Please be patient as the page loads
|
AIM2_HUMAN
|
||||||
| SwissProt Accessions | O14862 | Gene names | AIM2 | |||
|
Domain Architecture |
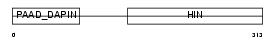
|
|||||
| Description | Interferon-inducible protein AIM2 (Absent in melanoma 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
AIM2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14862 | Gene names | AIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-inducible protein AIM2 (Absent in melanoma 2). | |||||
|
MNDA_HUMAN
|
||||||
| θ value | 7.51208e-47 (rank : 2) | NC score | 0.922491 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P41218 | Gene names | MNDA | |||
|
Domain Architecture |
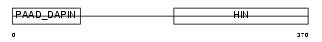
|
|||||
| Description | Myeloid cell nuclear differentiation antigen. | |||||
|
IF16_HUMAN
|
||||||
| θ value | 4.86918e-46 (rank : 3) | NC score | 0.893547 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
|
IFI3_MOUSE
|
||||||
| θ value | 1.05066e-40 (rank : 4) | NC score | 0.924964 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35368 | Gene names | Ifi203 | |||
|
Domain Architecture |
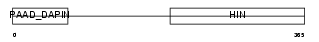
|
|||||
| Description | Interferon-activable protein 203 (Ifi-203) (Interferon-inducible protein p203). | |||||
|
IFI4_MOUSE
|
||||||
| θ value | 2.42216e-37 (rank : 5) | NC score | 0.905492 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15092 | Gene names | Ifi204 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-activable protein 204 (Ifi-204) (Interferon-inducible protein p204). | |||||
|
IFI5_MOUSE
|
||||||
| θ value | 5.0505e-35 (rank : 6) | NC score | 0.907836 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08619 | Gene names | Ifi205 | |||
|
Domain Architecture |
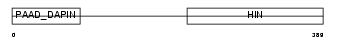
|
|||||
| Description | Interferon-activable protein 205 (IFI-205) (D3 protein). | |||||
|
II2A_MOUSE
|
||||||
| θ value | 3.27365e-34 (rank : 7) | NC score | 0.887107 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P15091 | Gene names | Ifi202a, Ifi202 | |||
|
Domain Architecture |
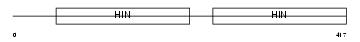
|
|||||
| Description | Interferon-activable protein 202a (Ifi-202a) (Interferon-inducible protein p202a). | |||||
|
II2B_MOUSE
|
||||||
| θ value | 5.584e-34 (rank : 8) | NC score | 0.886562 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R002 | Gene names | Ifi202b | |||
|
Domain Architecture |
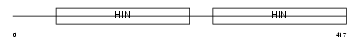
|
|||||
| Description | Interferon-activable protein 202b (Ifi-202b) (Interferon-inducible protein p202b). | |||||
|
MEFV_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 9) | NC score | 0.026706 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
NAL14_HUMAN
|
||||||
| θ value | 0.163984 (rank : 10) | NC score | 0.053943 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86W24, Q7RTR6 | Gene names | NALP14, NOD5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 14 (Nucleotide-binding oligomerization domain protein 5). | |||||
|
NALP2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.052373 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NX02, Q53FL5, Q59G09, Q8IXT0, Q9BVN5, Q9H6G6, Q9HAV9, Q9NWK3 | Gene names | NALP2, NBS1, PAN1, PYPAF2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 2 (PYRIN domain and NACHT domain-containing protein 1) (PYRIN-containing APAF1-like protein 2) (Nucleotide-binding site protein 1). | |||||
|
NALP4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.054156 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96MN2, Q86W87, Q96AY6 | Gene names | NALP4, PAN2, PYPAF4, RNH2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 4 (PYRIN-containing APAF1-like protein 4) (PAAD and NACHT-containing protein 2) (PYRIN and NACHT- containing protein 2) (Ribonuclease inhibitor 2). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 0.47712 (rank : 13) | NC score | 0.006501 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
BLMH_MOUSE
|
||||||
| θ value | 0.813845 (rank : 14) | NC score | 0.039818 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R016, Q3TJR8, Q8BLZ4, Q8BZH9, Q8C111, Q8CID9 | Gene names | Blmh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bleomycin hydrolase (EC 3.4.22.40) (BLM hydrolase) (BMH) (BH). | |||||
|
NAL12_HUMAN
|
||||||
| θ value | 1.81305 (rank : 15) | NC score | 0.045968 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P59046, Q8NEU4, Q9BY26 | Gene names | NALP12, PYPAF7, RNO | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 12 (PYRIN-containing APAF1-like protein 7) (Monarch-1) (Regulated by nitric oxide). | |||||
|
NALP6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 16) | NC score | 0.047471 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P59044 | Gene names | NALP6, PYPAF5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5). | |||||
|
PLXD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 17) | NC score | 0.017852 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4D7, Q6PJS9, Q8IZJ2, Q9BTQ2 | Gene names | PLXND1, KIAA0620 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-D1 precursor. | |||||
|
SURF6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 18) | NC score | 0.036130 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75683, Q9BRK9, Q9BTZ5, Q9UK24 | Gene names | SURF6, SURF-6 | |||
|
Domain Architecture |

|
|||||
| Description | Surfeit locus protein 6. | |||||
|
PLXC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.019938 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60486, Q59H25 | Gene names | PLXNC1, VESPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
|
PO6F2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.008096 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.017390 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
T53G5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.029829 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y2B4 | Gene names | TP53TG5, C20orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TP53-target gene 5 protein (TP53-inducible gene 5 protein). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.024476 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
ZRF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.012458 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
HRB2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.017061 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BGA5, Q8C029 | Gene names | Hrb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV-1 Rev-binding protein 2 homolog. | |||||
|
PLXC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.018711 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZC2, Q8CGW1 | Gene names | Plxnc1, Vespr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.009893 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.011051 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.004416 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
DDX55_MOUSE
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.003526 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZPL9, Q149H5, Q3U460, Q8BZR1 | Gene names | Ddx55, Kiaa1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
RA6I1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.015121 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IQ26, Q96GN3, Q9H6U7, Q9UFV0, Q9UPR1 | Gene names | RAB6IP1, KIAA1091 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
RA6I1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.015158 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PAL8, Q62146, Q8C829, Q8VDF6, Q9QYZ2 | Gene names | Rab6ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
SC11A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.003931 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.014604 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
AIM2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14862 | Gene names | AIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-inducible protein AIM2 (Absent in melanoma 2). | |||||
|
IFI3_MOUSE
|
||||||
| NC score | 0.924964 (rank : 2) | θ value | 1.05066e-40 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35368 | Gene names | Ifi203 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-activable protein 203 (Ifi-203) (Interferon-inducible protein p203). | |||||
|
MNDA_HUMAN
|
||||||
| NC score | 0.922491 (rank : 3) | θ value | 7.51208e-47 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P41218 | Gene names | MNDA | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid cell nuclear differentiation antigen. | |||||
|
IFI5_MOUSE
|
||||||
| NC score | 0.907836 (rank : 4) | θ value | 5.0505e-35 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q08619 | Gene names | Ifi205 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-activable protein 205 (IFI-205) (D3 protein). | |||||
|
IFI4_MOUSE
|
||||||
| NC score | 0.905492 (rank : 5) | θ value | 2.42216e-37 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15092 | Gene names | Ifi204 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-activable protein 204 (Ifi-204) (Interferon-inducible protein p204). | |||||
|
IF16_HUMAN
|
||||||
| NC score | 0.893547 (rank : 6) | θ value | 4.86918e-46 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
|
II2A_MOUSE
|
||||||
| NC score | 0.887107 (rank : 7) | θ value | 3.27365e-34 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P15091 | Gene names | Ifi202a, Ifi202 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-activable protein 202a (Ifi-202a) (Interferon-inducible protein p202a). | |||||
|
II2B_MOUSE
|
||||||
| NC score | 0.886562 (rank : 8) | θ value | 5.584e-34 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R002 | Gene names | Ifi202b | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-activable protein 202b (Ifi-202b) (Interferon-inducible protein p202b). | |||||
|
NALP4_HUMAN
|
||||||
| NC score | 0.054156 (rank : 9) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96MN2, Q86W87, Q96AY6 | Gene names | NALP4, PAN2, PYPAF4, RNH2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 4 (PYRIN-containing APAF1-like protein 4) (PAAD and NACHT-containing protein 2) (PYRIN and NACHT- containing protein 2) (Ribonuclease inhibitor 2). | |||||
|
NAL14_HUMAN
|
||||||
| NC score | 0.053943 (rank : 10) | θ value | 0.163984 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86W24, Q7RTR6 | Gene names | NALP14, NOD5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 14 (Nucleotide-binding oligomerization domain protein 5). | |||||
|
NALP2_HUMAN
|
||||||
| NC score | 0.052373 (rank : 11) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NX02, Q53FL5, Q59G09, Q8IXT0, Q9BVN5, Q9H6G6, Q9HAV9, Q9NWK3 | Gene names | NALP2, NBS1, PAN1, PYPAF2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 2 (PYRIN domain and NACHT domain-containing protein 1) (PYRIN-containing APAF1-like protein 2) (Nucleotide-binding site protein 1). | |||||
|
NALP6_HUMAN
|
||||||
| NC score | 0.047471 (rank : 12) | θ value | 2.36792 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P59044 | Gene names | NALP6, PYPAF5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5). | |||||
|
NAL12_HUMAN
|
||||||
| NC score | 0.045968 (rank : 13) | θ value | 1.81305 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P59046, Q8NEU4, Q9BY26 | Gene names | NALP12, PYPAF7, RNO | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 12 (PYRIN-containing APAF1-like protein 7) (Monarch-1) (Regulated by nitric oxide). | |||||
|
BLMH_MOUSE
|
||||||
| NC score | 0.039818 (rank : 14) | θ value | 0.813845 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R016, Q3TJR8, Q8BLZ4, Q8BZH9, Q8C111, Q8CID9 | Gene names | Blmh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bleomycin hydrolase (EC 3.4.22.40) (BLM hydrolase) (BMH) (BH). | |||||
|
SURF6_HUMAN
|
||||||
| NC score | 0.036130 (rank : 15) | θ value | 2.36792 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75683, Q9BRK9, Q9BTZ5, Q9UK24 | Gene names | SURF6, SURF-6 | |||
|
Domain Architecture |

|
|||||
| Description | Surfeit locus protein 6. | |||||
|
T53G5_HUMAN
|
||||||
| NC score | 0.029829 (rank : 16) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y2B4 | Gene names | TP53TG5, C20orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TP53-target gene 5 protein (TP53-inducible gene 5 protein). | |||||
|
MEFV_HUMAN
|
||||||
| NC score | 0.026706 (rank : 17) | θ value | 0.0563607 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.024476 (rank : 18) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
PLXC1_HUMAN
|
||||||
| NC score | 0.019938 (rank : 19) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60486, Q59H25 | Gene names | PLXNC1, VESPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
|
PLXC1_MOUSE
|
||||||
| NC score | 0.018711 (rank : 20) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZC2, Q8CGW1 | Gene names | Plxnc1, Vespr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-C1 precursor (Virus-encoded semaphorin protein receptor) (CD232 antigen). | |||||
|
PLXD1_HUMAN
|
||||||
| NC score | 0.017852 (rank : 21) | θ value | 2.36792 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4D7, Q6PJS9, Q8IZJ2, Q9BTQ2 | Gene names | PLXND1, KIAA0620 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-D1 precursor. | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.017390 (rank : 22) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
HRB2_MOUSE
|
||||||
| NC score | 0.017061 (rank : 23) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BGA5, Q8C029 | Gene names | Hrb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV-1 Rev-binding protein 2 homolog. | |||||
|
RA6I1_MOUSE
|
||||||
| NC score | 0.015158 (rank : 24) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PAL8, Q62146, Q8C829, Q8VDF6, Q9QYZ2 | Gene names | Rab6ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
RA6I1_HUMAN
|
||||||
| NC score | 0.015121 (rank : 25) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IQ26, Q96GN3, Q9H6U7, Q9UFV0, Q9UPR1 | Gene names | RAB6IP1, KIAA1091 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.014604 (rank : 26) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
ZRF1_HUMAN
|
||||||
| NC score | 0.012458 (rank : 27) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.011051 (rank : 28) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.009893 (rank : 29) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
PO6F2_MOUSE
|
||||||
| NC score | 0.008096 (rank : 30) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.006501 (rank : 31) | θ value | 0.47712 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.004416 (rank : 32) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
SC11A_MOUSE
|
||||||
| NC score | 0.003931 (rank : 33) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
DDX55_MOUSE
|
||||||
| NC score | 0.003526 (rank : 34) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZPL9, Q149H5, Q3U460, Q8BZR1 | Gene names | Ddx55, Kiaa1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||