||||||
| NC score | 0.317906 (rank : 50) | θ value | 0.000121331 (rank : 33) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q14563 | Gene names | SEMA3A | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3A precursor (Semaphorin III) (Sema III). | |||||
|
SEM6C_MOUSE
|
||||||
| NC score | 0.317295 (rank : 51) | θ value | 0.000270298 (rank : 38) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
SEM3D_HUMAN
|
||||||
| NC score | 0.315267 (rank : 52) | θ value | 0.00298849 (rank : 45) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O95025, Q6UW77, Q8NCQ1 | Gene names | SEMA3D | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3D precursor. | |||||
|
SEM3D_MOUSE
|
||||||
| NC score | 0.308199 (rank : 53) | θ value | 0.00228821 (rank : 44) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BH34 | Gene names | Sema3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-3D precursor. | |||||
|
MET_MOUSE
|
||||||
| NC score | 0.164738 (rank : 54) | θ value | 1.43324e-05 (rank : 29) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P16056, Q62125 | Gene names | Met | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor receptor precursor (EC 2.7.10.1) (HGF receptor) (Scatter factor receptor) (SF receptor) (HGF/SF receptor) (Met proto-oncogene tyrosine kinase) (c-Met). | |||||
|
MET_HUMAN
|
||||||
| NC score | 0.163321 (rank : 55) | θ value | 4.92598e-06 (rank : 26) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P08581, O60366, Q12875, Q9UDX7, Q9UPL8 | Gene names | MET | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor receptor precursor (EC 2.7.10.1) (HGF receptor) (Scatter factor receptor) (SF receptor) (HGF/SF receptor) (Met proto-oncogene tyrosine kinase) (c-Met). | |||||
|
PKHD1_HUMAN
|
||||||
| NC score | 0.139580 (rank : 56) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TCZ9 | Gene names | PKHD1, FCYT, TIGM1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney and hepatic disease 1 precursor (Fibrocystin) (Polyductin) (Tigmin). | |||||
|
RON_HUMAN
|
||||||
| NC score | 0.139099 (rank : 57) | θ value | 0.00665767 (rank : 49) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q04912 | Gene names | MST1R, RON | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage-stimulating protein receptor precursor (EC 2.7.10.1) (MSP receptor) (p185-Ron) (CDw136 antigen) [Contains: Macrophage- stimulating protein receptor alpha chain; Macrophage-stimulating protein receptor beta chain]. | |||||
|
PXDC1_MOUSE
|
||||||
| NC score | 0.135355 (rank : 58) | θ value | 0.0148317 (rank : 54) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91ZV7, Q8BM20, Q9CWV5 | Gene names | Plxdc1, Tem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 1 precursor (Tumor endothelial marker 7). | |||||
|
RON_MOUSE
|
||||||
| NC score | 0.134935 (rank : 59) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q62190, Q62555 | Gene names | Mst1r, Ron, Stk | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage-stimulating protein receptor precursor (EC 2.7.10.1) (MSP receptor) (p185-Ron) (Stem cell-derived tyrosine kinase) (CDw136 antigen) [Contains: Macrophage-stimulating protein receptor alpha chain; Macrophage-stimulating protein receptor beta chain]. | |||||
|
PXDC1_HUMAN
|
||||||
| NC score | 0.134440 (rank : 60) | θ value | 0.00509761 (rank : 47) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IUK5, Q5QCZ7, Q5QCZ8, Q5QCZ9, Q9HCT9 | Gene names | PLXDC1, TEM3, TEM7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 1 precursor (Tumor endothelial marker 7) (Tumor endothelial marker 3). | |||||
|
PXDC2_HUMAN
|
||||||
| NC score | 0.097240 (rank : 61) | θ value | 0.47712 (rank : 60) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6UX71, Q96E59, Q96PD9, Q96SU9 | Gene names | PLXDC2, TEM7R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 2 precursor (Tumor endothelial marker 7-related protein). | |||||
|
PXDC2_MOUSE
|
||||||
| NC score | 0.092308 (rank : 62) | θ value | 0.365318 (rank : 59) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9DC11, Q6PET5, Q91ZV6 | Gene names | Plxdc2, Tem7r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 2 precursor (Tumor endothelial marker 7-related protein). | |||||
|
EXOC2_HUMAN
|
||||||
| NC score | 0.081196 (rank : 63) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96KP1, Q5JPC8, Q96AN6, Q9NUZ8, Q9UJM7 | Gene names | EXOC2, SEC5, SEC5L1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 2 (Exocyst complex component Sec5). | |||||
|
EXOC2_MOUSE
|
||||||
| NC score | 0.076370 (rank : 64) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D4H1 | Gene names | Exoc2, Sec5, Sec5l1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 2 (Exocyst complex component Sec5). | |||||
|
ATRN_HUMAN
|
||||||
| NC score | 0.058477 (rank : 65) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
RASA3_HUMAN
|
||||||
| NC score | 0.053989 (rank : 66) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14644 | Gene names | RASA3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein). | |||||
|
MEGF8_HUMAN
|
||||||
| NC score | 0.052085 (rank : 67) | θ value | 0.813845 (rank : 65) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
ATRN_MOUSE
|
||||||
| NC score | 0.051354 (rank : 68) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9WU60, Q9R263, Q9WU77 | Gene names | Atrn, mg, Mgca | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany protein). | |||||
|
WAPL_HUMAN
|
||||||
| NC score | 0.045158 (rank : 69) | θ value | 0.47712 (rank : 61) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z5K2, Q5VSK5, Q8IX10, Q92549 | Gene names | WAPAL, FOE, KIAA0261, WAPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wings apart-like protein homolog (Friend of EBNA2 protein). | |||||
|
MEGF8_MOUSE
|
||||||
| NC score | 0.044835 (rank : 70) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P60882, Q80TR3, Q80V41, Q8BMN9, Q8JZW7, Q8K0J3 | Gene names | Megf8, Egfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
WAPL_MOUSE
|
||||||
| NC score | 0.044776 (rank : 71) | θ value | 0.47712 (rank : 62) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q65Z40, Q6ZQE9 | Gene names | Wapal, Kiaa0261, Wapl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wings apart-like protein homolog (Dioxin-inducible factor 2) (DIF-2). | |||||
|
CO020_HUMAN
|
||||||
| NC score | 0.023467 (rank : 72) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H611 | Gene names | C15orf20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf20. | |||||
|
AIM2_HUMAN
|
||||||
| NC score | 0.019938 (rank : 73) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14862 | Gene names | AIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-inducible protein AIM2 (Absent in melanoma 2). | |||||
|
ITB1_HUMAN
|
||||||
| NC score | 0.019270 (rank : 74) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P05556, P78466, P78467, Q13089, Q13090, Q13091, Q13212, Q14622, Q14647 | Gene names | ITGB1, FNRB | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
ITB1_MOUSE
|
||||||
| NC score | 0.019184 (rank : 75) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P09055 | Gene names | Itgb1 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
TRS85_HUMAN
|
||||||
| NC score | 0.018771 (rank : 76) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2L5, Q9H0L2 | Gene names | KIAA1012 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein TRS85 homolog. | |||||
|
ITB6_MOUSE
|
||||||
| NC score | 0.015921 (rank : 77) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z0T9 | Gene names | Itgb6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
SNX17_HUMAN
|
||||||
| NC score | 0.015761 (rank : 78) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15036, Q53HN7, Q6IAS3 | Gene names | SNX17, KIAA0064 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-17. | |||||
|
POTE1_HUMAN
|
||||||
| NC score | 0.015603 (rank : 79) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NUX5, O95018, Q9H662, Q9NW19, Q9UG95 | Gene names | POT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protection of telomeres 1 (hPot1) (POT1-like telomere end-binding protein). | |||||
|
MYCT_HUMAN
|
||||||
| NC score | 0.015597 (rank : 80) | θ value | 1.38821 (rank : 67) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96QE2 | Gene names | SLC2A13 | |||
|
Domain Architecture |

|
|||||
| Description | Proton myo-inositol cotransporter (H(+)-myo-inositol cotransporter) (Hmit). | |||||
|
CSMD3_HUMAN
|
||||||
| NC score | 0.015503 (rank : 81) | θ value | 0.0736092 (rank : 56) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z407, Q96PZ3 | Gene names | CSMD3, KIAA1894 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 3 precursor (CUB and sushi multiple domains protein 3). | |||||
|
SNX17_MOUSE
|
||||||
| NC score | 0.014766 (rank : 82) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVL3, Q8R0N8 | Gene names | Snx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-17. | |||||
|
RBP2_HUMAN
|
||||||
| NC score | 0.010954 (rank : 83) | θ value | 0.279714 (rank : 58) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49792, Q13074, Q15280, Q53TE2, Q59FH7 | Gene names | RANBP2, NUP358 | |||
|
Domain Architecture |

|
|||||
| Description | Ran-binding protein 2 (RanBP2) (Nuclear pore complex protein Nup358) (Nucleoporin Nup358) (358 kDa nucleoporin) (P270). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.010080 (rank : 84) | θ value | 0.813845 (rank : 64) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
ZDH15_HUMAN
|
||||||
| NC score | 0.009031 (rank : 85) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96MV8, Q3SY30, Q6UWH3 | Gene names | ZDHHC15 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC15 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 15) (DHHC-15). | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.008622 (rank : 86) | θ value | 0.813845 (rank : 63) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
TINF2_MOUSE
|
||||||
| NC score | 0.008182 (rank : 87) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QXG9 | Gene names | Tinf2, Tin2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TERF1-interacting nuclear factor 2 (TRF1-interacting nuclear protein 2). | |||||
|
ZDH15_MOUSE
|
||||||
| NC score | 0.007441 (rank : 88) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BGJ0, Q3TV63, Q8BMB7 | Gene names | Zdhhc15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Palmitoyltransferase ZDHHC15 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 15) (DHHC-15). | |||||
|
CASP_HUMAN
|
||||||
| NC score | 0.007196 (rank : 89) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CUBN_HUMAN
|
||||||
| NC score | 0.006628 (rank : 90) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60494, Q5VTA6, Q96RU9 | Gene names | CUBN, IFCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cubilin precursor (Intrinsic factor-cobalamin receptor) (Intrinsic factor-vitamin B12 receptor) (460 kDa receptor) (Intestinal intrinsic factor receptor). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.006620 (rank : 91) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
TPSNR_HUMAN
|
||||||
| NC score | 0.005257 (rank : 92) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BX59, Q9NWB8 | Gene names | TAPBPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tapasin-related protein precursor (TAPASIN-R) (Tapasin-like) (TAP- binding protein-related protein) (TAPBP-R) (TAP-binding protein-like). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.004741 (rank : 93) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.004718 (rank : 94) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
LIMA1_MOUSE
|
||||||
| NC score | 0.004301 (rank : 95) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ERG0, Q9ERG1 | Gene names | Lima1, D15Ertd366e, Eplin | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm) (mEPLIN). | |||||
|
WDR24_HUMAN
|
||||||
| NC score | 0.004295 (rank : 96) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96S15, Q96GC7, Q9H0B7 | Gene names | WDR24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 24. | |||||
|
DMBT1_MOUSE
|
||||||
| NC score | 0.004140 (rank : 97) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60997, Q80YC6, Q9JMJ9 | Gene names | Dmbt1, Crpd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (CRP-ductin) (Vomeroglandin). | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.003736 (rank : 98) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.002648 (rank : 99) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
HERC3_HUMAN
|
||||||
| NC score | 0.001645 (rank : 100) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 94 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15034 | Gene names | HERC3, KIAA0032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain protein 3. | |||||