Please be patient as the page loads
|
IF16_HUMAN
|
||||||
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
IF16_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 182 | |
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
|
MNDA_HUMAN
|
||||||
| θ value | 5.29683e-101 (rank : 2) | NC score | 0.937523 (rank : 2) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 22 | |
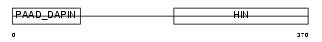
| SwissProt Accessions | P41218 | Gene names | MNDA | |||
|
Domain Architecture |
|
|||||
| Description | Myeloid cell nuclear differentiation antigen. | |||||
|
IFI4_MOUSE
|
||||||
| θ value | 1.00358e-83 (rank : 3) | NC score | 0.916608 (rank : 5) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P15092 | Gene names | Ifi204 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-activable protein 204 (Ifi-204) (Interferon-inducible protein p204). | |||||
|
IFI5_MOUSE
|
||||||
| θ value | 2.91998e-83 (rank : 4) | NC score | 0.925886 (rank : 3) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 9 | |
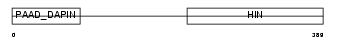
| SwissProt Accessions | Q08619 | Gene names | Ifi205 | |||
|
Domain Architecture |
|
|||||
| Description | Interferon-activable protein 205 (IFI-205) (D3 protein). | |||||
|
IFI3_MOUSE
|
||||||
| θ value | 3.58603e-65 (rank : 5) | NC score | 0.919201 (rank : 4) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 12 | |
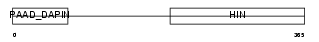
| SwissProt Accessions | O35368 | Gene names | Ifi203 | |||
|
Domain Architecture |
|
|||||
| Description | Interferon-activable protein 203 (Ifi-203) (Interferon-inducible protein p203). | |||||
|
II2A_MOUSE
|
||||||
| θ value | 5.7518e-47 (rank : 6) | NC score | 0.869803 (rank : 7) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 8 | |
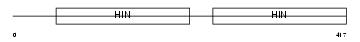
| SwissProt Accessions | P15091 | Gene names | Ifi202a, Ifi202 | |||
|
Domain Architecture |
|
|||||
| Description | Interferon-activable protein 202a (Ifi-202a) (Interferon-inducible protein p202a). | |||||
|
AIM2_HUMAN
|
||||||
| θ value | 4.86918e-46 (rank : 7) | NC score | 0.893547 (rank : 6) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14862 | Gene names | AIM2 | |||
|
Domain Architecture |
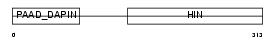
|
|||||
| Description | Interferon-inducible protein AIM2 (Absent in melanoma 2). | |||||
|
II2B_MOUSE
|
||||||
| θ value | 4.86918e-46 (rank : 8) | NC score | 0.868894 (rank : 8) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R002 | Gene names | Ifi202b | |||
|
Domain Architecture |
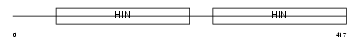
|
|||||
| Description | Interferon-activable protein 202b (Ifi-202b) (Interferon-inducible protein p202b). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 9) | NC score | 0.097776 (rank : 10) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 10) | NC score | 0.047478 (rank : 66) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 11) | NC score | 0.097887 (rank : 9) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 12) | NC score | 0.077494 (rank : 13) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 13) | NC score | 0.080299 (rank : 12) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 14) | NC score | 0.042140 (rank : 74) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 15) | NC score | 0.060062 (rank : 37) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NUCKS_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 16) | NC score | 0.072869 (rank : 16) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 0.125558 (rank : 17) | NC score | 0.057502 (rank : 48) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.071347 (rank : 20) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
SF3A1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 19) | NC score | 0.071336 (rank : 21) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15459 | Gene names | SF3A1, SAP114 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3 subunit 1 (Spliceosome-associated protein 114) (SAP 114) (SF3a120). | |||||
|
MARCS_MOUSE
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.071377 (rank : 19) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
EVL_MOUSE
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.058296 (rank : 42) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
SF3A1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.072177 (rank : 18) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8K4Z5, Q8C0M7, Q8C128, Q8C175, Q921T3 | Gene names | Sf3a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 3 subunit 1 (SF3a120). | |||||
|
SMAL1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.028175 (rank : 105) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BJL0, Q8BVK7, Q8BXW4, Q8K309, Q9EQK8, Q9QYC4 | Gene names | Smarcal1, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (mharp). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.054658 (rank : 55) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NOTC3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.015350 (rank : 148) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
SYN1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.055967 (rank : 53) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
AP3D1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.033400 (rank : 92) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.059234 (rank : 40) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.065828 (rank : 29) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.066949 (rank : 27) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.075351 (rank : 14) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.070281 (rank : 23) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TF3C1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.036068 (rank : 85) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12789, Q12838, Q9Y4W9 | Gene names | GTF3C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 1 (Transcription factor IIIC-subunit alpha) (TF3C-alpha) (TFIIIC 220 kDa subunit) (TFIIIC220) (TFIIIC box B-binding subunit). | |||||
|
ZBT20_HUMAN
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.004085 (rank : 189) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HC78 | Gene names | ZBTB20, DPZF, ZNF288 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 20 (Zinc finger protein 288) (Dendritic-derived BTB/POZ zinc finger protein). | |||||
|
ANKS6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.022509 (rank : 126) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
GAK5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.031701 (rank : 95) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GCAA_MOUSE
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.020515 (rank : 132) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P01863 | Gene names | ||||
|
Domain Architecture |
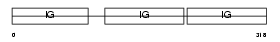
|
|||||
| Description | Ig gamma-2A chain C region, A allele. | |||||
|
GCAM_MOUSE
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.020424 (rank : 133) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P01865 | Gene names | Igh-1a | |||
|
Domain Architecture |
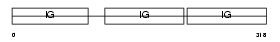
|
|||||
| Description | Ig gamma-2A chain C region, membrane-bound form. | |||||
|
H13_MOUSE
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.069432 (rank : 24) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P43277, Q8C6M4 | Gene names | Hist1h1d, H1f3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (H1 VAR.4) (H1d). | |||||
|
JAD1C_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.019675 (rank : 136) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P41229 | Gene names | SMCX, DXS1272E, JARID1C, XE169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (Protein SmcX) (Protein Xe169). | |||||
|
MYLK_MOUSE
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.009477 (rank : 176) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.056947 (rank : 50) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.056396 (rank : 51) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
ZCPW1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.056190 (rank : 52) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
DYNA_MOUSE
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.018986 (rank : 137) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
H12_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.062421 (rank : 33) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P16403 | Gene names | HIST1H1C, H1F2 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.2 (Histone H1d). | |||||
|
H12_MOUSE
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.062751 (rank : 31) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P15864 | Gene names | Hist1h1c, H1f2 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.2 (H1 VAR.1) (H1c). | |||||
|
KLF10_HUMAN
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.004346 (rank : 188) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13118 | Gene names | KLF10, TIEG, TIEG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 10 (Transforming growth factor-beta-inducible early growth response protein 1) (TGFB-inducible early growth response protein 1) (TIEG-1) (EGR-alpha). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.025979 (rank : 116) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MYO3B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.002486 (rank : 190) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WXR4, Q8IX64, Q8IX65, Q8IX66, Q8IX67, Q8IX68 | Gene names | MYO3B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin IIIB (EC 2.7.11.1). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.072252 (rank : 17) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PHF14_MOUSE
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.026330 (rank : 114) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.021675 (rank : 130) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.073549 (rank : 15) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.069379 (rank : 25) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.042871 (rank : 72) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
CASC3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.045391 (rank : 67) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
DYNA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.018385 (rank : 139) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
K0562_HUMAN
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.027304 (rank : 109) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60308, Q5JSQ3, Q5SR24, Q5SR25, Q6PKF5, Q86W32, Q86X14 | Gene names | KIAA0562 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0562. | |||||
|
KLDC4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.024532 (rank : 121) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921I2, Q8CIK0 | Gene names | Klhdc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch domain-containing protein 4. | |||||
|
RTN2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.025235 (rank : 120) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75298, O60509, Q7RTM6, Q7RTN1, Q7RTN2 | Gene names | RTN2, NSPL1 | |||
|
Domain Architecture |
|
|||||
| Description | Reticulon-2 (Neuroendocrine-specific protein-like 1) (NSP-like protein 1) (NSPLI). | |||||
|
SURF6_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.037304 (rank : 80) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P70279, Q8BPI4, Q8BRL7, Q8R0Q7 | Gene names | Surf6, Surf-6 | |||
|
Domain Architecture |
|
|||||
| Description | Surfeit locus protein 6. | |||||
|
COPD_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.033951 (rank : 89) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48444, Q52M80 | Gene names | ARCN1, COPD | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit delta (Delta-coat protein) (Delta-COP) (Archain). | |||||
|
EVL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.053337 (rank : 59) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UI08, O95884, Q7Z522, Q8TBV1, Q9UF25, Q9UIC2 | Gene names | EVL, RNB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
FOXC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.014483 (rank : 150) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99958 | Gene names | FOXC2, FKHL14, MFH1 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein C2 (Forkhead-related protein FKHL14) (Mesenchyme fork head protein 1) (MFH-1 protein) (Transcription factor FKH-14). | |||||
|
H14_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.066260 (rank : 28) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P10412 | Gene names | HIST1H1E, H1F4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (Histone H1b). | |||||
|
HXD9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.007995 (rank : 180) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P28356 | Gene names | HOXD9, HOX4C | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D9 (Hox-4C) (Hox-5.2). | |||||
|
MATR3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.025937 (rank : 117) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
NOTC3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.014221 (rank : 153) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.050938 (rank : 62) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
RFIP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.035212 (rank : 87) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6WKZ4, Q6AZK4, Q6WKZ2, Q6WKZ6, Q86YV4, Q8TDL1, Q9H642 | Gene names | RAB11FIP1, RCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
RGP2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.023343 (rank : 123) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P47736 | Gene names | RAP1GAP, RAP1GA1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-activating protein 1 (Rap1GAP). | |||||
|
SCMH1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.020235 (rank : 134) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K214, Q9JME0 | Gene names | Scmh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb protein SCMH1 (Sex comb on midleg homolog 1). | |||||
|
SYN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.050729 (rank : 63) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.045040 (rank : 69) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
FOXD3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.008805 (rank : 178) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJU5, Q9BYM2, Q9UDD1 | Gene names | FOXD3 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein D3 (HNF3/FH transcription factor genesis). | |||||
|
H11_MOUSE
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.057062 (rank : 49) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P43275 | Gene names | Hist1h1a, H1f1 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.1 (H1 VAR.3) (H1a). | |||||
|
LAD1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.086101 (rank : 11) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.034009 (rank : 88) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
TMC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.021894 (rank : 129) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TDI7, Q5JXT0, Q5JXT1, Q6UWW4, Q6ZS41, Q8N9F3, Q9BYN2, Q9BYN3, Q9BYN4, Q9BYN5 | Gene names | TMC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
|
ADRM1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.037419 (rank : 79) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16186, Q9H1P2 | Gene names | ADRM1, GP110 | |||
|
Domain Architecture |
|
|||||
| Description | Adhesion-regulating molecule 1 precursor (110 kDa cell membrane glycoprotein) (Gp110). | |||||
|
ATX2L_HUMAN
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.035241 (rank : 86) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.053712 (rank : 57) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CNOT4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.027229 (rank : 111) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
GAK11_HUMAN
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.030630 (rank : 97) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.030732 (rank : 96) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GP158_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.030266 (rank : 98) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
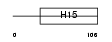
H13_HUMAN
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.058322 (rank : 41) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P16402 | Gene names | HIST1H1D, H1F3 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.3 (Histone H1c). | |||||
|
HXA3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.012994 (rank : 157) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43365 | Gene names | HOXA3, HOX1E | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1E). | |||||
|
LAMA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.008426 (rank : 179) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
PKHA5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.021943 (rank : 127) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.044700 (rank : 71) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
TAF3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.045003 (rank : 70) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
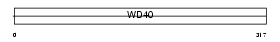
WDR22_MOUSE
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.012685 (rank : 159) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80T85, Q80VT3, Q80ZW6, Q8BIP8, Q8BVK5, Q8K0Q6 | Gene names | Wdr22, Kiaa1824 | |||
|
Domain Architecture |
|
|||||
| Description | WD repeat protein 22. | |||||
|
ZN516_HUMAN
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.005915 (rank : 185) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q92618 | Gene names | ZNF516, KIAA0222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 516. | |||||
|
ZN692_MOUSE
|
||||||
| θ value | 3.0926 (rank : 96) | NC score | 0.001794 (rank : 193) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3U381 | Gene names | Znf692, Zfp692 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 692. | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.027418 (rank : 108) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
BRPF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.016462 (rank : 142) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
CD2L5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.006369 (rank : 184) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1011 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q69ZA1, Q80V11, Q8BZG1, Q8K0A4 | Gene names | Cdc2l5, Kiaa1791 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5). | |||||
|
GAK6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.029927 (rank : 99) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.037023 (rank : 83) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.032680 (rank : 94) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
PAF49_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.042188 (rank : 73) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
PRP4B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.010179 (rank : 174) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1030 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13523, Q8TDP2, Q96QT7, Q9UEE6 | Gene names | PRPF4B, KIAA0536, PRP4, PRP4H, PRP4K | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (PRP4 kinase). | |||||
|
REXO1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.033349 (rank : 93) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.052897 (rank : 60) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.010552 (rank : 170) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 4.03905 (rank : 108) | NC score | 0.013676 (rank : 156) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
T2FA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 109) | NC score | 0.038281 (rank : 77) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 4.03905 (rank : 110) | NC score | 0.059869 (rank : 38) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TNKS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 111) | NC score | 0.016170 (rank : 144) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
ZC3H3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | 0.026937 (rank : 112) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.039618 (rank : 76) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
BMPR2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.001999 (rank : 191) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13873, Q16569 | Gene names | BMPR2, PPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein receptor type-2 precursor (EC 2.7.11.30) (Bone morphogenetic protein receptor type II) (BMP type II receptor) (BMPR-II). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.057975 (rank : 45) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
CO3A1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.016495 (rank : 141) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.013829 (rank : 154) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
COPD_MOUSE
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.028694 (rank : 103) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5XJY5 | Gene names | Arcn1, Copd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coatomer subunit delta (Delta-coat protein) (Delta-COP) (Archain). | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.022739 (rank : 125) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
ELOA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.036119 (rank : 84) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
HIF1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.009115 (rank : 177) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
HXA3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.011765 (rank : 163) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P02831, Q4V9Z8, Q61197 | Gene names | Hoxa3, Hox-1.5, Hoxa-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1.5) (MO-10). | |||||
|
KKCC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.000935 (rank : 194) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96RR4, O94883, Q8IUG2, Q8IUG3, Q8N3I4, Q8WY03, Q8WY04, Q8WY05, Q8WY06, Q96RP1, Q96RP2, Q96RR3, Q9BWE9, Q9UER3, Q9UES2, Q9Y5N2 | Gene names | CAMKK2, CAMKKB, KIAA0787 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase kinase 2 (EC 2.7.11.17) (Calcium/calmodulin-dependent protein kinase kinase beta) (CaM-kinase kinase beta) (CaM-KK beta) (CaMKK beta). | |||||
|
MYB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.010884 (rank : 168) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10242, P78391, P78392, P78525, P78526, Q14023, Q14024, Q9UE83 | Gene names | MYB | |||
|
Domain Architecture |
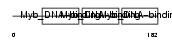
|
|||||
| Description | Myb proto-oncogene protein (C-myb). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.026518 (rank : 113) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
PHF14_HUMAN
|
||||||
| θ value | 5.27518 (rank : 126) | NC score | 0.021923 (rank : 128) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
RD23B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.025628 (rank : 119) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
SON_HUMAN
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.045269 (rank : 68) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TACC3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.026114 (rank : 115) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
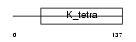
ZBTB9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.004693 (rank : 186) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CDC7, Q3U3B2, Q9CYQ0 | Gene names | Zbtb9 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger and BTB domain-containing protein 9. | |||||
|
ZN438_HUMAN
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.007815 (rank : 182) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7Z4V0, Q5T426, Q658Q4, Q6ZN65 | Gene names | ZNF438 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 438. | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.010538 (rank : 171) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
CIR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.023378 (rank : 122) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.011009 (rank : 167) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
DNL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.033711 (rank : 90) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P37913 | Gene names | Lig1, Lig-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
EDN3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.015224 (rank : 149) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P48299, Q543L0 | Gene names | Edn3 | |||
|
Domain Architecture |
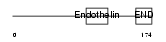
|
|||||
| Description | Endothelin-3 precursor (ET-3) (Preproendothelin-3) (PPET3). | |||||
|
FBLN2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.010448 (rank : 173) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
GAK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.028115 (rank : 106) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GCAB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.015676 (rank : 146) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01864 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig gamma-2A chain C region secreted form (B allele). | |||||
|
H14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.058023 (rank : 43) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P43274 | Gene names | Hist1h1e, H1f4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (H1 VAR.2) (H1e). | |||||
|
H15_HUMAN
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.057683 (rank : 46) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P16401, Q14529 | Gene names | HIST1H1B, H1F5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (Histone H1a). | |||||
|
INP4A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.009722 (rank : 175) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EPW0, Q8CBJ8 | Gene names | Inpp4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type I inositol-3,4-bisphosphate 4-phosphatase (EC 3.1.3.66) (Inositol polyphosphate 4-phosphatase type I). | |||||
|
MUCDL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.019705 (rank : 135) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
PAPOA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.012158 (rank : 161) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61183, Q61208, Q61209, Q8K4X2 | Gene names | Papola, Pap, Plap | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase). | |||||
|
PI4KB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.010485 (rank : 172) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UBF8, O15096, P78405, Q9BWR6 | Gene names | PIK4CB, PI4KB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta) (NPIK) (PI4K92). | |||||
|
PPRB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.037111 (rank : 82) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
PPRB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.037668 (rank : 78) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
PTMS_MOUSE
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.029092 (rank : 102) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
SALL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.006722 (rank : 183) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
SIA7A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.041727 (rank : 75) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NSC7, Q6UW90, Q9NSC6 | Gene names | ST6GALNAC1, SIAT7A | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
SIA7A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.025640 (rank : 118) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QZ39, Q9JJP5 | Gene names | St6galnac1, Siat7a | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
SURF6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.037198 (rank : 81) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75683, Q9BRK9, Q9BTZ5, Q9UK24 | Gene names | SURF6, SURF-6 | |||
|
Domain Architecture |

|
|||||
| Description | Surfeit locus protein 6. | |||||
|
SYNP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.016663 (rank : 140) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91YE8, Q8C592 | Gene names | Synpo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin). | |||||
|
TECT3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | 0.014358 (rank : 151) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R2Q6, Q8CDF3, Q9CRC5 | Gene names | Tect3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tectonic-3 precursor. | |||||
|
TJAP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 155) | NC score | 0.021607 (rank : 131) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DCD5, Q8CFL7, Q8R5I2 | Gene names | Tjap1, Pilt, Tjp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tight junction-associated protein 1 (Tight junction protein 4) (Protein incorporated later into tight junctions). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.012819 (rank : 158) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
COFA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.011137 (rank : 166) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P39059, Q5T6J4, Q9Y4W4 | Gene names | COL15A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV) (Restin) (Related to endostatin)]. | |||||
|
EDA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.012020 (rank : 162) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O54693, O35705, Q9QWJ8, Q9QZ01, Q9QZ02 | Gene names | Eda, Ed1, Ta | |||
|
Domain Architecture |

|
|||||
| Description | Ectodysplasin-A (EDA protein homolog) (Tabby protein) [Contains: Ectodysplasin-A, membrane form; Ectodysplasin-A, secreted form]. | |||||
|
ERRFI_HUMAN
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.013794 (rank : 155) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UJM3, Q9NTG9, Q9UD05 | Gene names | ERRFI1, MIG6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein) (Mig-6). | |||||
|
FEZ2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.015955 (rank : 145) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UHY8, Q76LN0, Q99690 | Gene names | FEZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fasciculation and elongation protein zeta 2 (Zygin-2) (Zygin II). | |||||
|
FEZ2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.015631 (rank : 147) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6TYB5 | Gene names | Fez2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fasciculation and elongation protein zeta 2 (Zygin-2) (Zygin II). | |||||
|
GAK10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.027238 (rank : 110) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GEMI5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.011311 (rank : 165) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 424 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BX17 | Gene names | Gemin5 | |||
|
Domain Architecture |

|
|||||
| Description | Gem-associated protein 5 (Gemin5). | |||||
|
JHD2C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.010716 (rank : 169) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15652, Q5SQZ8, Q5SQZ9, Q5SR00, Q7Z3E7, Q8N3U0, Q96KB9, Q9P2G7 | Gene names | JMJD1C, JHDM2C, KIAA1380, TRIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C) (Thyroid receptor-interacting protein 8) (TRIP-8). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.033432 (rank : 91) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
KAISO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.001861 (rank : 192) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86T24, O00319, Q7Z361, Q8IVP6, Q8N3P0 | Gene names | ZBTB33, KAISO, ZNF348 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional regulator Kaiso (Zinc finger and BTB domain-containing protein 33). | |||||
|
KI21A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.004546 (rank : 187) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
LIPS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.028557 (rank : 104) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
LSD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.014304 (rank : 152) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.048529 (rank : 65) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.067048 (rank : 26) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.027892 (rank : 107) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
NOP56_HUMAN
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.011527 (rank : 164) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00567, Q9NQ05 | Gene names | NOL5A, NOP56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
PKN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.000637 (rank : 195) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K045 | Gene names | Pkn3, Pknbeta | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N3 (EC 2.7.11.13) (Protein kinase PKN- beta) (Protein-kinase C-related kinase 3). | |||||
|
PODXL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 175) | NC score | 0.029217 (rank : 101) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00592 | Gene names | PODXL, PCLP, PCLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 176) | NC score | 0.071137 (rank : 22) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TJAP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 177) | NC score | 0.018608 (rank : 138) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5JTD0, Q5JTD1, Q5JWW1, Q68DB2, Q6P2P3, Q9H7V7 | Gene names | TJAP1, PILT, TJP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tight junction-associated protein 1 (Tight junction protein 4) (Protein incorporated later into tight junctions). | |||||
|
TTC16_HUMAN
|
||||||
| θ value | 8.99809 (rank : 178) | NC score | 0.029420 (rank : 100) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
YETS2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 179) | NC score | 0.023116 (rank : 124) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
ZC3H6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 180) | NC score | 0.016457 (rank : 143) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ZC3H8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 181) | NC score | 0.012190 (rank : 160) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 8.99809 (rank : 182) | NC score | 0.007909 (rank : 181) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
ALO17_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.058021 (rank : 44) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
BASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.060709 (rank : 36) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.054505 (rank : 56) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
GRIN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.053669 (rank : 58) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
H15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.050205 (rank : 64) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P43276, Q9CRM8 | Gene names | Hist1h1b, H1f5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (H1 VAR.5) (H1b). | |||||
|
KI67_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.052505 (rank : 61) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
LAD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.060967 (rank : 35) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
MBB1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.055021 (rank : 54) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.061286 (rank : 34) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
SELPL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.059602 (rank : 39) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.057561 (rank : 47) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.063606 (rank : 30) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.062512 (rank : 32) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
IF16_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 182 | |
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
|
MNDA_HUMAN
|
||||||
| NC score | 0.937523 (rank : 2) | θ value | 5.29683e-101 (rank : 2) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P41218 | Gene names | MNDA | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid cell nuclear differentiation antigen. | |||||
|
IFI5_MOUSE
|
||||||
| NC score | 0.925886 (rank : 3) | θ value | 2.91998e-83 (rank : 4) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08619 | Gene names | Ifi205 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-activable protein 205 (IFI-205) (D3 protein). | |||||
|
IFI3_MOUSE
|
||||||
| NC score | 0.919201 (rank : 4) | θ value | 3.58603e-65 (rank : 5) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35368 | Gene names | Ifi203 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-activable protein 203 (Ifi-203) (Interferon-inducible protein p203). | |||||
|
IFI4_MOUSE
|
||||||
| NC score | 0.916608 (rank : 5) | θ value | 1.00358e-83 (rank : 3) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P15092 | Gene names | Ifi204 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-activable protein 204 (Ifi-204) (Interferon-inducible protein p204). | |||||
|
AIM2_HUMAN
|
||||||
| NC score | 0.893547 (rank : 6) | θ value | 4.86918e-46 (rank : 7) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14862 | Gene names | AIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-inducible protein AIM2 (Absent in melanoma 2). | |||||
|
II2A_MOUSE
|
||||||
| NC score | 0.869803 (rank : 7) | θ value | 5.7518e-47 (rank : 6) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P15091 | Gene names | Ifi202a, Ifi202 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-activable protein 202a (Ifi-202a) (Interferon-inducible protein p202a). | |||||
|
II2B_MOUSE
|
||||||
| NC score | 0.868894 (rank : 8) | θ value | 4.86918e-46 (rank : 8) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R002 | Gene names | Ifi202b | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-activable protein 202b (Ifi-202b) (Interferon-inducible protein p202b). | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.097887 (rank : 9) | θ value | 0.0148317 (rank : 11) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.097776 (rank : 10) | θ value | 0.00665767 (rank : 9) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.086101 (rank : 11) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.080299 (rank : 12) | θ value | 0.0431538 (rank : 13) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.077494 (rank : 13) | θ value | 0.0252991 (rank : 12) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.075351 (rank : 14) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.073549 (rank : 15) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
NUCKS_HUMAN
|
||||||
| NC score | 0.072869 (rank : 16) | θ value | 0.0736092 (rank : 16) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.072252 (rank : 17) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SF3A1_MOUSE
|
||||||
| NC score | 0.072177 (rank : 18) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8K4Z5, Q8C0M7, Q8C128, Q8C175, Q921T3 | Gene names | Sf3a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 3 subunit 1 (SF3a120). | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.071377 (rank : 19) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.071347 (rank : 20) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
SF3A1_HUMAN
|
||||||
| NC score | 0.071336 (rank : 21) | θ value | 0.125558 (rank : 19) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15459 | Gene names | SF3A1, SAP114 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3 subunit 1 (Spliceosome-associated protein 114) (SAP 114) (SF3a120). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.071137 (rank : 22) | θ value | 8.99809 (rank : 176) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.070281 (rank : 23) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
H13_MOUSE
|
||||||
| NC score | 0.069432 (rank : 24) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P43277, Q8C6M4 | Gene names | Hist1h1d, H1f3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (H1 VAR.4) (H1d). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.069379 (rank : 25) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.067048 (rank : 26) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.066949 (rank : 27) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
H14_HUMAN
|
||||||
| NC score | 0.066260 (rank : 28) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P10412 | Gene names | HIST1H1E, H1F4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (Histone H1b). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.065828 (rank : 29) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.063606 (rank : 30) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
H12_MOUSE
|
||||||
| NC score | 0.062751 (rank : 31) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P15864 | Gene names | Hist1h1c, H1f2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.2 (H1 VAR.1) (H1c). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.062512 (rank : 32) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
H12_HUMAN
|
||||||
| NC score | 0.062421 (rank : 33) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P16403 | Gene names | HIST1H1C, H1F2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.2 (Histone H1d). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.061286 (rank : 34) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.060967 (rank : 35) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.060709 (rank : 36) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.060062 (rank : 37) | θ value | 0.0563607 (rank : 15) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 119 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.059869 (rank : 38) | θ value | 4.03905 (rank : 110) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.059602 (rank : 39) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.059234 (rank : 40) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
H13_HUMAN
|
||||||
| NC score | 0.058322 (rank : 41) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P16402 | Gene names | HIST1H1D, H1F3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (Histone H1c). | |||||
|
EVL_MOUSE
|
||||||
| NC score | 0.058296 (rank : 42) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
H14_MOUSE
|
||||||
| NC score | 0.058023 (rank : 43) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P43274 | Gene names | Hist1h1e, H1f4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (H1 VAR.2) (H1e). | |||||
|
ALO17_HUMAN
|
||||||
| NC score | 0.058021 (rank : 44) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.057975 (rank : 45) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
H15_HUMAN
|
||||||
| NC score | 0.057683 (rank : 46) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P16401, Q14529 | Gene names | HIST1H1B, H1F5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (Histone H1a). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.057561 (rank : 47) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.057502 (rank : 48) | θ value | 0.125558 (rank : 17) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
H11_MOUSE
|
||||||
| NC score | 0.057062 (rank : 49) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P43275 | Gene names | Hist1h1a, H1f1 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.1 (H1 VAR.3) (H1a). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.056947 (rank : 50) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.056396 (rank : 51) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
ZCPW1_MOUSE
|
||||||
| NC score | 0.056190 (rank : 52) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
SYN1_MOUSE
|
||||||
| NC score | 0.055967 (rank : 53) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
MBB1A_MOUSE
|
||||||
| NC score | 0.055021 (rank : 54) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.054658 (rank : 55) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.054505 (rank : 56) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.053712 (rank : 57) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
GRIN1_MOUSE
|
||||||
| NC score | 0.053669 (rank : 58) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
EVL_HUMAN
|
||||||
| NC score | 0.053337 (rank : 59) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UI08, O95884, Q7Z522, Q8TBV1, Q9UF25, Q9UIC2 | Gene names | EVL, RNB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.052897 (rank : 60) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.052505 (rank : 61) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.050938 (rank : 62) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SYN1_HUMAN
|
||||||
| NC score | 0.050729 (rank : 63) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
H15_MOUSE
|
||||||
| NC score | 0.050205 (rank : 64) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P43276, Q9CRM8 | Gene names | Hist1h1b, H1f5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (H1 VAR.5) (H1b). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.048529 (rank : 65) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.047478 (rank : 66) | θ value | 0.00869519 (rank : 10) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CASC3_MOUSE
|
||||||
| NC score | 0.045391 (rank : 67) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.045269 (rank : 68) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.045040 (rank : 69) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
TAF3_MOUSE
|
||||||
| NC score | 0.045003 (rank : 70) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.044700 (rank : 71) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.042871 (rank : 72) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
PAF49_HUMAN
|
||||||
| NC score | 0.042188 (rank : 73) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.042140 (rank : 74) | θ value | 0.0563607 (rank : 14) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SIA7A_HUMAN
|
||||||
| NC score | 0.041727 (rank : 75) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NSC7, Q6UW90, Q9NSC6 | Gene names | ST6GALNAC1, SIAT7A | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.039618 (rank : 76) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.038281 (rank : 77) | θ value | 4.03905 (rank : 109) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
PPRB_MOUSE
|
||||||
| NC score | 0.037668 (rank : 78) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
ADRM1_HUMAN
|
||||||
| NC score | 0.037419 (rank : 79) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16186, Q9H1P2 | Gene names | ADRM1, GP110 | |||
|
Domain Architecture |

|
|||||
| Description | Adhesion-regulating molecule 1 precursor (110 kDa cell membrane glycoprotein) (Gp110). | |||||
|
SURF6_MOUSE
|
||||||
| NC score | 0.037304 (rank : 80) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P70279, Q8BPI4, Q8BRL7, Q8R0Q7 | Gene names | Surf6, Surf-6 | |||
|
Domain Architecture |

|
|||||
| Description | Surfeit locus protein 6. | |||||
|
SURF6_HUMAN
|
||||||
| NC score | 0.037198 (rank : 81) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75683, Q9BRK9, Q9BTZ5, Q9UK24 | Gene names | SURF6, SURF-6 | |||
|
Domain Architecture |

|
|||||
| Description | Surfeit locus protein 6. | |||||
|
PPRB_HUMAN
|
||||||
| NC score | 0.037111 (rank : 82) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.037023 (rank : 83) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
ELOA1_HUMAN
|
||||||
| NC score | 0.036119 (rank : 84) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
TF3C1_HUMAN
|
||||||
| NC score | 0.036068 (rank : 85) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12789, Q12838, Q9Y4W9 | Gene names | GTF3C1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General transcription factor 3C polypeptide 1 (Transcription factor IIIC-subunit alpha) (TF3C-alpha) (TFIIIC 220 kDa subunit) (TFIIIC220) (TFIIIC box B-binding subunit). | |||||
|
ATX2L_HUMAN
|
||||||
| NC score | 0.035241 (rank : 86) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
RFIP1_HUMAN
|
||||||
| NC score | 0.035212 (rank : 87) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6WKZ4, Q6AZK4, Q6WKZ2, Q6WKZ6, Q86YV4, Q8TDL1, Q9H642 | Gene names | RAB11FIP1, RCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.034009 (rank : 88) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
COPD_HUMAN
|
||||||
| NC score | 0.033951 (rank : 89) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48444, Q52M80 | Gene names | ARCN1, COPD | |||
|
Domain Architecture |

|
|||||
| Description | Coatomer subunit delta (Delta-coat protein) (Delta-COP) (Archain). | |||||
|
DNL1_MOUSE
|
||||||
| NC score | 0.033711 (rank : 90) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P37913 | Gene names | Lig1, Lig-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.033432 (rank : 91) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
AP3D1_HUMAN
|
||||||
| NC score | 0.033400 (rank : 92) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O14617, O00202, O75262, Q59HF5, Q96G11, Q9H3C6 | Gene names | AP3D1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit delta-1 (Adapter-related protein complex 3 subunit delta-1) (Delta-adaptin 3) (AP-3 complex subunit delta) (Delta-adaptin). | |||||
|
REXO1_HUMAN
|
||||||
| NC score | 0.033349 (rank : 93) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.032680 (rank : 94) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
GAK5_HUMAN
|
||||||
| NC score | 0.031701 (rank : 95) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| NC score | 0.030732 (rank : 96) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK11_HUMAN
|
||||||
| NC score | 0.030630 (rank : 97) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GP158_HUMAN
|
||||||
| NC score | 0.030266 (rank : 98) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5T848, Q6QR81, Q9ULT3 | Gene names | GPR158, KIAA1136 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 158 precursor. | |||||
|
GAK6_HUMAN
|
||||||
| NC score | 0.029927 (rank : 99) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
TTC16_HUMAN
|
||||||
| NC score | 0.029420 (rank : 100) | θ value | 8.99809 (rank : 178) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
PODXL_HUMAN
|
||||||
| NC score | 0.029217 (rank : 101) | θ value | 8.99809 (rank : 175) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00592 | Gene names | PODXL, PCLP, PCLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
PTMS_MOUSE
|
||||||
| NC score | 0.029092 (rank : 102) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
COPD_MOUSE
|
||||||
| NC score | 0.028694 (rank : 103) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5XJY5 | Gene names | Arcn1, Copd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coatomer subunit delta (Delta-coat protein) (Delta-COP) (Archain). | |||||
|
LIPS_HUMAN
|
||||||
| NC score | 0.028557 (rank : 104) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q05469, Q3LRT2, Q6NSL7 | Gene names | LIPE | |||
|
Domain Architecture |

|
|||||
| Description | Hormone-sensitive lipase (EC 3.1.1.79) (HSL). | |||||
|
SMAL1_MOUSE
|
||||||
| NC score | 0.028175 (rank : 105) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BJL0, Q8BVK7, Q8BXW4, Q8K309, Q9EQK8, Q9QYC4 | Gene names | Smarcal1, Harp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (mharp). | |||||
|
GAK2_HUMAN
|
||||||
| NC score | 0.028115 (rank : 106) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.027892 (rank : 107) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.027418 (rank : 108) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
K0562_HUMAN
|
||||||
| NC score | 0.027304 (rank : 109) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60308, Q5JSQ3, Q5SR24, Q5SR25, Q6PKF5, Q86W32, Q86X14 | Gene names | KIAA0562 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0562. | |||||
|
GAK10_HUMAN
|
||||||
| NC score | 0.027238 (rank : 110) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
CNOT4_MOUSE
|
||||||
| NC score | 0.027229 (rank : 111) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
ZC3H3_HUMAN
|
||||||
| NC score | 0.026937 (rank : 112) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IXZ2, Q14163, Q8N4E2, Q9BUS4 | Gene names | ZC3H3, KIAA0150, ZC3HDC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 3. | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.026518 (rank : 113) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
PHF14_MOUSE
|
||||||
| NC score | 0.026330 (rank : 114) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
TACC3_HUMAN
|
||||||
| NC score | 0.026114 (rank : 115) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9Y6A5, Q9UMQ1 | Gene names | TACC3, ERIC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ERIC-1). | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.025979 (rank : 116) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MATR3_HUMAN
|
||||||
| NC score | 0.025937 (rank : 117) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
SIA7A_MOUSE
|
||||||
| NC score | 0.025640 (rank : 118) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QZ39, Q9JJP5 | Gene names | St6galnac1, Siat7a | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
RD23B_MOUSE
|
||||||
| NC score | 0.025628 (rank : 119) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
RTN2_HUMAN
|
||||||
| NC score | 0.025235 (rank : 120) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75298, O60509, Q7RTM6, Q7RTN1, Q7RTN2 | Gene names | RTN2, NSPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-2 (Neuroendocrine-specific protein-like 1) (NSP-like protein 1) (NSPLI). | |||||
|
KLDC4_MOUSE
|
||||||
| NC score | 0.024532 (rank : 121) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921I2, Q8CIK0 | Gene names | Klhdc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch domain-containing protein 4. | |||||
|
CIR_MOUSE
|
||||||
| NC score | 0.023378 (rank : 122) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
RGP2_HUMAN
|
||||||
| NC score | 0.023343 (rank : 123) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P47736 | Gene names | RAP1GAP, RAP1GA1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-activating protein 1 (Rap1GAP). | |||||
|
YETS2_HUMAN
|
||||||
| NC score | 0.023116 (rank : 124) | θ value | 8.99809 (rank : 179) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9ULM3, Q641P6, Q9NW96 | Gene names | YEATS2, KIAA1197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YEATS domain-containing protein 2. | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.022739 (rank : 125) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
ANKS6_MOUSE
|
||||||
| NC score | 0.022509 (rank : 126) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6GQX6 | Gene names | Anks6, Samd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 6 (Sterile alpha motif domain-containing protein 6). | |||||
|
PKHA5_HUMAN
|
||||||
| NC score | 0.021943 (rank : 127) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HAU0, Q8N3K6, Q96DY9, Q9BVR4, Q9C0H7, Q9H924, Q9NVK8 | Gene names | PLEKHA5, KIAA1686, PEPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 5 (Phosphoinositol 3-phosphate-binding protein 2) (PEPP-2). | |||||
|
PHF14_HUMAN
|
||||||
| NC score | 0.021923 (rank : 128) | θ value | 5.27518 (rank : 126) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
TMC2_HUMAN
|
||||||
| NC score | 0.021894 (rank : 129) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TDI7, Q5JXT0, Q5JXT1, Q6UWW4, Q6ZS41, Q8N9F3, Q9BYN2, Q9BYN3, Q9BYN4, Q9BYN5 | Gene names | TMC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane cochlear-expressed protein 2. | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.021675 (rank : 130) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
TJAP1_MOUSE
|
||||||
| NC score | 0.021607 (rank : 131) | θ value | 6.88961 (rank : 155) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DCD5, Q8CFL7, Q8R5I2 | Gene names | Tjap1, Pilt, Tjp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tight junction-associated protein 1 (Tight junction protein 4) (Protein incorporated later into tight junctions). | |||||
|
GCAA_MOUSE
|
||||||
| NC score | 0.020515 (rank : 132) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P01863 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig gamma-2A chain C region, A allele. | |||||
|
GCAM_MOUSE
|
||||||
| NC score | 0.020424 (rank : 133) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P01865 | Gene names | Igh-1a | |||
|
Domain Architecture |

|
|||||
| Description | Ig gamma-2A chain C region, membrane-bound form. | |||||
|
SCMH1_MOUSE
|
||||||
| NC score | 0.020235 (rank : 134) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K214, Q9JME0 | Gene names | Scmh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb protein SCMH1 (Sex comb on midleg homolog 1). | |||||
|
MUCDL_MOUSE
|
||||||
| NC score | 0.019705 (rank : 135) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
JAD1C_HUMAN
|
||||||
| NC score | 0.019675 (rank : 136) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P41229 | Gene names | SMCX, DXS1272E, JARID1C, XE169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (Protein SmcX) (Protein Xe169). | |||||
|
DYNA_MOUSE
|
||||||
| NC score | 0.018986 (rank : 137) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
TJAP1_HUMAN
|
||||||
| NC score | 0.018608 (rank : 138) | θ value | 8.99809 (rank : 177) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5JTD0, Q5JTD1, Q5JWW1, Q68DB2, Q6P2P3, Q9H7V7 | Gene names | TJAP1, PILT, TJP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tight junction-associated protein 1 (Tight junction protein 4) (Protein incorporated later into tight junctions). | |||||
|
DYNA_HUMAN
|
||||||
| NC score | 0.018385 (rank : 139) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
SYNP2_MOUSE
|
||||||
| NC score | 0.016663 (rank : 140) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91YE8, Q8C592 | Gene names | Synpo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin). | |||||
|
CO3A1_HUMAN
|
||||||
| NC score | 0.016495 (rank : 141) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
BRPF1_HUMAN
|
||||||
| NC score | 0.016462 (rank : 142) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
ZC3H6_HUMAN
|
||||||
| NC score | 0.016457 (rank : 143) | θ value | 8.99809 (rank : 180) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
TNKS1_HUMAN
|
||||||
| NC score | 0.016170 (rank : 144) | θ value | 4.03905 (rank : 111) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
FEZ2_HUMAN
|
||||||
| NC score | 0.015955 (rank : 145) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UHY8, Q76LN0, Q99690 | Gene names | FEZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fasciculation and elongation protein zeta 2 (Zygin-2) (Zygin II). | |||||
|
GCAB_MOUSE
|
||||||
| NC score | 0.015676 (rank : 146) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01864 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig gamma-2A chain C region secreted form (B allele). | |||||
|
FEZ2_MOUSE
|
||||||
| NC score | 0.015631 (rank : 147) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6TYB5 | Gene names | Fez2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fasciculation and elongation protein zeta 2 (Zygin-2) (Zygin II). | |||||
|
NOTC3_MOUSE
|
||||||
| NC score | 0.015350 (rank : 148) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
EDN3_MOUSE
|
||||||
| NC score | 0.015224 (rank : 149) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P48299, Q543L0 | Gene names | Edn3 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-3 precursor (ET-3) (Preproendothelin-3) (PPET3). | |||||
|
FOXC2_HUMAN
|
||||||
| NC score | 0.014483 (rank : 150) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99958 | Gene names | FOXC2, FKHL14, MFH1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C2 (Forkhead-related protein FKHL14) (Mesenchyme fork head protein 1) (MFH-1 protein) (Transcription factor FKH-14). | |||||
|
TECT3_MOUSE
|
||||||
| NC score | 0.014358 (rank : 151) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R2Q6, Q8CDF3, Q9CRC5 | Gene names | Tect3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tectonic-3 precursor. | |||||
|
LSD1_HUMAN
|
||||||
| NC score | 0.014304 (rank : 152) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
NOTC3_HUMAN
|
||||||
| NC score | 0.014221 (rank : 153) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.013829 (rank : 154) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
ERRFI_HUMAN
|
||||||
| NC score | 0.013794 (rank : 155) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UJM3, Q9NTG9, Q9UD05 | Gene names | ERRFI1, MIG6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein) (Mig-6). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.013676 (rank : 156) | θ value | 4.03905 (rank : 108) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
HXA3_HUMAN
|
||||||
| NC score | 0.012994 (rank : 157) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43365 | Gene names | HOXA3, HOX1E | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1E). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.012819 (rank : 158) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
WDR22_MOUSE
|
||||||
| NC score | 0.012685 (rank : 159) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80T85, Q80VT3, Q80ZW6, Q8BIP8, Q8BVK5, Q8K0Q6 | Gene names | Wdr22, Kiaa1824 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 22. | |||||
|
ZC3H8_HUMAN
|
||||||
| NC score | 0.012190 (rank : 160) | θ value | 8.99809 (rank : 181) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N5P1, Q9BZ75 | Gene names | ZC3H8, ZC3HDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8. | |||||
|
PAPOA_MOUSE
|
||||||
| NC score | 0.012158 (rank : 161) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61183, Q61208, Q61209, Q8K4X2 | Gene names | Papola, Pap, Plap | |||
|
Domain Architecture |

|
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase). | |||||
|
EDA_MOUSE
|
||||||
| NC score | 0.012020 (rank : 162) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O54693, O35705, Q9QWJ8, Q9QZ01, Q9QZ02 | Gene names | Eda, Ed1, Ta | |||
|
Domain Architecture |

|
|||||
| Description | Ectodysplasin-A (EDA protein homolog) (Tabby protein) [Contains: Ectodysplasin-A, membrane form; Ectodysplasin-A, secreted form]. | |||||
|
HXA3_MOUSE
|
||||||
| NC score | 0.011765 (rank : 163) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 558 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P02831, Q4V9Z8, Q61197 | Gene names | Hoxa3, Hox-1.5, Hoxa-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A3 (Hox-1.5) (MO-10). | |||||
|
NOP56_HUMAN
|
||||||
| NC score | 0.011527 (rank : 164) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00567, Q9NQ05 | Gene names | NOL5A, NOP56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
GEMI5_MOUSE
|
||||||
| NC score | 0.011311 (rank : 165) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 424 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BX17 | Gene names | Gemin5 | |||
|
Domain Architecture |

|
|||||
| Description | Gem-associated protein 5 (Gemin5). | |||||
|
COFA1_HUMAN
|
||||||
| NC score | 0.011137 (rank : 166) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P39059, Q5T6J4, Q9Y4W4 | Gene names | COL15A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV) (Restin) (Related to endostatin)]. | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.011009 (rank : 167) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
MYB_HUMAN
|
||||||
| NC score | 0.010884 (rank : 168) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10242, P78391, P78392, P78525, P78526, Q14023, Q14024, Q9UE83 | Gene names | MYB | |||
|
Domain Architecture |
|
|||||
| Description | Myb proto-oncogene protein (C-myb). | |||||
|
JHD2C_HUMAN
|
||||||
| NC score | 0.010716 (rank : 169) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15652, Q5SQZ8, Q5SQZ9, Q5SR00, Q7Z3E7, Q8N3U0, Q96KB9, Q9P2G7 | Gene names | JMJD1C, JHDM2C, KIAA1380, TRIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C) (Thyroid receptor-interacting protein 8) (TRIP-8). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.010552 (rank : 170) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.010538 (rank : 171) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
PI4KB_HUMAN
|
||||||
| NC score | 0.010485 (rank : 172) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UBF8, O15096, P78405, Q9BWR6 | Gene names | PIK4CB, PI4KB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta) (NPIK) (PI4K92). | |||||
|
FBLN2_MOUSE
|
||||||
| NC score | 0.010448 (rank : 173) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
PRP4B_HUMAN
|
||||||
| NC score | 0.010179 (rank : 174) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1030 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13523, Q8TDP2, Q96QT7, Q9UEE6 | Gene names | PRPF4B, KIAA0536, PRP4, PRP4H, PRP4K | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (PRP4 kinase). | |||||
|
INP4A_MOUSE
|
||||||
| NC score | 0.009722 (rank : 175) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EPW0, Q8CBJ8 | Gene names | Inpp4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Type I inositol-3,4-bisphosphate 4-phosphatase (EC 3.1.3.66) (Inositol polyphosphate 4-phosphatase type I). | |||||
|
MYLK_MOUSE
|
||||||
| NC score | 0.009477 (rank : 176) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
HIF1A_HUMAN
|
||||||
| NC score | 0.009115 (rank : 177) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q16665, Q96PT9, Q9UPB1 | Gene names | HIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hypoxia-inducible factor 1 alpha (HIF-1 alpha) (HIF1 alpha) (ARNT- interacting protein) (Member of PAS protein 1) (MOP1). | |||||
|
FOXD3_HUMAN
|
||||||
| NC score | 0.008805 (rank : 178) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJU5, Q9BYM2, Q9UDD1 | Gene names | FOXD3 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D3 (HNF3/FH transcription factor genesis). | |||||
|
LAMA2_HUMAN
|
||||||
| NC score | 0.008426 (rank : 179) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
HXD9_HUMAN
|
||||||
| NC score | 0.007995 (rank : 180) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P28356 | Gene names | HOXD9, HOX4C | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D9 (Hox-4C) (Hox-5.2). | |||||
|
ZN516_MOUSE
|
||||||
| NC score | 0.007909 (rank : 181) | θ value | 8.99809 (rank : 182) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
ZN438_HUMAN
|
||||||
| NC score | 0.007815 (rank : 182) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7Z4V0, Q5T426, Q658Q4, Q6ZN65 | Gene names | ZNF438 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 438. | |||||
|
SALL3_HUMAN
|
||||||
| NC score | 0.006722 (rank : 183) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1066 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9BXA9, Q9UGH1 | Gene names | SALL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 3 (Zinc finger protein SALL3) (hSALL3). | |||||
|
CD2L5_MOUSE
|
||||||
| NC score | 0.006369 (rank : 184) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1011 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q69ZA1, Q80V11, Q8BZG1, Q8K0A4 | Gene names | Cdc2l5, Kiaa1791 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5). | |||||
|
ZN516_HUMAN
|
||||||
| NC score | 0.005915 (rank : 185) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 854 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q92618 | Gene names | ZNF516, KIAA0222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 516. | |||||
|
ZBTB9_MOUSE
|
||||||
| NC score | 0.004693 (rank : 186) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8CDC7, Q3U3B2, Q9CYQ0 | Gene names | Zbtb9 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 9. | |||||
|
KI21A_MOUSE
|
||||||
| NC score | 0.004546 (rank : 187) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
KLF10_HUMAN
|
||||||
| NC score | 0.004346 (rank : 188) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13118 | Gene names | KLF10, TIEG, TIEG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 10 (Transforming growth factor-beta-inducible early growth response protein 1) (TGFB-inducible early growth response protein 1) (TIEG-1) (EGR-alpha). | |||||
|
ZBT20_HUMAN
|
||||||
| NC score | 0.004085 (rank : 189) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9HC78 | Gene names | ZBTB20, DPZF, ZNF288 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 20 (Zinc finger protein 288) (Dendritic-derived BTB/POZ zinc finger protein). | |||||
|
MYO3B_HUMAN
|
||||||
| NC score | 0.002486 (rank : 190) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WXR4, Q8IX64, Q8IX65, Q8IX66, Q8IX67, Q8IX68 | Gene names | MYO3B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin IIIB (EC 2.7.11.1). | |||||
|
BMPR2_HUMAN
|
||||||
| NC score | 0.001999 (rank : 191) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13873, Q16569 | Gene names | BMPR2, PPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein receptor type-2 precursor (EC 2.7.11.30) (Bone morphogenetic protein receptor type II) (BMP type II receptor) (BMPR-II). | |||||
|
KAISO_HUMAN
|
||||||
| NC score | 0.001861 (rank : 192) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86T24, O00319, Q7Z361, Q8IVP6, Q8N3P0 | Gene names | ZBTB33, KAISO, ZNF348 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional regulator Kaiso (Zinc finger and BTB domain-containing protein 33). | |||||
|
ZN692_MOUSE
|
||||||
| NC score | 0.001794 (rank : 193) | θ value | 3.0926 (rank : 96) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3U381 | Gene names | Znf692, Zfp692 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 692. | |||||
|
KKCC2_HUMAN
|
||||||
| NC score | 0.000935 (rank : 194) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96RR4, O94883, Q8IUG2, Q8IUG3, Q8N3I4, Q8WY03, Q8WY04, Q8WY05, Q8WY06, Q96RP1, Q96RP2, Q96RR3, Q9BWE9, Q9UER3, Q9UES2, Q9Y5N2 | Gene names | CAMKK2, CAMKKB, KIAA0787 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase kinase 2 (EC 2.7.11.17) (Calcium/calmodulin-dependent protein kinase kinase beta) (CaM-kinase kinase beta) (CaM-KK beta) (CaMKK beta). | |||||
|
PKN3_MOUSE
|
||||||
| NC score | 0.000637 (rank : 195) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 182 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K045 | Gene names | Pkn3, Pknbeta | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N3 (EC 2.7.11.13) (Protein kinase PKN- beta) (Protein-kinase C-related kinase 3). | |||||