Please be patient as the page loads
|
SIA7A_HUMAN
|
||||||
| SwissProt Accessions | Q9NSC7, Q6UW90, Q9NSC6 | Gene names | ST6GALNAC1, SIAT7A | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SIA7A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 177 | |
| SwissProt Accessions | Q9NSC7, Q6UW90, Q9NSC6 | Gene names | ST6GALNAC1, SIAT7A | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
SIA7A_MOUSE
|
||||||
| θ value | 2.32313e-181 (rank : 2) | NC score | 0.953413 (rank : 2) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QZ39, Q9JJP5 | Gene names | St6galnac1, Siat7a | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
SIA7B_MOUSE
|
||||||
| θ value | 1.31681e-67 (rank : 3) | NC score | 0.922086 (rank : 3) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70277, Q3UWG0, Q9DC24 | Gene names | St6galnac2, Siat7, Siat7b | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 (EC 2.4.99.-) (GalNAc alpha-2,6-sialyltransferase II) (ST6GalNAc II) (Gal-beta-1,3-GalNAc alpha-2,6-sialyltransferase) (Sialyltransferase 7B). | |||||
|
SIA7B_HUMAN
|
||||||
| θ value | 6.53529e-67 (rank : 4) | NC score | 0.922032 (rank : 4) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UJ37, Q12971 | Gene names | ST6GALNAC2, SIAT7B, STHM | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 (EC 2.4.99.-) (GalNAc alpha-2,6-sialyltransferase II) (ST6GalNAc II) (Sialyltransferase 7B) (SThM). | |||||
|
SIAT6_HUMAN
|
||||||
| θ value | 4.58923e-20 (rank : 5) | NC score | 0.747036 (rank : 10) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q11203, Q86UR6, Q86UR7, Q86UR8, Q86UR9, Q86US0, Q86US1, Q86US2, Q8IX41, Q8IX42, Q8IX43, Q8IX44, Q8IX45, Q8IX46, Q8IX47, Q8IX48, Q8IX49, Q8IX50, Q8IX51, Q8IX52, Q8IX53, Q8IX54, Q8IX55, Q8IX56, Q8IX57, Q8IX58 | Gene names | ST3GAL3, SIAT6 | |||
|
Domain Architecture |

|
|||||
| Description | CMP-N-acetylneuraminate-beta-1,4-galactoside alpha-2,3- sialyltransferase (EC 2.4.99.6) (N-acetyllactosaminide alpha-2,3- sialyltransferase) (Gal beta-1,3(4) GlcNAc alpha-2,3 sialyltransferase) (ST3N) (ST3GalIII) (Sialyltransferase 6). | |||||
|
SIAT6_MOUSE
|
||||||
| θ value | 2.27762e-19 (rank : 6) | NC score | 0.750491 (rank : 9) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97325 | Gene names | St3gal3, Siat3, Siat6 | |||
|
Domain Architecture |

|
|||||
| Description | CMP-N-acetylneuraminate-beta-1,4-galactoside alpha-2,3- sialyltransferase (EC 2.4.99.6) (N-acetyllactosaminide alpha-2,3- sialyltransferase) (Gal beta-1,3(4) GlcNAc alpha-2,3 sialyltransferase) (ST3N) (ST3GalIII) (Sialyltransferase 6). | |||||
|
SIA4A_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 7) | NC score | 0.768936 (rank : 5) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q11201, O60677, Q9UN51 | Gene names | ST3GAL1, SIAT4, SIAT4A | |||
|
Domain Architecture |

|
|||||
| Description | CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3-sialyltransferase (EC 2.4.99.4) (Beta-galactoside alpha-2,3-sialyltransferase) (Alpha 2,3-ST) (Gal-NAc6S) (Gal-beta-1,3-GalNAc-alpha-2,3-sialyltransferase) (ST3GalIA) (ST3O) (ST3GalA.1) (SIAT4-A) (ST3Gal I) (SIATFL). | |||||
|
SIA4B_MOUSE
|
||||||
| θ value | 8.95645e-16 (rank : 8) | NC score | 0.756627 (rank : 7) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q11204 | Gene names | St3gal2, Siat4b, Siat5 | |||
|
Domain Architecture |

|
|||||
| Description | CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3-sialyltransferase (EC 2.4.99.-) (Beta-galactoside alpha-2,3-sialyltransferase) (Alpha 2,3-ST) (Gal-NAc6S) (Gal-beta-1,3-GalNAc-alpha-2,3-sialyltransferase) (ST3GalA.2) (SIAT4-B) (ST3Gal II). | |||||
|
SIA4C_HUMAN
|
||||||
| θ value | 8.95645e-16 (rank : 9) | NC score | 0.735622 (rank : 11) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q11206, O60497, Q96QQ9 | Gene names | ST3GAL4, SIAT4C, STZ | |||
|
Domain Architecture |

|
|||||
| Description | CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3-sialyltransferase (EC 2.4.99.-) (Beta-galactoside alpha-2,3-sialyltransferase) (Alpha 2,3-sialyltransferase IV) (Alpha 2,3-ST) (Gal-NAc6S) (STZ) (SIAT4-C) (ST3Gal III) (SAT-3) (ST-4). | |||||
|
SIA8B_MOUSE
|
||||||
| θ value | 1.99529e-15 (rank : 10) | NC score | 0.702267 (rank : 17) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O35696 | Gene names | St8sia2, Siat8b, Stx | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-2,8-sialyltransferase 8B (EC 2.4.99.-) (ST8Sia II) (Sialyltransferase X) (STX) (Polysialic acid synthase). | |||||
|
SIA4B_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 11) | NC score | 0.756272 (rank : 8) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q16842, O00654 | Gene names | ST3GAL2, SIAT4B | |||
|
Domain Architecture |

|
|||||
| Description | CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3-sialyltransferase (EC 2.4.99.-) (Beta-galactoside alpha-2,3-sialyltransferase) (Alpha 2,3-ST) (Gal-NAc6S) (Gal-beta-1,3-GalNAc-alpha-2,3-sialyltransferase) (ST3GalA.2) (SIAT4-B) (ST3Gal II). | |||||
|
SIA8F_MOUSE
|
||||||
| θ value | 2.60593e-15 (rank : 12) | NC score | 0.680190 (rank : 23) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K4T1 | Gene names | St8sia6, Siat8f | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-2,8-sialyltransferase 8F (EC 2.4.99.-) (ST8Sia VI). | |||||
|
SIA4C_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 13) | NC score | 0.732414 (rank : 12) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q91Y74 | Gene names | St3gal4, Siat4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3-sialyltransferase (EC 2.4.99.-) (Beta-galactoside alpha-2,3-sialyltransferase) (Alpha 2,3-sialyltransferase IV) (Alpha 2,3-ST) (SIAT4-C). | |||||
|
SIA8C_HUMAN
|
||||||
| θ value | 4.44505e-15 (rank : 14) | NC score | 0.721532 (rank : 14) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O43173, Q9NS41 | Gene names | ST8SIA3, SIAT8C | |||
|
Domain Architecture |

|
|||||
| Description | Sia-alpha-2,3-Gal-beta-1,4-GlcNAc-R:alpha 2,8-sialyltransferase (EC 2.4.99.-) (ST8 alpha-N-acetyl-neuraminide alpha-2,8- sialyltransferase 3) (Alpha-2,8-sialyltransferase 8C) (Alpha-2,8- sialyltransferase III) (ST8Sia III). | |||||
|
SIA4A_MOUSE
|
||||||
| θ value | 5.8054e-15 (rank : 15) | NC score | 0.762936 (rank : 6) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P54751, Q11202 | Gene names | St3gal1, Siat4, Siat4a | |||
|
Domain Architecture |
|
|||||
| Description | CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3-sialyltransferase (EC 2.4.99.4) (Beta-galactoside alpha-2,3-sialyltransferase) (Alpha 2,3-ST) (Gal-NAc6S) (Gal-beta-1,3-GalNAc-alpha-2,3-sialyltransferase) (ST3GalIA) (ST3O) (ST3GalA.1) (SIAT4-A) (ST3Gal I). | |||||
|
SIAT9_MOUSE
|
||||||
| θ value | 7.58209e-15 (rank : 16) | NC score | 0.728641 (rank : 13) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O88829 | Gene names | St3gal5, Siat9 | |||
|
Domain Architecture |
|
|||||
| Description | Lactosylceramide alpha-2,3-sialyltransferase (EC 2.4.99.9) (CMP- NeuAc:lactosylceramide alpha-2,3-sialyltransferase) (Ganglioside GM3 synthase) (ST3Gal V) (Sialyltransferase 9). | |||||
|
SIA8B_HUMAN
|
||||||
| θ value | 9.90251e-15 (rank : 17) | NC score | 0.695796 (rank : 21) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q92186, Q4VAZ0, Q92470, Q92746 | Gene names | ST8SIA2, SIAT8B, STX | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-2,8-sialyltransferase 8B (EC 2.4.99.-) (ST8Sia II) (Sialyltransferase X) (STX). | |||||
|
SIA8C_MOUSE
|
||||||
| θ value | 3.76295e-14 (rank : 18) | NC score | 0.714446 (rank : 16) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64689 | Gene names | St8sia3, Siat8c | |||
|
Domain Architecture |

|
|||||
| Description | Sia-alpha-2,3-Gal-beta-1,4-GlcNAc-R:alpha 2,8-sialyltransferase (EC 2.4.99.-) (ST8 alpha-N-acetyl-neuraminide alpha-2,8- sialyltransferase 3) (Alpha-2,8-sialyltransferase 8C) (Alpha-2,8- sialyltransferase III) (ST8Sia III). | |||||
|
SIAT9_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 19) | NC score | 0.718571 (rank : 15) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UNP4, O94902, Q53QU1 | Gene names | ST3GAL5, SIAT9 | |||
|
Domain Architecture |
|
|||||
| Description | Lactosylceramide alpha-2,3-sialyltransferase (EC 2.4.99.9) (CMP- NeuAc:lactosylceramide alpha-2,3-sialyltransferase) (Ganglioside GM3 synthase) (ST3Gal V) (Sialyltransferase 9). | |||||
|
SIA8D_HUMAN
|
||||||
| θ value | 2.0648e-12 (rank : 20) | NC score | 0.697757 (rank : 19) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q92187, Q92693 | Gene names | ST8SIA4, PST, PST1, SIAT8D | |||
|
Domain Architecture |
|
|||||
| Description | CMP-N-acetylneuraminate-poly-alpha-2,8-sialyltransferase (EC 2.4.99.-) (Alpha-2,8-sialyltransferase 8D) (ST8Sia IV) (Polysialyltransferase- 1). | |||||
|
SIA8D_MOUSE
|
||||||
| θ value | 2.69671e-12 (rank : 21) | NC score | 0.695329 (rank : 22) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64692 | Gene names | St8sia4, Pst, Siat8d | |||
|
Domain Architecture |
|
|||||
| Description | CMP-N-acetylneuraminate-poly-alpha-2,8-sialyltransferase (EC 2.4.99.-) (Alpha-2,8-sialyltransferase 8D) (ST8Sia IV) (Polysialyltransferase- 1). | |||||
|
SIA8F_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 22) | NC score | 0.660595 (rank : 28) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P61647 | Gene names | ST8SIA6, SIAT8F | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-2,8-sialyltransferase 8F (EC 2.4.99.-) (ST8Sia VI). | |||||
|
SIA8E_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 23) | NC score | 0.661794 (rank : 27) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70126, P70127, P70128 | Gene names | St8sia5, Siat8e, St8siav | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-2,8-sialyltransferase 8E (EC 2.4.99.-) (ST8Sia V). | |||||
|
SIAT1_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 24) | NC score | 0.678229 (rank : 24) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64685 | Gene names | St6gal1, Siat1 | |||
|
Domain Architecture |
|
|||||
| Description | CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,6-sialyltransferase (EC 2.4.99.1) (Beta-galactoside alpha-2,6-sialyltransferase) (Alpha 2,6-ST) (Sialyltransferase 1) (ST6Gal I). | |||||
|
SIA10_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 25) | NC score | 0.696998 (rank : 20) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y274 | Gene names | ST3GAL6, SIAT10 | |||
|
Domain Architecture |
|
|||||
| Description | Type 2 lactosamine alpha-2,3-sialyltransferase (EC 2.4.99.-) (CMP- NeuAc:beta-galactoside alpha-2,3-sialyltransferase VI) (ST3Gal VI) (Sialyltransferase 10). | |||||
|
SIA8E_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 26) | NC score | 0.665413 (rank : 26) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15466 | Gene names | ST8SIA5, SIAT8E | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2,8-sialyltransferase 8E (EC 2.4.99.-) (ST8Sia V). | |||||
|
SIA8A_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 27) | NC score | 0.642792 (rank : 29) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q92185, Q93064 | Gene names | ST8SIA1, SIAT8, SIAT8A | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylneuraminide alpha-2,8-sialyltransferase (EC 2.4.99.8) (Ganglioside GD3 synthase) (Ganglioside GT3 synthase) (Alpha-2,8- sialyltransferase 8A) (ST8Sia I). | |||||
|
SIA10_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 28) | NC score | 0.701770 (rank : 18) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8VIB3, Q3TRN3, Q80UR7, Q9WVG2 | Gene names | St3gal6, Siat10 | |||
|
Domain Architecture |

|
|||||
| Description | Type 2 lactosamine alpha-2,3-sialyltransferase (EC 2.4.99.-) (CMP- NeuAc:beta-galactoside alpha-2,3-sialyltransferase VI) (ST3Gal VI) (Sialyltransferase 10). | |||||
|
SIAT1_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 29) | NC score | 0.670792 (rank : 25) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P15907 | Gene names | ST6GAL1, SIAT1 | |||
|
Domain Architecture |
|
|||||
| Description | CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,6-sialyltransferase (EC 2.4.99.1) (Beta-galactoside alpha-2,6-sialyltransferase) (Alpha 2,6-ST) (Sialyltransferase 1) (ST6Gal I) (B-cell antigen CD75). | |||||
|
SIA8A_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 30) | NC score | 0.635238 (rank : 30) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64687 | Gene names | St8sia1, Siat8, Siat8a | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylneuraminide alpha-2,8-sialyltransferase (EC 2.4.99.8) (Ganglioside GD3 synthase) (Ganglioside GT3 synthase) (Alpha-2,8- sialyltransferase 8A) (ST8Sia I). | |||||
|
SIA7C_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 31) | NC score | 0.530380 (rank : 35) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9WUV2, Q9JHP5 | Gene names | St6galnac3, Siat7c | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 (EC 2.4.99.-) (GalNAc alpha-2,6-sialyltransferase III) (ST6GalNAc III) (Sialyltransferase 7C) (STY). | |||||
|
SIA7D_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 32) | NC score | 0.576137 (rank : 32) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9R2B6, O88725, Q9JHP0, Q9QUP9, Q9R2B5 | Gene names | St6galnac4, Siat7d | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3-N-acetyl- galactosaminide alpha-2,6-sialyltransferase (EC 2.4.99.7) (NeuAc- alpha-2,3-Gal-beta-1,3-GalNAc-alpha-2,6-sialyltransferase) (ST6GalNAc IV) (Sialyltransferase 7D). | |||||
|
SIA7E_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 33) | NC score | 0.562344 (rank : 34) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QYJ1, Q9R0K6 | Gene names | St6galnac5, Siat7e | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 (EC 2.4.99.-) (GalNAc alpha-2,6-sialyltransferase V) (ST6GalNAc V) (GD1 alpha synthase) (Sialyltransferase 7E). | |||||
|
SIA7E_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 34) | NC score | 0.583018 (rank : 31) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BVH7 | Gene names | ST6GALNAC5, SIAT7E | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 (EC 2.4.99.-) (GalNAc alpha-2,6-sialyltransferase V) (ST6GalNAc V) (GD1 alpha synthase) (Sialyltransferase 7E). | |||||
|
SIA7C_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 35) | NC score | 0.527705 (rank : 36) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8NDV1, Q6UX29, Q8N259 | Gene names | ST6GALNAC3, SIAT7C | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 (EC 2.4.99.-) (GalNAc alpha-2,6-sialyltransferase III) (ST6GalNAc III) (Sialyltransferase 7C) (STY). | |||||
|
SIA7D_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 36) | NC score | 0.567811 (rank : 33) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H4F1, Q9NWU6, Q9UKU1, Q9ULB9, Q9Y3G3, Q9Y3G4 | Gene names | ST6GALNAC4, SIAT3C, SIAT7D | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3-N-acetyl- galactosaminide alpha-2,6-sialyltransferase (EC 2.4.99.7) (NeuAc- alpha-2,3-Gal-beta-1,3-GalNAc-alpha-2,6-sialyltransferase) (ST6GalNAc IV) (Sialyltransferase 7D) (Sialyltransferase 3C). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 37) | NC score | 0.114552 (rank : 37) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 38) | NC score | 0.108166 (rank : 39) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 39) | NC score | 0.107799 (rank : 40) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
ATF2_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 40) | NC score | 0.080687 (rank : 48) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 41) | NC score | 0.091896 (rank : 43) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 42) | NC score | 0.087491 (rank : 44) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
ATF2_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 43) | NC score | 0.079147 (rank : 49) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 44) | NC score | 0.041291 (rank : 101) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
AAK1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 45) | NC score | 0.018637 (rank : 146) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1077 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q2M2I8, Q4ZFZ3, Q53RX6, Q9UPV4 | Gene names | AAK1, KIAA1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 46) | NC score | 0.081643 (rank : 47) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 47) | NC score | 0.084230 (rank : 45) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 48) | NC score | 0.105807 (rank : 41) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
CIZ1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 49) | NC score | 0.077597 (rank : 50) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
H14_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 50) | NC score | 0.064233 (rank : 69) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P10412 | Gene names | HIST1H1E, H1F4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (Histone H1b). | |||||
|
HCN1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 51) | NC score | 0.023298 (rank : 134) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 52) | NC score | 0.073943 (rank : 54) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 53) | NC score | 0.042127 (rank : 99) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 54) | NC score | 0.073373 (rank : 55) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
PARM1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 55) | NC score | 0.072164 (rank : 59) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 56) | NC score | 0.069534 (rank : 60) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 57) | NC score | 0.082215 (rank : 46) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
H13_MOUSE
|
||||||
| θ value | 0.125558 (rank : 58) | NC score | 0.072281 (rank : 58) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P43277, Q8C6M4 | Gene names | Hist1h1d, H1f3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (H1 VAR.4) (H1d). | |||||
|
LAD1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 59) | NC score | 0.113401 (rank : 38) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.163984 (rank : 60) | NC score | 0.066843 (rank : 66) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 61) | NC score | 0.039817 (rank : 104) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
LAD1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 62) | NC score | 0.104550 (rank : 42) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 0.279714 (rank : 63) | NC score | 0.077018 (rank : 51) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
SRP72_HUMAN
|
||||||
| θ value | 0.279714 (rank : 64) | NC score | 0.047944 (rank : 93) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O76094 | Gene names | SRP72 | |||
|
Domain Architecture |

|
|||||
| Description | Signal recognition particle 72 kDa protein (SRP72). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 0.365318 (rank : 65) | NC score | 0.061887 (rank : 75) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
BLNK_HUMAN
|
||||||
| θ value | 0.47712 (rank : 66) | NC score | 0.047596 (rank : 94) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WV28, O75498, O75499 | Gene names | BLNK, BASH, SLP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (SLP-65). | |||||
|
HUCE1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 67) | NC score | 0.037324 (rank : 106) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43159, Q7KZ78, Q9BVM6 | Gene names | KIAA0409 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerebral protein 1. | |||||
|
IL16_HUMAN
|
||||||
| θ value | 0.47712 (rank : 68) | NC score | 0.020591 (rank : 141) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14005, Q16435, Q9UP18 | Gene names | IL16 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-16 precursor (IL-16) (Lymphocyte chemoattractant factor) (LCF). | |||||
|
SAM14_MOUSE
|
||||||
| θ value | 0.47712 (rank : 69) | NC score | 0.048877 (rank : 92) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K070, Q5SWB7, Q8BHE2, Q8C8N5 | Gene names | Samd14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 14. | |||||
|
MINT_MOUSE
|
||||||
| θ value | 0.62314 (rank : 70) | NC score | 0.056112 (rank : 82) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.62314 (rank : 71) | NC score | 0.059571 (rank : 78) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SMAL1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 72) | NC score | 0.025821 (rank : 127) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 73) | NC score | 0.076586 (rank : 52) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
ADDB_HUMAN
|
||||||
| θ value | 0.813845 (rank : 74) | NC score | 0.025573 (rank : 130) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 75) | NC score | 0.061151 (rank : 76) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 76) | NC score | 0.018543 (rank : 147) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
PAF49_MOUSE
|
||||||
| θ value | 0.813845 (rank : 77) | NC score | 0.034401 (rank : 108) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q76KJ5, Q8K0Y9 | Gene names | Cd3eap, Ase1, Paf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein). | |||||
|
PININ_HUMAN
|
||||||
| θ value | 0.813845 (rank : 78) | NC score | 0.027490 (rank : 123) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
PPRB_MOUSE
|
||||||
| θ value | 0.813845 (rank : 79) | NC score | 0.063935 (rank : 70) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 0.813845 (rank : 80) | NC score | 0.006103 (rank : 179) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
CF010_HUMAN
|
||||||
| θ value | 1.06291 (rank : 81) | NC score | 0.040448 (rank : 103) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
MCES_HUMAN
|
||||||
| θ value | 1.06291 (rank : 82) | NC score | 0.033959 (rank : 111) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43148, O94996, Q9UIJ9 | Gene names | RNMT, KIAA0398 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA cap guanine-N7 methyltransferase (EC 2.1.1.56) (mRNA (guanine- N(7)-)-methyltransferase) (RG7MT1) (mRNA cap methyltransferase) (hcm1p) (hCMT1) (hMet). | |||||
|
MILK2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 83) | NC score | 0.026393 (rank : 125) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 1.06291 (rank : 84) | NC score | 0.067779 (rank : 63) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PKHA4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 85) | NC score | 0.025782 (rank : 129) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
SYN1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 86) | NC score | 0.031011 (rank : 116) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 87) | NC score | 0.032058 (rank : 114) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
BASP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 88) | NC score | 0.072735 (rank : 57) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 1.38821 (rank : 89) | NC score | 0.069495 (rank : 61) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
DJC12_HUMAN
|
||||||
| θ value | 1.38821 (rank : 90) | NC score | 0.019688 (rank : 144) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKB3, Q9UKB2 | Gene names | DNAJC12, JDP1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
FOS_HUMAN
|
||||||
| θ value | 1.38821 (rank : 91) | NC score | 0.028886 (rank : 120) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P01100, P18849 | Gene names | FOS, G0S7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos) (G0/G1 switch regulatory protein 7). | |||||
|
H14_MOUSE
|
||||||
| θ value | 1.38821 (rank : 92) | NC score | 0.053692 (rank : 84) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P43274 | Gene names | Hist1h1e, H1f4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (H1 VAR.2) (H1e). | |||||
|
MAGD1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 93) | NC score | 0.012305 (rank : 162) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y5V3, Q5VSH6, Q8IZ84, Q8WY92, Q9H352, Q9HBT4, Q9UF36 | Gene names | MAGED1, NRAGE | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (MAGE tumor antigen CCF). | |||||
|
PPRB_HUMAN
|
||||||
| θ value | 1.38821 (rank : 94) | NC score | 0.062726 (rank : 73) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
WBP4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 95) | NC score | 0.029984 (rank : 119) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75554 | Gene names | WBP4, FBP21, FNBP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 4 (WBP-4) (Formin-binding protein 21). | |||||
|
CD2L5_MOUSE
|
||||||
| θ value | 1.81305 (rank : 96) | NC score | 0.002367 (rank : 183) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1011 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q69ZA1, Q80V11, Q8BZG1, Q8K0A4 | Gene names | Cdc2l5, Kiaa1791 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 97) | NC score | 0.028822 (rank : 121) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 98) | NC score | 0.032839 (rank : 113) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 1.81305 (rank : 99) | NC score | 0.020415 (rank : 142) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
SELPL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 100) | NC score | 0.036676 (rank : 107) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14242, Q12775 | Gene names | SELPLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand) (CD162 antigen). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 1.81305 (rank : 101) | NC score | 0.072830 (rank : 56) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
UBP51_HUMAN
|
||||||
| θ value | 1.81305 (rank : 102) | NC score | 0.008829 (rank : 171) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
IFI4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 103) | NC score | 0.021300 (rank : 139) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P15092 | Gene names | Ifi204 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-activable protein 204 (Ifi-204) (Interferon-inducible protein p204). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 104) | NC score | 0.076574 (rank : 53) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
SAM14_HUMAN
|
||||||
| θ value | 2.36792 (rank : 105) | NC score | 0.040661 (rank : 102) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IZD0, Q8N2X0 | Gene names | SAMD14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 14. | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 106) | NC score | 0.039441 (rank : 105) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SMC3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 107) | NC score | 0.009879 (rank : 168) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
SMC3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 108) | NC score | 0.009892 (rank : 167) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
TTF2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 109) | NC score | 0.030699 (rank : 118) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
ATF7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 110) | NC score | 0.052280 (rank : 87) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
CAMKV_HUMAN
|
||||||
| θ value | 3.0926 (rank : 111) | NC score | 0.006313 (rank : 178) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
CCD71_MOUSE
|
||||||
| θ value | 3.0926 (rank : 112) | NC score | 0.022272 (rank : 137) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VEG0, Q3UDX1 | Gene names | Ccdc71 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 71. | |||||
|
DZIP3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 113) | NC score | 0.009946 (rank : 166) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86Y13, O75162, Q6P3R9, Q6PH82, Q86Y14, Q86Y15, Q86Y16, Q8IWI0, Q96RS9 | Gene names | DZIP3, KIAA0675 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein DZIP3 (EC 6.3.2.-) (DAZ-interacting protein 3) (RNA-binding ubiquitin ligase of 138 kDa) (hRUL138). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 114) | NC score | 0.015797 (rank : 151) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
RANB3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 115) | NC score | 0.024143 (rank : 133) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H6Z4, O60405, O75759, O75760, Q9BT47, Q9UG74 | Gene names | RANBP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 3 (RanBP3). | |||||
|
SHAN2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 116) | NC score | 0.030864 (rank : 117) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UPX8, Q3Y8G9, Q52LK2, Q9UKP1 | Gene names | SHANK2, KIAA1022 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2). | |||||
|
SHAN2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 117) | NC score | 0.025964 (rank : 126) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80Z38, Q3UTK4, Q5DU07 | Gene names | Shank2, Kiaa1022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2) (Cortactin- binding protein 1) (CortBP1). | |||||
|
SSH2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 118) | NC score | 0.012235 (rank : 164) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
UTS2B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 119) | NC score | 0.046522 (rank : 96) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q765I0 | Gene names | UTS2D, URP, UTS2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Urotensin-2B precursor (Urotensin-IIB) (U-IIB) (UIIB) (Urotensin II- related peptide) (Urotensin 2 domain-containing protein). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | 0.066875 (rank : 65) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
CK024_HUMAN
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.052373 (rank : 86) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
FOG2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 122) | NC score | 0.001875 (rank : 185) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 731 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WW38, Q9NPL7, Q9NPS4, Q9UNI5 | Gene names | ZFPM2, FOG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM2 (Zinc finger protein multitype 2) (Friend of GATA protein 2) (FOG-2) (hFOG-2). | |||||
|
H15_HUMAN
|
||||||
| θ value | 4.03905 (rank : 123) | NC score | 0.059457 (rank : 79) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P16401, Q14529 | Gene names | HIST1H1B, H1F5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (Histone H1a). | |||||
|
IFI5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 124) | NC score | 0.020787 (rank : 140) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08619 | Gene names | Ifi205 | |||
|
Domain Architecture |
|
|||||
| Description | Interferon-activable protein 205 (IFI-205) (D3 protein). | |||||
|
KSR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 125) | NC score | -0.000221 (rank : 186) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IVT5, Q13476 | Gene names | KSR1, KSR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras). | |||||
|
REXO1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 126) | NC score | 0.034056 (rank : 110) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
BXDC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 127) | NC score | 0.015102 (rank : 153) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TDN6, Q8N453, Q96DH1 | Gene names | BXDC2, BRIX | |||
|
Domain Architecture |
|
|||||
| Description | Brix domain-containing protein 2 (Ribosome biogenesis protein Brix). | |||||
|
DAF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.006439 (rank : 177) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08174, P09679, P78361 | Gene names | CD55, CR, DAF | |||
|
Domain Architecture |
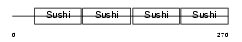
|
|||||
| Description | Complement decay-accelerating factor precursor (CD55 antigen). | |||||
|
DEK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.027280 (rank : 124) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35659, Q5TGV4, Q5TGV5 | Gene names | DEK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
MSH6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.014523 (rank : 158) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54276, O54710 | Gene names | Msh6, Gtmbp | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
SCEL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.014711 (rank : 156) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EQG3, Q9CTT9 | Gene names | Scel | |||
|
Domain Architecture |

|
|||||
| Description | Sciellin. | |||||
|
SCG3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.016545 (rank : 150) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P47867 | Gene names | Scg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretogranin-3 precursor (Secretogranin III) (SgIII). | |||||
|
TGON2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.034269 (rank : 109) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
3BP5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.007238 (rank : 176) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60239 | Gene names | SH3BP5, SAB | |||
|
Domain Architecture |
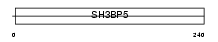
|
|||||
| Description | SH3 domain-binding protein 5 (SH3 domain-binding protein that preferentially associates with BTK). | |||||
|
AAK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.012418 (rank : 161) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.055445 (rank : 83) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BLNK_MOUSE
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.042679 (rank : 97) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QUN3, O88504 | Gene names | Blnk, Bash, Ly57, Slp65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (Slp-65) (Lymphocyte antigen 57). | |||||
|
CN092_MOUSE
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.017141 (rank : 149) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
EAF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.023178 (rank : 136) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96JC9, Q8IW10 | Gene names | EAF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELL-associated factor 1. | |||||
|
EAF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.025025 (rank : 132) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D4C5 | Gene names | Eaf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELL-associated factor 1. | |||||
|
GLI3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | -0.002936 (rank : 191) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61602 | Gene names | Gli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
GP178_HUMAN
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.020058 (rank : 143) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P2C4, Q5VTU1 | Gene names | GPR178, KIAA1423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integral membrane protein GPR178. | |||||
|
IF16_HUMAN
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.041727 (rank : 100) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
|
ILDR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.013097 (rank : 160) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86SU0, Q6ZP61, Q7Z578 | Gene names | ILDR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immunoglobulin-like domain-containing receptor 1 precursor. | |||||
|
M3K10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | -0.002343 (rank : 189) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02779, Q12761, Q14871 | Gene names | MAP3K10, MLK2, MST | |||
|
Domain Architecture |
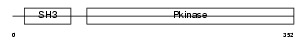
|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 10 (EC 2.7.11.25) (Mixed lineage kinase 2) (Protein kinase MST). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.064768 (rank : 67) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MAST3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.001903 (rank : 184) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1128 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O60307, Q7LDZ8, Q9UPI0 | Gene names | MAST3, KIAA0561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 3 (EC 2.7.11.1). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.047228 (rank : 95) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PK3CD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.004378 (rank : 182) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35904 | Gene names | Pik3cd | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PO6F2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.014707 (rank : 157) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
RPGF6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.007579 (rank : 175) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TEU7, Q8NI21, Q8TEU6, Q96PC1 | Gene names | RAPGEF6, PDZGEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 6 (PDZ domain-containing guanine nucleotide exchange factor 2) (PDZ-GEF2) (RA-GEF-2). | |||||
|
SALL2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | -0.000274 (rank : 187) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QX96 | Gene names | Sall2, Sal2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Spalt-like protein 2) (MSal-2). | |||||
|
UT14C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.008941 (rank : 170) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5TAP6, Q5FWG3, Q92555 | Gene names | UTP14C, KIAA0266 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog C. | |||||
|
ZFP91_HUMAN
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | -0.002582 (rank : 190) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96JP5, Q86V47, Q96JP4, Q96QA3 | Gene names | ZFP91 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 91 homolog (Zfp-91). | |||||
|
ZN207_HUMAN
|
||||||
| θ value | 6.88961 (rank : 155) | NC score | 0.027648 (rank : 122) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
AFAD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.012238 (rank : 163) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
APC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.025458 (rank : 131) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
CCNK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.017208 (rank : 148) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CU025_HUMAN
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.011055 (rank : 165) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y426, Q5R2V7, Q6AHX8, Q9NSE6 | Gene names | C21orf25, C21orf258 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf25 precursor. | |||||
|
DEN2A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.004496 (rank : 181) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C4S8, Q3TJU2, Q3TUG3, Q3UXA3 | Gene names | Dennd2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
FBXW7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | -0.001239 (rank : 188) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VBV4, Q8CCS5, Q8VHP4 | Gene names | Fbxw7, Fbw7, Fbwd6, Fbxw6 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 7 (F-box and WD-40 domain protein 7) (F-box protein FBW7) (F-box protein Fbxw6) (F-box-WD40 repeat protein 6) (SEL-10). | |||||
|
FYB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.033592 (rank : 112) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
GP152_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.014759 (rank : 155) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
H12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.042545 (rank : 98) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P16403 | Gene names | HIST1H1C, H1F2 | |||
|
Domain Architecture |
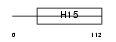
|
|||||
| Description | Histone H1.2 (Histone H1d). | |||||
|
HCN2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.018822 (rank : 145) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
K0690_MOUSE
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.008981 (rank : 169) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P5B0, Q7TMI5, Q80TU2 | Gene names | Kiaa0690 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0690. | |||||
|
KCNH7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.008258 (rank : 172) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
KCNK4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.005688 (rank : 180) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.025804 (rank : 128) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
NCKX1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.013715 (rank : 159) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.022167 (rank : 138) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.023200 (rank : 135) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.014984 (rank : 154) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
PARD3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.007789 (rank : 174) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TEW0, Q8TEW1, Q8TEW2, Q8TEW3, Q96K28, Q96RM6, Q96RM7, Q9BY57, Q9BY58, Q9HC48, Q9NWL4, Q9NYE6 | Gene names | PARD3, PAR3, PAR3A | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (CTCL tumor antigen se2- 5) (PAR3-alpha). | |||||
|
RFX1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 175) | NC score | 0.015635 (rank : 152) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P22670 | Gene names | RFX1 | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II regulatory factor RFX1 (RFX) (Enhancer factor C) (EF-C). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 8.99809 (rank : 176) | NC score | 0.008065 (rank : 173) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SYNPO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 177) | NC score | 0.031865 (rank : 115) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.050333 (rank : 90) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.052578 (rank : 85) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
H11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.051132 (rank : 89) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q02539 | Gene names | HIST1H1A, H1F1 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.1. | |||||
|
H15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.063161 (rank : 71) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P43276, Q9CRM8 | Gene names | Hist1h1b, H1f5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (H1 VAR.5) (H1b). | |||||
|
KI67_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.060370 (rank : 77) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.051292 (rank : 88) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
MDC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.068761 (rank : 62) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.056693 (rank : 81) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
RL1D1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.067506 (rank : 64) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.058857 (rank : 80) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.050050 (rank : 91) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
TARA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.062806 (rank : 72) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.064753 (rank : 68) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.062711 (rank : 74) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
SIA7A_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 177 | |
| SwissProt Accessions | Q9NSC7, Q6UW90, Q9NSC6 | Gene names | ST6GALNAC1, SIAT7A | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
SIA7A_MOUSE
|
||||||
| NC score | 0.953413 (rank : 2) | θ value | 2.32313e-181 (rank : 2) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9QZ39, Q9JJP5 | Gene names | St6galnac1, Siat7a | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
SIA7B_MOUSE
|
||||||
| NC score | 0.922086 (rank : 3) | θ value | 1.31681e-67 (rank : 3) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70277, Q3UWG0, Q9DC24 | Gene names | St6galnac2, Siat7, Siat7b | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 (EC 2.4.99.-) (GalNAc alpha-2,6-sialyltransferase II) (ST6GalNAc II) (Gal-beta-1,3-GalNAc alpha-2,6-sialyltransferase) (Sialyltransferase 7B). | |||||
|
SIA7B_HUMAN
|
||||||
| NC score | 0.922032 (rank : 4) | θ value | 6.53529e-67 (rank : 4) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UJ37, Q12971 | Gene names | ST6GALNAC2, SIAT7B, STHM | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 (EC 2.4.99.-) (GalNAc alpha-2,6-sialyltransferase II) (ST6GalNAc II) (Sialyltransferase 7B) (SThM). | |||||
|
SIA4A_HUMAN
|
||||||
| NC score | 0.768936 (rank : 5) | θ value | 2.7842e-17 (rank : 7) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q11201, O60677, Q9UN51 | Gene names | ST3GAL1, SIAT4, SIAT4A | |||
|
Domain Architecture |

|
|||||
| Description | CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3-sialyltransferase (EC 2.4.99.4) (Beta-galactoside alpha-2,3-sialyltransferase) (Alpha 2,3-ST) (Gal-NAc6S) (Gal-beta-1,3-GalNAc-alpha-2,3-sialyltransferase) (ST3GalIA) (ST3O) (ST3GalA.1) (SIAT4-A) (ST3Gal I) (SIATFL). | |||||
|
SIA4A_MOUSE
|
||||||
| NC score | 0.762936 (rank : 6) | θ value | 5.8054e-15 (rank : 15) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P54751, Q11202 | Gene names | St3gal1, Siat4, Siat4a | |||
|
Domain Architecture |

|
|||||
| Description | CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3-sialyltransferase (EC 2.4.99.4) (Beta-galactoside alpha-2,3-sialyltransferase) (Alpha 2,3-ST) (Gal-NAc6S) (Gal-beta-1,3-GalNAc-alpha-2,3-sialyltransferase) (ST3GalIA) (ST3O) (ST3GalA.1) (SIAT4-A) (ST3Gal I). | |||||
|
SIA4B_MOUSE
|
||||||
| NC score | 0.756627 (rank : 7) | θ value | 8.95645e-16 (rank : 8) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q11204 | Gene names | St3gal2, Siat4b, Siat5 | |||
|
Domain Architecture |

|
|||||
| Description | CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3-sialyltransferase (EC 2.4.99.-) (Beta-galactoside alpha-2,3-sialyltransferase) (Alpha 2,3-ST) (Gal-NAc6S) (Gal-beta-1,3-GalNAc-alpha-2,3-sialyltransferase) (ST3GalA.2) (SIAT4-B) (ST3Gal II). | |||||
|
SIA4B_HUMAN
|
||||||
| NC score | 0.756272 (rank : 8) | θ value | 2.60593e-15 (rank : 11) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q16842, O00654 | Gene names | ST3GAL2, SIAT4B | |||
|
Domain Architecture |

|
|||||
| Description | CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3-sialyltransferase (EC 2.4.99.-) (Beta-galactoside alpha-2,3-sialyltransferase) (Alpha 2,3-ST) (Gal-NAc6S) (Gal-beta-1,3-GalNAc-alpha-2,3-sialyltransferase) (ST3GalA.2) (SIAT4-B) (ST3Gal II). | |||||
|
SIAT6_MOUSE
|
||||||
| NC score | 0.750491 (rank : 9) | θ value | 2.27762e-19 (rank : 6) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97325 | Gene names | St3gal3, Siat3, Siat6 | |||
|
Domain Architecture |

|
|||||
| Description | CMP-N-acetylneuraminate-beta-1,4-galactoside alpha-2,3- sialyltransferase (EC 2.4.99.6) (N-acetyllactosaminide alpha-2,3- sialyltransferase) (Gal beta-1,3(4) GlcNAc alpha-2,3 sialyltransferase) (ST3N) (ST3GalIII) (Sialyltransferase 6). | |||||
|
SIAT6_HUMAN
|
||||||
| NC score | 0.747036 (rank : 10) | θ value | 4.58923e-20 (rank : 5) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q11203, Q86UR6, Q86UR7, Q86UR8, Q86UR9, Q86US0, Q86US1, Q86US2, Q8IX41, Q8IX42, Q8IX43, Q8IX44, Q8IX45, Q8IX46, Q8IX47, Q8IX48, Q8IX49, Q8IX50, Q8IX51, Q8IX52, Q8IX53, Q8IX54, Q8IX55, Q8IX56, Q8IX57, Q8IX58 | Gene names | ST3GAL3, SIAT6 | |||
|
Domain Architecture |

|
|||||
| Description | CMP-N-acetylneuraminate-beta-1,4-galactoside alpha-2,3- sialyltransferase (EC 2.4.99.6) (N-acetyllactosaminide alpha-2,3- sialyltransferase) (Gal beta-1,3(4) GlcNAc alpha-2,3 sialyltransferase) (ST3N) (ST3GalIII) (Sialyltransferase 6). | |||||
|
SIA4C_HUMAN
|
||||||
| NC score | 0.735622 (rank : 11) | θ value | 8.95645e-16 (rank : 9) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q11206, O60497, Q96QQ9 | Gene names | ST3GAL4, SIAT4C, STZ | |||
|
Domain Architecture |

|
|||||
| Description | CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3-sialyltransferase (EC 2.4.99.-) (Beta-galactoside alpha-2,3-sialyltransferase) (Alpha 2,3-sialyltransferase IV) (Alpha 2,3-ST) (Gal-NAc6S) (STZ) (SIAT4-C) (ST3Gal III) (SAT-3) (ST-4). | |||||
|
SIA4C_MOUSE
|
||||||
| NC score | 0.732414 (rank : 12) | θ value | 3.40345e-15 (rank : 13) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q91Y74 | Gene names | St3gal4, Siat4c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3-sialyltransferase (EC 2.4.99.-) (Beta-galactoside alpha-2,3-sialyltransferase) (Alpha 2,3-sialyltransferase IV) (Alpha 2,3-ST) (SIAT4-C). | |||||
|
SIAT9_MOUSE
|
||||||
| NC score | 0.728641 (rank : 13) | θ value | 7.58209e-15 (rank : 16) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O88829 | Gene names | St3gal5, Siat9 | |||
|
Domain Architecture |
|
|||||
| Description | Lactosylceramide alpha-2,3-sialyltransferase (EC 2.4.99.9) (CMP- NeuAc:lactosylceramide alpha-2,3-sialyltransferase) (Ganglioside GM3 synthase) (ST3Gal V) (Sialyltransferase 9). | |||||
|
SIA8C_HUMAN
|
||||||
| NC score | 0.721532 (rank : 14) | θ value | 4.44505e-15 (rank : 14) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O43173, Q9NS41 | Gene names | ST8SIA3, SIAT8C | |||
|
Domain Architecture |

|
|||||
| Description | Sia-alpha-2,3-Gal-beta-1,4-GlcNAc-R:alpha 2,8-sialyltransferase (EC 2.4.99.-) (ST8 alpha-N-acetyl-neuraminide alpha-2,8- sialyltransferase 3) (Alpha-2,8-sialyltransferase 8C) (Alpha-2,8- sialyltransferase III) (ST8Sia III). | |||||
|
SIAT9_HUMAN
|
||||||
| NC score | 0.718571 (rank : 15) | θ value | 4.16044e-13 (rank : 19) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UNP4, O94902, Q53QU1 | Gene names | ST3GAL5, SIAT9 | |||
|
Domain Architecture |
|
|||||
| Description | Lactosylceramide alpha-2,3-sialyltransferase (EC 2.4.99.9) (CMP- NeuAc:lactosylceramide alpha-2,3-sialyltransferase) (Ganglioside GM3 synthase) (ST3Gal V) (Sialyltransferase 9). | |||||
|
SIA8C_MOUSE
|
||||||
| NC score | 0.714446 (rank : 16) | θ value | 3.76295e-14 (rank : 18) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64689 | Gene names | St8sia3, Siat8c | |||
|
Domain Architecture |

|
|||||
| Description | Sia-alpha-2,3-Gal-beta-1,4-GlcNAc-R:alpha 2,8-sialyltransferase (EC 2.4.99.-) (ST8 alpha-N-acetyl-neuraminide alpha-2,8- sialyltransferase 3) (Alpha-2,8-sialyltransferase 8C) (Alpha-2,8- sialyltransferase III) (ST8Sia III). | |||||
|
SIA8B_MOUSE
|
||||||
| NC score | 0.702267 (rank : 17) | θ value | 1.99529e-15 (rank : 10) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O35696 | Gene names | St8sia2, Siat8b, Stx | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2,8-sialyltransferase 8B (EC 2.4.99.-) (ST8Sia II) (Sialyltransferase X) (STX) (Polysialic acid synthase). | |||||
|
SIA10_MOUSE
|
||||||
| NC score | 0.701770 (rank : 18) | θ value | 1.06045e-08 (rank : 28) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8VIB3, Q3TRN3, Q80UR7, Q9WVG2 | Gene names | St3gal6, Siat10 | |||
|
Domain Architecture |

|
|||||
| Description | Type 2 lactosamine alpha-2,3-sialyltransferase (EC 2.4.99.-) (CMP- NeuAc:beta-galactoside alpha-2,3-sialyltransferase VI) (ST3Gal VI) (Sialyltransferase 10). | |||||
|
SIA8D_HUMAN
|
||||||
| NC score | 0.697757 (rank : 19) | θ value | 2.0648e-12 (rank : 20) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q92187, Q92693 | Gene names | ST8SIA4, PST, PST1, SIAT8D | |||
|
Domain Architecture |

|
|||||
| Description | CMP-N-acetylneuraminate-poly-alpha-2,8-sialyltransferase (EC 2.4.99.-) (Alpha-2,8-sialyltransferase 8D) (ST8Sia IV) (Polysialyltransferase- 1). | |||||
|
SIA10_HUMAN
|
||||||
| NC score | 0.696998 (rank : 20) | θ value | 1.63604e-09 (rank : 25) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y274 | Gene names | ST3GAL6, SIAT10 | |||
|
Domain Architecture |

|
|||||
| Description | Type 2 lactosamine alpha-2,3-sialyltransferase (EC 2.4.99.-) (CMP- NeuAc:beta-galactoside alpha-2,3-sialyltransferase VI) (ST3Gal VI) (Sialyltransferase 10). | |||||
|
SIA8B_HUMAN
|
||||||
| NC score | 0.695796 (rank : 21) | θ value | 9.90251e-15 (rank : 17) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q92186, Q4VAZ0, Q92470, Q92746 | Gene names | ST8SIA2, SIAT8B, STX | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2,8-sialyltransferase 8B (EC 2.4.99.-) (ST8Sia II) (Sialyltransferase X) (STX). | |||||
|
SIA8D_MOUSE
|
||||||
| NC score | 0.695329 (rank : 22) | θ value | 2.69671e-12 (rank : 21) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64692 | Gene names | St8sia4, Pst, Siat8d | |||
|
Domain Architecture |

|
|||||
| Description | CMP-N-acetylneuraminate-poly-alpha-2,8-sialyltransferase (EC 2.4.99.-) (Alpha-2,8-sialyltransferase 8D) (ST8Sia IV) (Polysialyltransferase- 1). | |||||
|
SIA8F_MOUSE
|
||||||
| NC score | 0.680190 (rank : 23) | θ value | 2.60593e-15 (rank : 12) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K4T1 | Gene names | St8sia6, Siat8f | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2,8-sialyltransferase 8F (EC 2.4.99.-) (ST8Sia VI). | |||||
|
SIAT1_MOUSE
|
||||||
| NC score | 0.678229 (rank : 24) | θ value | 4.30538e-10 (rank : 24) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64685 | Gene names | St6gal1, Siat1 | |||
|
Domain Architecture |

|
|||||
| Description | CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,6-sialyltransferase (EC 2.4.99.1) (Beta-galactoside alpha-2,6-sialyltransferase) (Alpha 2,6-ST) (Sialyltransferase 1) (ST6Gal I). | |||||
|
SIAT1_HUMAN
|
||||||
| NC score | 0.670792 (rank : 25) | θ value | 1.38499e-08 (rank : 29) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P15907 | Gene names | ST6GAL1, SIAT1 | |||
|
Domain Architecture |

|
|||||
| Description | CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,6-sialyltransferase (EC 2.4.99.1) (Beta-galactoside alpha-2,6-sialyltransferase) (Alpha 2,6-ST) (Sialyltransferase 1) (ST6Gal I) (B-cell antigen CD75). | |||||
|
SIA8E_HUMAN
|
||||||
| NC score | 0.665413 (rank : 26) | θ value | 1.63604e-09 (rank : 26) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O15466 | Gene names | ST8SIA5, SIAT8E | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2,8-sialyltransferase 8E (EC 2.4.99.-) (ST8Sia V). | |||||
|
SIA8E_MOUSE
|
||||||
| NC score | 0.661794 (rank : 27) | θ value | 1.47974e-10 (rank : 23) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70126, P70127, P70128 | Gene names | St8sia5, Siat8e, St8siav | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2,8-sialyltransferase 8E (EC 2.4.99.-) (ST8Sia V). | |||||
|
SIA8F_HUMAN
|
||||||
| NC score | 0.660595 (rank : 28) | θ value | 7.84624e-12 (rank : 22) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P61647 | Gene names | ST8SIA6, SIAT8F | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-2,8-sialyltransferase 8F (EC 2.4.99.-) (ST8Sia VI). | |||||
|
SIA8A_HUMAN
|
||||||
| NC score | 0.642792 (rank : 29) | θ value | 4.76016e-09 (rank : 27) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q92185, Q93064 | Gene names | ST8SIA1, SIAT8, SIAT8A | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylneuraminide alpha-2,8-sialyltransferase (EC 2.4.99.8) (Ganglioside GD3 synthase) (Ganglioside GT3 synthase) (Alpha-2,8- sialyltransferase 8A) (ST8Sia I). | |||||
|
SIA8A_MOUSE
|
||||||
| NC score | 0.635238 (rank : 30) | θ value | 1.80886e-08 (rank : 30) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q64687 | Gene names | St8sia1, Siat8, Siat8a | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylneuraminide alpha-2,8-sialyltransferase (EC 2.4.99.8) (Ganglioside GD3 synthase) (Ganglioside GT3 synthase) (Alpha-2,8- sialyltransferase 8A) (ST8Sia I). | |||||
|
SIA7E_HUMAN
|
||||||
| NC score | 0.583018 (rank : 31) | θ value | 8.40245e-06 (rank : 34) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BVH7 | Gene names | ST6GALNAC5, SIAT7E | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 (EC 2.4.99.-) (GalNAc alpha-2,6-sialyltransferase V) (ST6GalNAc V) (GD1 alpha synthase) (Sialyltransferase 7E). | |||||
|
SIA7D_MOUSE
|
||||||
| NC score | 0.576137 (rank : 32) | θ value | 3.77169e-06 (rank : 32) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9R2B6, O88725, Q9JHP0, Q9QUP9, Q9R2B5 | Gene names | St6galnac4, Siat7d | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3-N-acetyl- galactosaminide alpha-2,6-sialyltransferase (EC 2.4.99.7) (NeuAc- alpha-2,3-Gal-beta-1,3-GalNAc-alpha-2,6-sialyltransferase) (ST6GalNAc IV) (Sialyltransferase 7D). | |||||
|
SIA7D_HUMAN
|
||||||
| NC score | 0.567811 (rank : 33) | θ value | 9.29e-05 (rank : 36) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H4F1, Q9NWU6, Q9UKU1, Q9ULB9, Q9Y3G3, Q9Y3G4 | Gene names | ST6GALNAC4, SIAT3C, SIAT7D | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3-N-acetyl- galactosaminide alpha-2,6-sialyltransferase (EC 2.4.99.7) (NeuAc- alpha-2,3-Gal-beta-1,3-GalNAc-alpha-2,6-sialyltransferase) (ST6GalNAc IV) (Sialyltransferase 7D) (Sialyltransferase 3C). | |||||
|
SIA7E_MOUSE
|
||||||
| NC score | 0.562344 (rank : 34) | θ value | 6.43352e-06 (rank : 33) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QYJ1, Q9R0K6 | Gene names | St6galnac5, Siat7e | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 (EC 2.4.99.-) (GalNAc alpha-2,6-sialyltransferase V) (ST6GalNAc V) (GD1 alpha synthase) (Sialyltransferase 7E). | |||||
|
SIA7C_MOUSE
|
||||||
| NC score | 0.530380 (rank : 35) | θ value | 1.69304e-06 (rank : 31) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9WUV2, Q9JHP5 | Gene names | St6galnac3, Siat7c | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 (EC 2.4.99.-) (GalNAc alpha-2,6-sialyltransferase III) (ST6GalNAc III) (Sialyltransferase 7C) (STY). | |||||
|
SIA7C_HUMAN
|
||||||
| NC score | 0.527705 (rank : 36) | θ value | 1.87187e-05 (rank : 35) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8NDV1, Q6UX29, Q8N259 | Gene names | ST6GALNAC3, SIAT7C | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 (EC 2.4.99.-) (GalNAc alpha-2,6-sialyltransferase III) (ST6GalNAc III) (Sialyltransferase 7C) (STY). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.114552 (rank : 37) | θ value | 0.000461057 (rank : 37) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.113401 (rank : 38) | θ value | 0.125558 (rank : 59) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.108166 (rank : 39) | θ value | 0.000786445 (rank : 38) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.107799 (rank : 40) | θ value | 0.00134147 (rank : 39) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.105807 (rank : 41) | θ value | 0.0330416 (rank : 48) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.104550 (rank : 42) | θ value | 0.279714 (rank : 62) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.091896 (rank : 43) | θ value | 0.00298849 (rank : 41) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.087491 (rank : 44) | θ value | 0.00298849 (rank : 42) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.084230 (rank : 45) | θ value | 0.0330416 (rank : 47) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.082215 (rank : 46) | θ value | 0.0961366 (rank : 57) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.081643 (rank : 47) | θ value | 0.0193708 (rank : 46) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
ATF2_MOUSE
|
||||||
| NC score | 0.080687 (rank : 48) | θ value | 0.00175202 (rank : 40) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P16951, Q64089, Q64090, Q64091 | Gene names | Atf2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (MXBP protein). | |||||
|
ATF2_HUMAN
|
||||||
| NC score | 0.079147 (rank : 49) | θ value | 0.00390308 (rank : 43) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P15336, Q13000 | Gene names | ATF2, CREB2, CREBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-2 (Activating transcription factor 2) (cAMP response element-binding protein CRE- BP1) (HB16). | |||||
|
CIZ1_HUMAN
|
||||||
| NC score | 0.077597 (rank : 50) | θ value | 0.0563607 (rank : 49) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.077018 (rank : 51) | θ value | 0.279714 (rank : 63) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.076586 (rank : 52) | θ value | 0.62314 (rank : 73) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.076574 (rank : 53) | θ value | 2.36792 (rank : 104) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.073943 (rank : 54) | θ value | 0.0563607 (rank : 52) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.073373 (rank : 55) | θ value | 0.0961366 (rank : 54) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.072830 (rank : 56) | θ value | 1.81305 (rank : 101) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.072735 (rank : 57) | θ value | 1.38821 (rank : 88) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
H13_MOUSE
|
||||||
| NC score | 0.072281 (rank : 58) | θ value | 0.125558 (rank : 58) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P43277, Q8C6M4 | Gene names | Hist1h1d, H1f3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (H1 VAR.4) (H1d). | |||||
|
PARM1_MOUSE
|
||||||
| NC score | 0.072164 (rank : 59) | θ value | 0.0961366 (rank : 55) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.069534 (rank : 60) | θ value | 0.0961366 (rank : 56) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.069495 (rank : 61) | θ value | 1.38821 (rank : 89) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.068761 (rank : 62) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.067779 (rank : 63) | θ value | 1.06291 (rank : 84) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
RL1D1_HUMAN
|
||||||
| NC score | 0.067506 (rank : 64) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.066875 (rank : 65) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.066843 (rank : 66) | θ value | 0.163984 (rank : 60) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.064768 (rank : 67) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.064753 (rank : 68) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
H14_HUMAN
|
||||||
| NC score | 0.064233 (rank : 69) | θ value | 0.0563607 (rank : 50) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P10412 | Gene names | HIST1H1E, H1F4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (Histone H1b). | |||||
|
PPRB_MOUSE
|
||||||
| NC score | 0.063935 (rank : 70) | θ value | 0.813845 (rank : 79) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
H15_MOUSE
|
||||||
| NC score | 0.063161 (rank : 71) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P43276, Q9CRM8 | Gene names | Hist1h1b, H1f5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (H1 VAR.5) (H1b). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.062806 (rank : 72) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
PPRB_HUMAN
|
||||||
| NC score | 0.062726 (rank : 73) | θ value | 1.38821 (rank : 94) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.062711 (rank : 74) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.061887 (rank : 75) | θ value | 0.365318 (rank : 65) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.061151 (rank : 76) | θ value | 0.813845 (rank : 75) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.060370 (rank : 77) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.059571 (rank : 78) | θ value | 0.62314 (rank : 71) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
H15_HUMAN
|
||||||
| NC score | 0.059457 (rank : 79) | θ value | 4.03905 (rank : 123) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P16401, Q14529 | Gene names | HIST1H1B, H1F5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (Histone H1a). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.058857 (rank : 80) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.056693 (rank : 81) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.056112 (rank : 82) | θ value | 0.62314 (rank : 70) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.055445 (rank : 83) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
H14_MOUSE
|
||||||
| NC score | 0.053692 (rank : 84) | θ value | 1.38821 (rank : 92) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P43274 | Gene names | Hist1h1e, H1f4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (H1 VAR.2) (H1e). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.052578 (rank : 85) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.052373 (rank : 86) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
ATF7_HUMAN
|
||||||
| NC score | 0.052280 (rank : 87) | θ value | 3.0926 (rank : 110) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.051292 (rank : 88) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
H11_HUMAN
|
||||||
| NC score | 0.051132 (rank : 89) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q02539 | Gene names | HIST1H1A, H1F1 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.1. | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.050333 (rank : 90) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.050050 (rank : 91) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
SAM14_MOUSE
|
||||||
| NC score | 0.048877 (rank : 92) | θ value | 0.47712 (rank : 69) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K070, Q5SWB7, Q8BHE2, Q8C8N5 | Gene names | Samd14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 14. | |||||
|
SRP72_HUMAN
|
||||||
| NC score | 0.047944 (rank : 93) | θ value | 0.279714 (rank : 64) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O76094 | Gene names | SRP72 | |||
|
Domain Architecture |

|
|||||
| Description | Signal recognition particle 72 kDa protein (SRP72). | |||||
|
BLNK_HUMAN
|
||||||
| NC score | 0.047596 (rank : 94) | θ value | 0.47712 (rank : 66) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WV28, O75498, O75499 | Gene names | BLNK, BASH, SLP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (SLP-65). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.047228 (rank : 95) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
UTS2B_HUMAN
|
||||||
| NC score | 0.046522 (rank : 96) | θ value | 3.0926 (rank : 119) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q765I0 | Gene names | UTS2D, URP, UTS2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Urotensin-2B precursor (Urotensin-IIB) (U-IIB) (UIIB) (Urotensin II- related peptide) (Urotensin 2 domain-containing protein). | |||||
|
BLNK_MOUSE
|
||||||
| NC score | 0.042679 (rank : 97) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QUN3, O88504 | Gene names | Blnk, Bash, Ly57, Slp65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (Slp-65) (Lymphocyte antigen 57). | |||||
|
H12_HUMAN
|
||||||
| NC score | 0.042545 (rank : 98) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P16403 | Gene names | HIST1H1C, H1F2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.2 (Histone H1d). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.042127 (rank : 99) | θ value | 0.0563607 (rank : 53) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
IF16_HUMAN
|
||||||
| NC score | 0.041727 (rank : 100) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q16666, Q9UH78 | Gene names | IFI16, IFNGIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-interferon-inducible protein Ifi-16 (Interferon-inducible myeloid differentiation transcriptional activator) (IFI 16). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.041291 (rank : 101) | θ value | 0.0113563 (rank : 44) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SAM14_HUMAN
|
||||||
| NC score | 0.040661 (rank : 102) | θ value | 2.36792 (rank : 105) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IZD0, Q8N2X0 | Gene names | SAMD14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 14. | |||||
|
CF010_HUMAN
|
||||||
| NC score | 0.040448 (rank : 103) | θ value | 1.06291 (rank : 81) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.039817 (rank : 104) | θ value | 0.21417 (rank : 61) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.039441 (rank : 105) | θ value | 2.36792 (rank : 106) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
HUCE1_HUMAN
|
||||||
| NC score | 0.037324 (rank : 106) | θ value | 0.47712 (rank : 67) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43159, Q7KZ78, Q9BVM6 | Gene names | KIAA0409 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerebral protein 1. | |||||
|
SELPL_HUMAN
|
||||||
| NC score | 0.036676 (rank : 107) | θ value | 1.81305 (rank : 100) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14242, Q12775 | Gene names | SELPLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand) (CD162 antigen). | |||||
|
PAF49_MOUSE
|
||||||
| NC score | 0.034401 (rank : 108) | θ value | 0.813845 (rank : 77) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q76KJ5, Q8K0Y9 | Gene names | Cd3eap, Ase1, Paf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein). | |||||
|
TGON2_MOUSE
|
||||||
| NC score | 0.034269 (rank : 109) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
REXO1_HUMAN
|
||||||
| NC score | 0.034056 (rank : 110) | θ value | 4.03905 (rank : 126) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
MCES_HUMAN
|
||||||
| NC score | 0.033959 (rank : 111) | θ value | 1.06291 (rank : 82) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43148, O94996, Q9UIJ9 | Gene names | RNMT, KIAA0398 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA cap guanine-N7 methyltransferase (EC 2.1.1.56) (mRNA (guanine- N(7)-)-methyltransferase) (RG7MT1) (mRNA cap methyltransferase) (hcm1p) (hCMT1) (hMet). | |||||
|
FYB_HUMAN
|
||||||
| NC score | 0.033592 (rank : 112) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.032839 (rank : 113) | θ value | 1.81305 (rank : 98) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.032058 (rank : 114) | θ value | 1.38821 (rank : 87) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
SYNPO_HUMAN
|
||||||
| NC score | 0.031865 (rank : 115) | θ value | 8.99809 (rank : 177) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
SYN1_MOUSE
|
||||||
| NC score | 0.031011 (rank : 116) | θ value | 1.06291 (rank : 86) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
SHAN2_HUMAN
|
||||||
| NC score | 0.030864 (rank : 117) | θ value | 3.0926 (rank : 116) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UPX8, Q3Y8G9, Q52LK2, Q9UKP1 | Gene names | SHANK2, KIAA1022 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2). | |||||
|
TTF2_MOUSE
|
||||||
| NC score | 0.030699 (rank : 118) | θ value | 2.36792 (rank : 109) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5NC05, Q5M924 | Gene names | Ttf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2). | |||||
|
WBP4_HUMAN
|
||||||
| NC score | 0.029984 (rank : 119) | θ value | 1.38821 (rank : 95) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75554 | Gene names | WBP4, FBP21, FNBP21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 4 (WBP-4) (Formin-binding protein 21). | |||||
|
FOS_HUMAN
|
||||||
| NC score | 0.028886 (rank : 120) | θ value | 1.38821 (rank : 91) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P01100, P18849 | Gene names | FOS, G0S7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene protein c-fos (Cellular oncogene fos) (G0/G1 switch regulatory protein 7). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.028822 (rank : 121) | θ value | 1.81305 (rank : 97) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
ZN207_HUMAN
|
||||||
| NC score | 0.027648 (rank : 122) | θ value | 6.88961 (rank : 155) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.027490 (rank : 123) | θ value | 0.813845 (rank : 78) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
DEK_HUMAN
|
||||||
| NC score | 0.027280 (rank : 124) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35659, Q5TGV4, Q5TGV5 | Gene names | DEK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
MILK2_HUMAN
|
||||||
| NC score | 0.026393 (rank : 125) | θ value | 1.06291 (rank : 83) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
SHAN2_MOUSE
|
||||||
| NC score | 0.025964 (rank : 126) | θ value | 3.0926 (rank : 117) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80Z38, Q3UTK4, Q5DU07 | Gene names | Shank2, Kiaa1022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2) (Cortactin- binding protein 1) (CortBP1). | |||||
|
SMAL1_HUMAN
|
||||||
| NC score | 0.025821 (rank : 127) | θ value | 0.62314 (rank : 72) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.025804 (rank : 128) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
PKHA4_HUMAN
|
||||||
| NC score | 0.025782 (rank : 129) | θ value | 1.06291 (rank : 85) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H4M7, Q8N4M8, Q8N658 | Gene names | PLEKHA4, PEPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 4 (Phosphoinositol 3-phosphate-binding protein 1) (PEPP-1). | |||||
|
ADDB_HUMAN
|
||||||
| NC score | 0.025573 (rank : 130) | θ value | 0.813845 (rank : 74) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
|
APC_HUMAN
|
||||||
| NC score | 0.025458 (rank : 131) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
EAF1_MOUSE
|
||||||
| NC score | 0.025025 (rank : 132) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D4C5 | Gene names | Eaf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELL-associated factor 1. | |||||
|
RANB3_HUMAN
|
||||||
| NC score | 0.024143 (rank : 133) | θ value | 3.0926 (rank : 115) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H6Z4, O60405, O75759, O75760, Q9BT47, Q9UG74 | Gene names | RANBP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 3 (RanBP3). | |||||
|
HCN1_MOUSE
|
||||||
| NC score | 0.023298 (rank : 134) | θ value | 0.0563607 (rank : 51) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.023200 (rank : 135) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
EAF1_HUMAN
|
||||||
| NC score | 0.023178 (rank : 136) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96JC9, Q8IW10 | Gene names | EAF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELL-associated factor 1. | |||||
|
CCD71_MOUSE
|
||||||
| NC score | 0.022272 (rank : 137) | θ value | 3.0926 (rank : 112) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VEG0, Q3UDX1 | Gene names | Ccdc71 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 71. | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.022167 (rank : 138) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
IFI4_MOUSE
|
||||||
| NC score | 0.021300 (rank : 139) | θ value | 2.36792 (rank : 103) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P15092 | Gene names | Ifi204 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-activable protein 204 (Ifi-204) (Interferon-inducible protein p204). | |||||
|
IFI5_MOUSE
|
||||||
| NC score | 0.020787 (rank : 140) | θ value | 4.03905 (rank : 124) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08619 | Gene names | Ifi205 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-activable protein 205 (IFI-205) (D3 protein). | |||||
|
IL16_HUMAN
|
||||||
| NC score | 0.020591 (rank : 141) | θ value | 0.47712 (rank : 68) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14005, Q16435, Q9UP18 | Gene names | IL16 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-16 precursor (IL-16) (Lymphocyte chemoattractant factor) (LCF). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.020415 (rank : 142) | θ value | 1.81305 (rank : 99) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
GP178_HUMAN
|
||||||
| NC score | 0.020058 (rank : 143) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P2C4, Q5VTU1 | Gene names | GPR178, KIAA1423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integral membrane protein GPR178. | |||||
|
DJC12_HUMAN
|
||||||
| NC score | 0.019688 (rank : 144) | θ value | 1.38821 (rank : 90) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKB3, Q9UKB2 | Gene names | DNAJC12, JDP1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily C member 12 (J domain-containing protein 1). | |||||
|
HCN2_MOUSE
|
||||||
| NC score | 0.018822 (rank : 145) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
AAK1_HUMAN
|
||||||
| NC score | 0.018637 (rank : 146) | θ value | 0.0193708 (rank : 45) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1077 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q2M2I8, Q4ZFZ3, Q53RX6, Q9UPV4 | Gene names | AAK1, KIAA1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.018543 (rank : 147) | θ value | 0.813845 (rank : 76) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.017208 (rank : 148) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CN092_MOUSE
|
||||||
| NC score | 0.017141 (rank : 149) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BU11, Q80UI2, Q99PN9, Q9CS16 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Epidermal Langerhans cell protein LCP1. | |||||
|
SCG3_MOUSE
|
||||||
| NC score | 0.016545 (rank : 150) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P47867 | Gene names | Scg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretogranin-3 precursor (Secretogranin III) (SgIII). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.015797 (rank : 151) | θ value | 3.0926 (rank : 114) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
RFX1_HUMAN
|
||||||
| NC score | 0.015635 (rank : 152) | θ value | 8.99809 (rank : 175) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P22670 | Gene names | RFX1 | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II regulatory factor RFX1 (RFX) (Enhancer factor C) (EF-C). | |||||
|
BXDC2_HUMAN
|
||||||
| NC score | 0.015102 (rank : 153) | θ value | 5.27518 (rank : 127) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TDN6, Q8N453, Q96DH1 | Gene names | BXDC2, BRIX | |||
|
Domain Architecture |

|
|||||
| Description | Brix domain-containing protein 2 (Ribosome biogenesis protein Brix). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.014984 (rank : 154) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
GP152_HUMAN
|
||||||
| NC score | 0.014759 (rank : 155) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
SCEL_MOUSE
|
||||||
| NC score | 0.014711 (rank : 156) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9EQG3, Q9CTT9 | Gene names | Scel | |||
|
Domain Architecture |

|
|||||
| Description | Sciellin. | |||||
|
PO6F2_MOUSE
|
||||||
| NC score | 0.014707 (rank : 157) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
MSH6_MOUSE
|
||||||
| NC score | 0.014523 (rank : 158) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54276, O54710 | Gene names | Msh6, Gtmbp | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
NCKX1_HUMAN
|
||||||
| NC score | 0.013715 (rank : 159) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
ILDR1_HUMAN
|
||||||
| NC score | 0.013097 (rank : 160) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86SU0, Q6ZP61, Q7Z578 | Gene names | ILDR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immunoglobulin-like domain-containing receptor 1 precursor. | |||||
|
AAK1_MOUSE
|
||||||
| NC score | 0.012418 (rank : 161) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q3UHJ0, Q3TY53, Q6ZPZ6, Q80XP6 | Gene names | Aak1, Kiaa1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
MAGD1_HUMAN
|
||||||
| NC score | 0.012305 (rank : 162) | θ value | 1.38821 (rank : 93) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y5V3, Q5VSH6, Q8IZ84, Q8WY92, Q9H352, Q9HBT4, Q9UF36 | Gene names | MAGED1, NRAGE | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (MAGE tumor antigen CCF). | |||||
|
AFAD_HUMAN
|
||||||
| NC score | 0.012238 (rank : 163) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
SSH2_HUMAN
|
||||||
| NC score | 0.012235 (rank : 164) | θ value | 3.0926 (rank : 118) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q76I76, Q8TDB5, Q8WYL1, Q8WYL2, Q96F40, Q96H36, Q9C0D8 | Gene names | SSH2, KIAA1725, SSH2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 2 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-2L) (hSSH-2L). | |||||
|
CU025_HUMAN
|
||||||
| NC score | 0.011055 (rank : 165) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y426, Q5R2V7, Q6AHX8, Q9NSE6 | Gene names | C21orf25, C21orf258 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C21orf25 precursor. | |||||
|
DZIP3_HUMAN
|
||||||
| NC score | 0.009946 (rank : 166) | θ value | 3.0926 (rank : 113) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86Y13, O75162, Q6P3R9, Q6PH82, Q86Y14, Q86Y15, Q86Y16, Q8IWI0, Q96RS9 | Gene names | DZIP3, KIAA0675 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein DZIP3 (EC 6.3.2.-) (DAZ-interacting protein 3) (RNA-binding ubiquitin ligase of 138 kDa) (hRUL138). | |||||
|
SMC3_MOUSE
|
||||||
| NC score | 0.009892 (rank : 167) | θ value | 2.36792 (rank : 108) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
SMC3_HUMAN
|
||||||
| NC score | 0.009879 (rank : 168) | θ value | 2.36792 (rank : 107) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
K0690_MOUSE
|
||||||
| NC score | 0.008981 (rank : 169) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P5B0, Q7TMI5, Q80TU2 | Gene names | Kiaa0690 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0690. | |||||
|
UT14C_HUMAN
|
||||||
| NC score | 0.008941 (rank : 170) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5TAP6, Q5FWG3, Q92555 | Gene names | UTP14C, KIAA0266 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog C. | |||||
|
UBP51_HUMAN
|
||||||
| NC score | 0.008829 (rank : 171) | θ value | 1.81305 (rank : 102) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q70EK9, Q8IWJ8 | Gene names | USP51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 51 (EC 3.1.2.15) (Ubiquitin thioesterase 51) (Ubiquitin-specific-processing protease 51) (Deubiquitinating enzyme 51). | |||||
|
KCNH7_HUMAN
|
||||||
| NC score | 0.008258 (rank : 172) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.008065 (rank : 173) | θ value | 8.99809 (rank : 176) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
PARD3_HUMAN
|
||||||
| NC score | 0.007789 (rank : 174) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TEW0, Q8TEW1, Q8TEW2, Q8TEW3, Q96K28, Q96RM6, Q96RM7, Q9BY57, Q9BY58, Q9HC48, Q9NWL4, Q9NYE6 | Gene names | PARD3, PAR3, PAR3A | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (CTCL tumor antigen se2- 5) (PAR3-alpha). | |||||
|
RPGF6_HUMAN
|
||||||
| NC score | 0.007579 (rank : 175) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TEU7, Q8NI21, Q8TEU6, Q96PC1 | Gene names | RAPGEF6, PDZGEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 6 (PDZ domain-containing guanine nucleotide exchange factor 2) (PDZ-GEF2) (RA-GEF-2). | |||||
|
3BP5_HUMAN
|
||||||
| NC score | 0.007238 (rank : 176) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60239 | Gene names | SH3BP5, SAB | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 5 (SH3 domain-binding protein that preferentially associates with BTK). | |||||
|
DAF_HUMAN
|
||||||
| NC score | 0.006439 (rank : 177) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08174, P09679, P78361 | Gene names | CD55, CR, DAF | |||
|
Domain Architecture |

|
|||||
| Description | Complement decay-accelerating factor precursor (CD55 antigen). | |||||
|
CAMKV_HUMAN
|
||||||
| NC score | 0.006313 (rank : 178) | θ value | 3.0926 (rank : 111) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
ZN516_MOUSE
|
||||||
| NC score | 0.006103 (rank : 179) | θ value | 0.813845 (rank : 80) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
KCNK4_HUMAN
|
||||||
| NC score | 0.005688 (rank : 180) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
DEN2A_MOUSE
|
||||||
| NC score | 0.004496 (rank : 181) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C4S8, Q3TJU2, Q3TUG3, Q3UXA3 | Gene names | Dennd2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
PK3CD_MOUSE
|
||||||
| NC score | 0.004378 (rank : 182) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35904 | Gene names | Pik3cd | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
CD2L5_MOUSE
|
||||||
| NC score | 0.002367 (rank : 183) | θ value | 1.81305 (rank : 96) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1011 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q69ZA1, Q80V11, Q8BZG1, Q8K0A4 | Gene names | Cdc2l5, Kiaa1791 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 5 (EC 2.7.11.22) (CDC2- related protein kinase 5). | |||||
|
MAST3_HUMAN
|
||||||
| NC score | 0.001903 (rank : 184) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1128 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O60307, Q7LDZ8, Q9UPI0 | Gene names | MAST3, KIAA0561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 3 (EC 2.7.11.1). | |||||
|
FOG2_HUMAN
|
||||||
| NC score | 0.001875 (rank : 185) | θ value | 4.03905 (rank : 122) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 731 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WW38, Q9NPL7, Q9NPS4, Q9UNI5 | Gene names | ZFPM2, FOG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM2 (Zinc finger protein multitype 2) (Friend of GATA protein 2) (FOG-2) (hFOG-2). | |||||
|
KSR1_HUMAN
|
||||||
| NC score | -0.000221 (rank : 186) | θ value | 4.03905 (rank : 125) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IVT5, Q13476 | Gene names | KSR1, KSR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras). | |||||
|
SALL2_MOUSE
|
||||||
| NC score | -0.000274 (rank : 187) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QX96 | Gene names | Sall2, Sal2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Spalt-like protein 2) (MSal-2). | |||||
|
FBXW7_MOUSE
|
||||||
| NC score | -0.001239 (rank : 188) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VBV4, Q8CCS5, Q8VHP4 | Gene names | Fbxw7, Fbw7, Fbwd6, Fbxw6 | |||
|
Domain Architecture |

|
|||||
| Description | F-box/WD repeat protein 7 (F-box and WD-40 domain protein 7) (F-box protein FBW7) (F-box protein Fbxw6) (F-box-WD40 repeat protein 6) (SEL-10). | |||||
|
M3K10_HUMAN
|
||||||
| NC score | -0.002343 (rank : 189) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02779, Q12761, Q14871 | Gene names | MAP3K10, MLK2, MST | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 10 (EC 2.7.11.25) (Mixed lineage kinase 2) (Protein kinase MST). | |||||
|
ZFP91_HUMAN
|
||||||
| NC score | -0.002582 (rank : 190) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96JP5, Q86V47, Q96JP4, Q96QA3 | Gene names | ZFP91 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 91 homolog (Zfp-91). | |||||
|
GLI3_MOUSE
|
||||||
| NC score | -0.002936 (rank : 191) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61602 | Gene names | Gli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||