Please be patient as the page loads
|
KCNK4_HUMAN
|
||||||
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
KCNK4_HUMAN
|
||||||
| θ value | 1.56915e-153 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
KCNK4_MOUSE
|
||||||
| θ value | 3.28708e-135 (rank : 2) | NC score | 0.984754 (rank : 2) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O88454 | Gene names | Kcnk4, Traak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK). | |||||
|
KCNK2_MOUSE
|
||||||
| θ value | 5.15425e-80 (rank : 3) | NC score | 0.965325 (rank : 4) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P97438 | Gene names | Kcnk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (Two-pore potassium channel TPKC1) (TREK-1 K(+) channel subunit). | |||||
|
KCNK2_HUMAN
|
||||||
| θ value | 9.7205e-79 (rank : 4) | NC score | 0.965343 (rank : 3) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O95069, Q9UNE3 | Gene names | KCNK2, TREK, TREK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (TREK-1 K(+) channel subunit) (Two-pore potassium channel TPKC1). | |||||
|
KCNKA_HUMAN
|
||||||
| θ value | 2.73936e-73 (rank : 5) | NC score | 0.954537 (rank : 5) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P57789, Q8TDK7, Q8TDK8, Q9HB59 | Gene names | KCNK10, TREK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 10 (Outward rectifying potassium channel protein TREK-2) (TREK-2 K(+) channel subunit). | |||||
|
KCNK5_HUMAN
|
||||||
| θ value | 1.16164e-39 (rank : 6) | NC score | 0.930460 (rank : 7) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O95279 | Gene names | KCNK5, TASK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 5 (Acid-sensitive potassium channel protein TASK-2) (TWIK-related acid-sensitive K(+) channel 2). | |||||
|
KCNKG_HUMAN
|
||||||
| θ value | 2.19075e-38 (rank : 7) | NC score | 0.944972 (rank : 6) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q96T55, Q5TCF3, Q9H591 | Gene names | KCNK16, TALK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 16 (TWIK-related alkaline pH- activated K(+) channel 1) (2P domain potassium channel Talk-1). | |||||
|
KCNKH_HUMAN
|
||||||
| θ value | 4.27553e-34 (rank : 8) | NC score | 0.923935 (rank : 8) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96T54, Q5TCF4, Q8TAW4, Q9BXD1, Q9H592 | Gene names | KCNK17, TALK2, TASK4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 17 (TWIK-related alkaline pH- activated K(+) channel 2) (2P domain potassium channel Talk-2) (TWIK- related acid-sensitive K(+) channel 4) (TASK-4). | |||||
|
KCNK6_HUMAN
|
||||||
| θ value | 1.16434e-31 (rank : 9) | NC score | 0.900880 (rank : 9) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
KCNK1_HUMAN
|
||||||
| θ value | 4.57862e-28 (rank : 10) | NC score | 0.882945 (rank : 10) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O00180, Q13307 | Gene names | KCNK1, HOHO1, KCNO1, TWIK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1) (Potassium channel KCNO1). | |||||
|
KCNK1_MOUSE
|
||||||
| θ value | 1.92365e-26 (rank : 11) | NC score | 0.880561 (rank : 11) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O08581 | Gene names | Kcnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1). | |||||
|
KCNK9_MOUSE
|
||||||
| θ value | 1.92365e-26 (rank : 12) | NC score | 0.857268 (rank : 12) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q3LS21 | Gene names | Kcnk9, Task3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3). | |||||
|
KCNK9_HUMAN
|
||||||
| θ value | 1.05554e-24 (rank : 13) | NC score | 0.852204 (rank : 14) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NPC2, Q2M290, Q540F2 | Gene names | KCNK9, TASK3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3) (Two pore potassium channel KT3.2). | |||||
|
KCNK3_MOUSE
|
||||||
| θ value | 1.80048e-24 (rank : 14) | NC score | 0.847984 (rank : 16) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O35111, O35163 | Gene names | Kcnk3, Ctbak, Task, Task1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Cardiac two-pore background K(+) channel) (cTBAK-1) (Two pore potassium channel KT3.1). | |||||
|
KCNK3_HUMAN
|
||||||
| θ value | 9.87957e-23 (rank : 15) | NC score | 0.848987 (rank : 15) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O14649 | Gene names | KCNK3, TASK, TASK1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Two pore potassium channel KT3.1). | |||||
|
KCNKF_HUMAN
|
||||||
| θ value | 1.5773e-20 (rank : 16) | NC score | 0.853435 (rank : 13) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9H427, Q9HBC8 | Gene names | KCNK15, TASK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 15 (Acid-sensitive potassium channel protein TASK-5) (TWIK-related acid-sensitive K(+) channel 5) (Two pore potassium channel KT3.3). | |||||
|
KCNKD_MOUSE
|
||||||
| θ value | 6.20254e-17 (rank : 17) | NC score | 0.794823 (rank : 20) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8R1P5 | Gene names | Kcnk13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCNKD_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 18) | NC score | 0.786144 (rank : 21) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HB14, Q96E79 | Gene names | KCNK13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCNK7_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 19) | NC score | 0.837063 (rank : 17) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z2T1, Q9QXY0, Q9QYE8, Q9R1V1, Q9R242 | Gene names | Kcnk7, Dpkch3, Kcnk6, Kcnk8, Knot1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7 (Putative potassium channel DP3) (Double-pore K(+) channel 3) (Neuromuscular two p domain potassium channel). | |||||
|
KCNKC_HUMAN
|
||||||
| θ value | 8.95645e-16 (rank : 20) | NC score | 0.800163 (rank : 19) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HB15 | Gene names | KCNK12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 12 (Tandem pore domain halothane- inhibited potassium channel 2) (THIK-2). | |||||
|
KCNK7_HUMAN
|
||||||
| θ value | 6.41864e-14 (rank : 21) | NC score | 0.821433 (rank : 18) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y2U2, Q9Y2U3, Q9Y2U4 | Gene names | KCNK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7. | |||||
|
KCMA1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 22) | NC score | 0.200495 (rank : 25) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q12791, Q12886, Q12917, Q12921, Q12960, Q13150, Q96LG8, Q9UBB0, Q9UQK6 | Gene names | KCNMA1, KCNMA | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (hSlo). | |||||
|
KCMA1_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 23) | NC score | 0.200731 (rank : 24) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q08460, Q64703, Q8VHF1, Q9R196 | Gene names | Kcnma1, Kcnma | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (mSlo) (mSlo1). | |||||
|
KCNH5_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 24) | NC score | 0.206307 (rank : 22) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
KCNH5_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 25) | NC score | 0.205968 (rank : 23) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q920E3, Q8C035 | Gene names | Kcnh5, Eag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (Eag2). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 26) | NC score | 0.038098 (rank : 110) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
UBP6_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 27) | NC score | 0.023478 (rank : 119) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35125, Q15634, Q86WP6, Q8IWT4 | Gene names | USP6, TRE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 6 (EC 3.1.2.15) (Ubiquitin thioesterase 6) (Ubiquitin-specific-processing protease 6) (Deubiquitinating enzyme 6) (Proto-oncogene TRE-2). | |||||
|
KCNH1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 28) | NC score | 0.197175 (rank : 27) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O95259, O76035 | Gene names | KCNH1, EAG, EAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (hEAG1) (h-eag). | |||||
|
KCNH1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 29) | NC score | 0.197514 (rank : 26) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
KCNH3_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 30) | NC score | 0.168936 (rank : 32) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
COA2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 31) | NC score | 0.030386 (rank : 114) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00763, Q16852 | Gene names | ACACB, ACC2, ACCB | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 2 (EC 6.4.1.2) (ACC-beta) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
KCNH4_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 32) | NC score | 0.159814 (rank : 38) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
KCNH8_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 33) | NC score | 0.172892 (rank : 30) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96L42 | Gene names | KCNH8 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 8 (Voltage-gated potassium channel subunit Kv12.1) (Ether-a-go-go-like potassium channel 3) (ELK channel 3) (ELK3) (ELK1) (hElk1). | |||||
|
CG1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 34) | NC score | 0.051077 (rank : 105) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 35) | NC score | 0.021845 (rank : 121) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
NRAM1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 36) | NC score | 0.059047 (rank : 87) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49279 | Gene names | SLC11A1, NRAMP, NRAMP1 | |||
|
Domain Architecture |
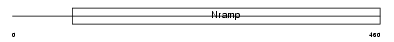
|
|||||
| Description | Natural resistance-associated macrophage protein 1 (NRAMP 1). | |||||
|
IF4B_HUMAN
|
||||||
| θ value | 0.21417 (rank : 37) | NC score | 0.037155 (rank : 111) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P23588 | Gene names | EIF4B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
KCNH3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 38) | NC score | 0.166641 (rank : 35) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 39) | NC score | 0.030647 (rank : 113) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
KCND2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 40) | NC score | 0.110471 (rank : 42) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 41) | NC score | 0.110496 (rank : 41) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCNH7_HUMAN
|
||||||
| θ value | 0.279714 (rank : 42) | NC score | 0.168897 (rank : 33) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
KCNH7_MOUSE
|
||||||
| θ value | 0.279714 (rank : 43) | NC score | 0.167222 (rank : 34) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
KCNQ4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 44) | NC score | 0.124746 (rank : 39) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
PCD12_MOUSE
|
||||||
| θ value | 0.279714 (rank : 45) | NC score | 0.005103 (rank : 146) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O55134 | Gene names | Pcdh12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-12 precursor (Vascular cadherin-2) (Vascular endothelial cadherin-2) (VE-cadherin-2) (VE-cad-2). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 46) | NC score | 0.020594 (rank : 122) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
KCNH6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 47) | NC score | 0.170444 (rank : 31) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
KCNH2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 48) | NC score | 0.163786 (rank : 36) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
KCNH2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 49) | NC score | 0.163263 (rank : 37) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.47712 (rank : 50) | NC score | 0.008849 (rank : 142) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NRAM1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 51) | NC score | 0.058175 (rank : 89) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P41251 | Gene names | Slc11a1, Bcg, Ity, Lsh, Nramp1 | |||
|
Domain Architecture |
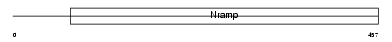
|
|||||
| Description | Natural resistance-associated macrophage protein 1 (NRAMP 1). | |||||
|
KCND3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.109882 (rank : 44) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 53) | NC score | 0.109901 (rank : 43) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
ATX7_MOUSE
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.026187 (rank : 118) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
KCND1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.110788 (rank : 40) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCNQ5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.105899 (rank : 46) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 57) | NC score | 0.007521 (rank : 143) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
DOK1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 58) | NC score | 0.018359 (rank : 124) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97465, Q9R213 | Gene names | Dok1, Dok | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 1 (Downstream of tyrosine kinase 1) (p62(dok)). | |||||
|
AL2S4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.023462 (rank : 120) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3V0J1, Q3TIS2 | Gene names | Als2cr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 4 protein homolog. | |||||
|
KCNQ3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.099153 (rank : 49) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNT1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.182189 (rank : 28) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5JUK3, Q9P2C5 | Gene names | KCNT1, KIAA1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1 (KCa4.1). | |||||
|
KCNT1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.173019 (rank : 29) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6ZPR4, Q8C3E7 | Gene names | Kcnt1, Kiaa1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1. | |||||
|
YIF1A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.030269 (rank : 115) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95070, Q96G83, Q9BVD0 | Gene names | YIF1A, 54TM, HYIF1P, YIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein YIF1A (YIP1-interacting factor homolog A) (54TMp). | |||||
|
KCNN4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.096141 (rank : 52) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O15554 | Gene names | KCNN4, IK1, IKCA1, KCA4, SK4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1) (IKCa1) (Putative Gardos channel). | |||||
|
KCNQ3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.098512 (rank : 50) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.014113 (rank : 131) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.016161 (rank : 129) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.028316 (rank : 116) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.009140 (rank : 140) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
KCNC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.072822 (rank : 69) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.072854 (rank : 68) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.075841 (rank : 61) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNQ2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.102554 (rank : 47) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.101994 (rank : 48) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
NRAM2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.050524 (rank : 106) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49281, O43288, O60932, Q96J35 | Gene names | SLC11A2, NRAMP2 | |||
|
Domain Architecture |
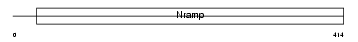
|
|||||
| Description | Natural resistance-associated macrophage protein 2 (NRAMP 2) (Divalent metal transporter 1) (DMT1). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.016897 (rank : 126) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
R4RL2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.002874 (rank : 150) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7M6Z0, Q3UQ62 | Gene names | Rtn4rl2, Ngrl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-4 receptor-like 2 precursor (Nogo-66 receptor homolog 1) (Nogo-66 receptor-related protein 2) (NgR2) (Nogo receptor-like 3). | |||||
|
TMM20_HUMAN
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.038967 (rank : 109) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q2M3R5, Q86YG5, Q8NBA5 | Gene names | TMEM20, C10orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 20. | |||||
|
CSPG5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.019501 (rank : 123) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95196, Q71M39, Q71M40 | Gene names | CSPG5, CALEB, NGC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
GA2L2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.012880 (rank : 133) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
GLT12_MOUSE
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.004147 (rank : 149) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BGT9 | Gene names | Galnt12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 12 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 12) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 12) (Polypeptide GalNAc transferase 12) (GalNAc-T12) (pp-GaNTase 12). | |||||
|
KCND1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.107314 (rank : 45) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
NRAM2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.049340 (rank : 108) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49282, O54903 | Gene names | Slc11a2, Nramp2 | |||
|
Domain Architecture |
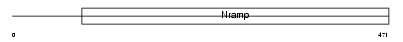
|
|||||
| Description | Natural resistance-associated macrophage protein 2 (NRAMP 2). | |||||
|
SAPS2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.014487 (rank : 130) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R3Q2, Q9CXB7 | Gene names | Saps2, Kiaa0685 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 2. | |||||
|
SCYL2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.004530 (rank : 147) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CFE4, Q3UT57, Q3UWU9, Q8K0M4 | Gene names | Scyl2, Cvak104, D10Ertd802e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCY1-like protein 2 (Coated vesicle-associated kinase of 104 kDa). | |||||
|
ZFHX2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.005710 (rank : 144) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
FREA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.004442 (rank : 148) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43638, Q96D28 | Gene names | FKHL18, FREAC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead-related transcription factor 10 (FREAC-10) (Forkhead-like 18 protein). | |||||
|
IF4B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.032807 (rank : 112) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
KCNN4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.096521 (rank : 51) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O89109, Q3U1J8, Q9CY39 | Gene names | Kcnn4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1). | |||||
|
KCNV2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.075341 (rank : 63) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | -0.000287 (rank : 153) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.013072 (rank : 132) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.016670 (rank : 128) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.018154 (rank : 125) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
DLX5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.000891 (rank : 152) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70396, O54876, O54877, O54878, Q9JJ45 | Gene names | Dlx5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-5. | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.012321 (rank : 134) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
KCNC3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.067641 (rank : 72) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.063105 (rank : 80) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
TMM20_MOUSE
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.026780 (rank : 117) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BY79 | Gene names | Tmem20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 20. | |||||
|
ERBB2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | -0.003128 (rank : 154) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.012029 (rank : 135) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.016866 (rank : 127) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.009026 (rank : 141) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
RUNX2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.010481 (rank : 138) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13950, O14614, O14615, O95181 | Gene names | RUNX2, AML3, CBFA1, OSF2, PEBP2A | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
RUNX2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.010660 (rank : 136) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q08775, O35183, Q08776, Q9JLN0, Q9QUQ6, Q9QY29, Q9R0U4, Q9Z2J7 | Gene names | Runx2, Aml3, Cbfa1, Osf2, Pebp2a | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
TAF6L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.009417 (rank : 139) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R2K4, Q8C5T2 | Gene names | Taf6l, Paf65a | |||
|
Domain Architecture |
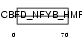
|
|||||
| Description | TAF6-like RNA polymerase II p300/CBP-associated factor-associated factor 65 kDa subunit 6L (PCAF-associated factor 65 alpha) (PAF65- alpha). | |||||
|
CAC1H_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.010533 (rank : 137) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
LRTM3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.001552 (rank : 151) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BZ81, Q3TQA3, Q8BGJ7 | Gene names | Lrrtm3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat transmembrane neuronal protein 3 precursor. | |||||
|
SIA7A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.005688 (rank : 145) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NSC7, Q6UW90, Q9NSC6 | Gene names | ST6GALNAC1, SIAT7A | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
CNGA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.055936 (rank : 91) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.055383 (rank : 96) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.059568 (rank : 85) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62398 | Gene names | Cnga2, Cncg2, Cncg4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated olfactory channel (Cyclic nucleotide-gated cation channel 2) (CNG channel 2) (CNG-2) (CNG2). | |||||
|
CNGA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.055875 (rank : 92) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q16281, Q9UP64 | Gene names | CNGA3, CNCG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.058258 (rank : 88) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JJZ8, Q9WV01 | Gene names | Cnga3, Cng3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
HCN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.053767 (rank : 100) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
|
HCN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.053270 (rank : 101) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.059275 (rank : 86) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
HCN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.057885 (rank : 90) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
HCN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.051879 (rank : 103) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
HCN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.051681 (rank : 104) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
HCN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.055553 (rank : 95) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
HCN4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.054740 (rank : 99) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
KCNA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.065968 (rank : 76) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.065978 (rank : 75) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.070790 (rank : 71) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.070798 (rank : 70) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.072881 (rank : 67) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.074811 (rank : 64) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.086438 (rank : 55) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.075429 (rank : 62) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.079735 (rank : 56) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.076228 (rank : 60) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.079357 (rank : 57) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.076954 (rank : 59) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.065028 (rank : 78) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.064726 (rank : 79) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCNB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.065685 (rank : 77) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.072940 (rank : 66) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.073239 (rank : 65) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.077744 (rank : 58) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCNG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.061042 (rank : 83) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.062129 (rank : 82) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.050518 (rank : 107) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.060025 (rank : 84) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q92952 | Gene names | KCNN1, SK | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCNN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.062355 (rank : 81) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9EQR3, Q99JA9 | Gene names | Kcnn1, Sk1 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCNN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.055707 (rank : 94) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H2S1 | Gene names | KCNN2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.055782 (rank : 93) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P58390 | Gene names | Kcnn2, Sk2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.055128 (rank : 97) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UGI6, O43517 | Gene names | KCNN3, K3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3) (SKCa3). | |||||
|
KCNN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.055083 (rank : 98) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P58391 | Gene names | Kcnn3, Sk3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3). | |||||
|
KCNQ1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.093349 (rank : 53) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.091591 (rank : 54) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.052468 (rank : 102) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.067230 (rank : 74) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.067245 (rank : 73) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNK4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.56915e-153 (rank : 1) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
KCNK4_MOUSE
|
||||||
| NC score | 0.984754 (rank : 2) | θ value | 3.28708e-135 (rank : 2) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O88454 | Gene names | Kcnk4, Traak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK). | |||||
|
KCNK2_HUMAN
|
||||||
| NC score | 0.965343 (rank : 3) | θ value | 9.7205e-79 (rank : 4) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O95069, Q9UNE3 | Gene names | KCNK2, TREK, TREK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (TREK-1 K(+) channel subunit) (Two-pore potassium channel TPKC1). | |||||
|
KCNK2_MOUSE
|
||||||
| NC score | 0.965325 (rank : 4) | θ value | 5.15425e-80 (rank : 3) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P97438 | Gene names | Kcnk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (Two-pore potassium channel TPKC1) (TREK-1 K(+) channel subunit). | |||||
|
KCNKA_HUMAN
|
||||||
| NC score | 0.954537 (rank : 5) | θ value | 2.73936e-73 (rank : 5) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P57789, Q8TDK7, Q8TDK8, Q9HB59 | Gene names | KCNK10, TREK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 10 (Outward rectifying potassium channel protein TREK-2) (TREK-2 K(+) channel subunit). | |||||
|
KCNKG_HUMAN
|
||||||
| NC score | 0.944972 (rank : 6) | θ value | 2.19075e-38 (rank : 7) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q96T55, Q5TCF3, Q9H591 | Gene names | KCNK16, TALK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 16 (TWIK-related alkaline pH- activated K(+) channel 1) (2P domain potassium channel Talk-1). | |||||
|
KCNK5_HUMAN
|
||||||
| NC score | 0.930460 (rank : 7) | θ value | 1.16164e-39 (rank : 6) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O95279 | Gene names | KCNK5, TASK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 5 (Acid-sensitive potassium channel protein TASK-2) (TWIK-related acid-sensitive K(+) channel 2). | |||||
|
KCNKH_HUMAN
|
||||||
| NC score | 0.923935 (rank : 8) | θ value | 4.27553e-34 (rank : 8) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96T54, Q5TCF4, Q8TAW4, Q9BXD1, Q9H592 | Gene names | KCNK17, TALK2, TASK4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 17 (TWIK-related alkaline pH- activated K(+) channel 2) (2P domain potassium channel Talk-2) (TWIK- related acid-sensitive K(+) channel 4) (TASK-4). | |||||
|
KCNK6_HUMAN
|
||||||
| NC score | 0.900880 (rank : 9) | θ value | 1.16434e-31 (rank : 9) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
KCNK1_HUMAN
|
||||||
| NC score | 0.882945 (rank : 10) | θ value | 4.57862e-28 (rank : 10) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O00180, Q13307 | Gene names | KCNK1, HOHO1, KCNO1, TWIK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1) (Potassium channel KCNO1). | |||||
|
KCNK1_MOUSE
|
||||||
| NC score | 0.880561 (rank : 11) | θ value | 1.92365e-26 (rank : 11) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O08581 | Gene names | Kcnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1). | |||||
|
KCNK9_MOUSE
|
||||||
| NC score | 0.857268 (rank : 12) | θ value | 1.92365e-26 (rank : 12) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q3LS21 | Gene names | Kcnk9, Task3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3). | |||||
|
KCNKF_HUMAN
|
||||||
| NC score | 0.853435 (rank : 13) | θ value | 1.5773e-20 (rank : 16) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9H427, Q9HBC8 | Gene names | KCNK15, TASK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 15 (Acid-sensitive potassium channel protein TASK-5) (TWIK-related acid-sensitive K(+) channel 5) (Two pore potassium channel KT3.3). | |||||
|
KCNK9_HUMAN
|
||||||
| NC score | 0.852204 (rank : 14) | θ value | 1.05554e-24 (rank : 13) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NPC2, Q2M290, Q540F2 | Gene names | KCNK9, TASK3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3) (Two pore potassium channel KT3.2). | |||||
|
KCNK3_HUMAN
|
||||||
| NC score | 0.848987 (rank : 15) | θ value | 9.87957e-23 (rank : 15) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O14649 | Gene names | KCNK3, TASK, TASK1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Two pore potassium channel KT3.1). | |||||
|
KCNK3_MOUSE
|
||||||
| NC score | 0.847984 (rank : 16) | θ value | 1.80048e-24 (rank : 14) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O35111, O35163 | Gene names | Kcnk3, Ctbak, Task, Task1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Cardiac two-pore background K(+) channel) (cTBAK-1) (Two pore potassium channel KT3.1). | |||||
|
KCNK7_MOUSE
|
||||||
| NC score | 0.837063 (rank : 17) | θ value | 4.02038e-16 (rank : 19) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z2T1, Q9QXY0, Q9QYE8, Q9R1V1, Q9R242 | Gene names | Kcnk7, Dpkch3, Kcnk6, Kcnk8, Knot1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7 (Putative potassium channel DP3) (Double-pore K(+) channel 3) (Neuromuscular two p domain potassium channel). | |||||
|
KCNK7_HUMAN
|
||||||
| NC score | 0.821433 (rank : 18) | θ value | 6.41864e-14 (rank : 21) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y2U2, Q9Y2U3, Q9Y2U4 | Gene names | KCNK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7. | |||||
|
KCNKC_HUMAN
|
||||||
| NC score | 0.800163 (rank : 19) | θ value | 8.95645e-16 (rank : 20) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HB15 | Gene names | KCNK12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 12 (Tandem pore domain halothane- inhibited potassium channel 2) (THIK-2). | |||||
|
KCNKD_MOUSE
|
||||||
| NC score | 0.794823 (rank : 20) | θ value | 6.20254e-17 (rank : 17) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8R1P5 | Gene names | Kcnk13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCNKD_HUMAN
|
||||||
| NC score | 0.786144 (rank : 21) | θ value | 1.38178e-16 (rank : 18) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HB14, Q96E79 | Gene names | KCNK13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCNH5_HUMAN
|
||||||
| NC score | 0.206307 (rank : 22) | θ value | 0.00298849 (rank : 24) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
KCNH5_MOUSE
|
||||||
| NC score | 0.205968 (rank : 23) | θ value | 0.00298849 (rank : 25) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q920E3, Q8C035 | Gene names | Kcnh5, Eag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (Eag2). | |||||
|
KCMA1_MOUSE
|
||||||
| NC score | 0.200731 (rank : 24) | θ value | 0.000121331 (rank : 23) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q08460, Q64703, Q8VHF1, Q9R196 | Gene names | Kcnma1, Kcnma | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (mSlo) (mSlo1). | |||||
|
KCMA1_HUMAN
|
||||||
| NC score | 0.200495 (rank : 25) | θ value | 0.000121331 (rank : 22) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q12791, Q12886, Q12917, Q12921, Q12960, Q13150, Q96LG8, Q9UBB0, Q9UQK6 | Gene names | KCNMA1, KCNMA | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (hSlo). | |||||
|
KCNH1_MOUSE
|
||||||
| NC score | 0.197514 (rank : 26) | θ value | 0.0431538 (rank : 29) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
KCNH1_HUMAN
|
||||||
| NC score | 0.197175 (rank : 27) | θ value | 0.0431538 (rank : 28) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O95259, O76035 | Gene names | KCNH1, EAG, EAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (hEAG1) (h-eag). | |||||
|
KCNT1_HUMAN
|
||||||
| NC score | 0.182189 (rank : 28) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5JUK3, Q9P2C5 | Gene names | KCNT1, KIAA1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1 (KCa4.1). | |||||
|
KCNT1_MOUSE
|
||||||
| NC score | 0.173019 (rank : 29) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6ZPR4, Q8C3E7 | Gene names | Kcnt1, Kiaa1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1. | |||||
|
KCNH8_HUMAN
|
||||||
| NC score | 0.172892 (rank : 30) | θ value | 0.0563607 (rank : 33) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96L42 | Gene names | KCNH8 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 8 (Voltage-gated potassium channel subunit Kv12.1) (Ether-a-go-go-like potassium channel 3) (ELK channel 3) (ELK3) (ELK1) (hElk1). | |||||
|
KCNH6_HUMAN
|
||||||
| NC score | 0.170444 (rank : 31) | θ value | 0.365318 (rank : 47) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
KCNH3_MOUSE
|
||||||
| NC score | 0.168936 (rank : 32) | θ value | 0.0431538 (rank : 30) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
KCNH7_HUMAN
|
||||||
| NC score | 0.168897 (rank : 33) | θ value | 0.279714 (rank : 42) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
KCNH7_MOUSE
|
||||||
| NC score | 0.167222 (rank : 34) | θ value | 0.279714 (rank : 43) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
KCNH3_HUMAN
|
||||||
| NC score | 0.166641 (rank : 35) | θ value | 0.21417 (rank : 38) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
KCNH2_HUMAN
|
||||||
| NC score | 0.163786 (rank : 36) | θ value | 0.47712 (rank : 48) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
KCNH2_MOUSE
|
||||||
| NC score | 0.163263 (rank : 37) | θ value | 0.47712 (rank : 49) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
KCNH4_HUMAN
|
||||||
| NC score | 0.159814 (rank : 38) | θ value | 0.0563607 (rank : 32) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
KCNQ4_HUMAN
|
||||||
| NC score | 0.124746 (rank : 39) | θ value | 0.279714 (rank : 44) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
KCND1_HUMAN
|
||||||
| NC score | 0.110788 (rank : 40) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCND2_MOUSE
|
||||||
| NC score | 0.110496 (rank : 41) | θ value | 0.279714 (rank : 41) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_HUMAN
|
||||||
| NC score | 0.110471 (rank : 42) | θ value | 0.279714 (rank : 40) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND3_MOUSE
|
||||||
| NC score | 0.109901 (rank : 43) | θ value | 0.62314 (rank : 53) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_HUMAN
|
||||||
| NC score | 0.109882 (rank : 44) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND1_MOUSE
|
||||||
| NC score | 0.107314 (rank : 45) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCNQ5_HUMAN
|
||||||
| NC score | 0.105899 (rank : 46) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
KCNQ2_HUMAN
|
||||||
| NC score | 0.102554 (rank : 47) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ2_MOUSE
|
||||||
| NC score | 0.101994 (rank : 48) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ3_HUMAN
|
||||||
| NC score | 0.099153 (rank : 49) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ3_MOUSE
|
||||||
| NC score | 0.098512 (rank : 50) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNN4_MOUSE
|
||||||
| NC score | 0.096521 (rank : 51) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O89109, Q3U1J8, Q9CY39 | Gene names | Kcnn4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1). | |||||
|
KCNN4_HUMAN
|
||||||
| NC score | 0.096141 (rank : 52) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O15554 | Gene names | KCNN4, IK1, IKCA1, KCA4, SK4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1) (IKCa1) (Putative Gardos channel). | |||||
|
KCNQ1_HUMAN
|
||||||
| NC score | 0.093349 (rank : 53) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ1_MOUSE
|
||||||
| NC score | 0.091591 (rank : 54) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNA4_HUMAN
|
||||||
| NC score | 0.086438 (rank : 55) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNA5_HUMAN
|
||||||
| NC score | 0.079735 (rank : 56) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNA6_HUMAN
|
||||||
| NC score | 0.079357 (rank : 57) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNF1_HUMAN
|
||||||
| NC score | 0.077744 (rank : 58) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCNA6_MOUSE
|
||||||
| NC score | 0.076954 (rank : 59) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNA5_MOUSE
|
||||||
| NC score | 0.076228 (rank : 60) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNC3_HUMAN
|
||||||
| NC score | 0.075841 (rank : 61) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNA4_MOUSE
|
||||||
| NC score | 0.075429 (rank : 62) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNV2_HUMAN
|
||||||
| NC score | 0.075341 (rank : 63) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
KCNA3_MOUSE
|
||||||
| NC score | 0.074811 (rank : 64) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNC4_MOUSE
|
||||||
| NC score | 0.073239 (rank : 65) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNC4_HUMAN
|
||||||
| NC score | 0.072940 (rank : 66) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNA3_HUMAN
|
||||||
| NC score | 0.072881 (rank : 67) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNC1_MOUSE
|
||||||
| NC score | 0.072854 (rank : 68) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC1_HUMAN
|
||||||
| NC score | 0.072822 (rank : 69) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNA2_MOUSE
|
||||||
| NC score | 0.070798 (rank : 70) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNA2_HUMAN
|
||||||
| NC score | 0.070790 (rank : 71) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNC3_MOUSE
|
||||||
| NC score | 0.067641 (rank : 72) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNS2_MOUSE
|
||||||
| NC score | 0.067245 (rank : 73) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_HUMAN
|
||||||
| NC score | 0.067230 (rank : 74) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNA1_MOUSE
|
||||||
| NC score | 0.065978 (rank : 75) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA1_HUMAN
|
||||||
| NC score | 0.065968 (rank : 76) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNB2_HUMAN
|
||||||
| NC score | 0.065685 (rank : 77) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNB1_HUMAN
|
||||||
| NC score | 0.065028 (rank : 78) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNB1_MOUSE
|
||||||
| NC score | 0.064726 (rank : 79) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCNS1_MOUSE
|
||||||
| NC score | 0.063105 (rank : 80) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNN1_MOUSE
|
||||||
| NC score | 0.062355 (rank : 81) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9EQR3, Q99JA9 | Gene names | Kcnn1, Sk1 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCNG2_HUMAN
|
||||||
| NC score | 0.062129 (rank : 82) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNG1_HUMAN
|
||||||
| NC score | 0.061042 (rank : 83) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNN1_HUMAN
|
||||||
| NC score | 0.060025 (rank : 84) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q92952 | Gene names | KCNN1, SK | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
CNGA2_MOUSE
|
||||||
| NC score | 0.059568 (rank : 85) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62398 | Gene names | Cnga2, Cncg2, Cncg4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated olfactory channel (Cyclic nucleotide-gated cation channel 2) (CNG channel 2) (CNG-2) (CNG2). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.059275 (rank : 86) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
NRAM1_HUMAN
|
||||||
| NC score | 0.059047 (rank : 87) | θ value | 0.163984 (rank : 36) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49279 | Gene names | SLC11A1, NRAMP, NRAMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Natural resistance-associated macrophage protein 1 (NRAMP 1). | |||||
|
CNGA3_MOUSE
|
||||||
| NC score | 0.058258 (rank : 88) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JJZ8, Q9WV01 | Gene names | Cnga3, Cng3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
NRAM1_MOUSE
|
||||||
| NC score | 0.058175 (rank : 89) | θ value | 0.47712 (rank : 51) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P41251 | Gene names | Slc11a1, Bcg, Ity, Lsh, Nramp1 | |||
|
Domain Architecture |

|
|||||
| Description | Natural resistance-associated macrophage protein 1 (NRAMP 1). | |||||
|
HCN2_MOUSE
|
||||||
| NC score | 0.057885 (rank : 90) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
CNGA1_HUMAN
|
||||||
| NC score | 0.055936 (rank : 91) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA3_HUMAN
|
||||||
| NC score | 0.055875 (rank : 92) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q16281, Q9UP64 | Gene names | CNGA3, CNCG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
KCNN2_MOUSE
|
||||||
| NC score | 0.055782 (rank : 93) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P58390 | Gene names | Kcnn2, Sk2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN2_HUMAN
|
||||||
| NC score | 0.055707 (rank : 94) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H2S1 | Gene names | KCNN2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.055553 (rank : 95) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
CNGA1_MOUSE
|
||||||
| NC score | 0.055383 (rank : 96) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
KCNN3_HUMAN
|
||||||
| NC score | 0.055128 (rank : 97) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UGI6, O43517 | Gene names | KCNN3, K3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3) (SKCa3). | |||||
|
KCNN3_MOUSE
|
||||||
| NC score | 0.055083 (rank : 98) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P58391 | Gene names | Kcnn3, Sk3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3). | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.054740 (rank : 99) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
HCN1_HUMAN
|
||||||
| NC score | 0.053767 (rank : 100) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
|
HCN1_MOUSE
|
||||||
| NC score | 0.053270 (rank : 101) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
KCNS1_HUMAN
|
||||||
| NC score | 0.052468 (rank : 102) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
HCN3_HUMAN
|
||||||
| NC score | 0.051879 (rank : 103) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
HCN3_MOUSE
|
||||||
| NC score | 0.051681 (rank : 104) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
CG1_HUMAN
|
||||||
| NC score | 0.051077 (rank : 105) | θ value | 0.0961366 (rank : 34) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
NRAM2_HUMAN
|
||||||
| NC score | 0.050524 (rank : 106) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49281, O43288, O60932, Q96J35 | Gene names | SLC11A2, NRAMP2 | |||
|
Domain Architecture |

|
|||||
| Description | Natural resistance-associated macrophage protein 2 (NRAMP 2) (Divalent metal transporter 1) (DMT1). | |||||
|
KCNG4_HUMAN
|
||||||
| NC score | 0.050518 (rank : 107) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
NRAM2_MOUSE
|
||||||
| NC score | 0.049340 (rank : 108) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49282, O54903 | Gene names | Slc11a2, Nramp2 | |||
|
Domain Architecture |

|
|||||
| Description | Natural resistance-associated macrophage protein 2 (NRAMP 2). | |||||
|
TMM20_HUMAN
|
||||||
| NC score | 0.038967 (rank : 109) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q2M3R5, Q86YG5, Q8NBA5 | Gene names | TMEM20, C10orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 20. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.038098 (rank : 110) | θ value | 0.00390308 (rank : 26) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
IF4B_HUMAN
|
||||||
| NC score | 0.037155 (rank : 111) | θ value | 0.21417 (rank : 37) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P23588 | Gene names | EIF4B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
IF4B_MOUSE
|
||||||
| NC score | 0.032807 (rank : 112) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.030647 (rank : 113) | θ value | 0.21417 (rank : 39) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
COA2_HUMAN
|
||||||
| NC score | 0.030386 (rank : 114) | θ value | 0.0563607 (rank : 31) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00763, Q16852 | Gene names | ACACB, ACC2, ACCB | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 2 (EC 6.4.1.2) (ACC-beta) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
YIF1A_HUMAN
|
||||||
| NC score | 0.030269 (rank : 115) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95070, Q96G83, Q9BVD0 | Gene names | YIF1A, 54TM, HYIF1P, YIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein YIF1A (YIP1-interacting factor homolog A) (54TMp). | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.028316 (rank : 116) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
TMM20_MOUSE
|
||||||
| NC score | 0.026780 (rank : 117) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BY79 | Gene names | Tmem20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 20. | |||||
|
ATX7_MOUSE
|
||||||
| NC score | 0.026187 (rank : 118) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
UBP6_HUMAN
|
||||||
| NC score | 0.023478 (rank : 119) | θ value | 0.0113563 (rank : 27) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35125, Q15634, Q86WP6, Q8IWT4 | Gene names | USP6, TRE2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 6 (EC 3.1.2.15) (Ubiquitin thioesterase 6) (Ubiquitin-specific-processing protease 6) (Deubiquitinating enzyme 6) (Proto-oncogene TRE-2). | |||||
|
AL2S4_MOUSE
|
||||||
| NC score | 0.023462 (rank : 120) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3V0J1, Q3TIS2 | Gene names | Als2cr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 4 protein homolog. | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.021845 (rank : 121) | θ value | 0.0961366 (rank : 35) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.020594 (rank : 122) | θ value | 0.365318 (rank : 46) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
CSPG5_HUMAN
|
||||||
| NC score | 0.019501 (rank : 123) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95196, Q71M39, Q71M40 | Gene names | CSPG5, CALEB, NGC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
DOK1_MOUSE
|
||||||
| NC score | 0.018359 (rank : 124) | θ value | 1.06291 (rank : 58) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97465, Q9R213 | Gene names | Dok1, Dok | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 1 (Downstream of tyrosine kinase 1) (p62(dok)). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.018154 (rank : 125) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.016897 (rank : 126) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.016866 (rank : 127) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.016670 (rank : 128) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.016161 (rank : 129) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
SAPS2_MOUSE
|
||||||
| NC score | 0.014487 (rank : 130) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R3Q2, Q9CXB7 | Gene names | Saps2, Kiaa0685 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 2. | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.014113 (rank : 131) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.013072 (rank : 132) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
GA2L2_MOUSE
|
||||||
| NC score | 0.012880 (rank : 133) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.012321 (rank : 134) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.012029 (rank : 135) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
RUNX2_MOUSE
|
||||||
| NC score | 0.010660 (rank : 136) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q08775, O35183, Q08776, Q9JLN0, Q9QUQ6, Q9QY29, Q9R0U4, Q9Z2J7 | Gene names | Runx2, Aml3, Cbfa1, Osf2, Pebp2a | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
CAC1H_MOUSE
|
||||||
| NC score | 0.010533 (rank : 137) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
RUNX2_HUMAN
|
||||||
| NC score | 0.010481 (rank : 138) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13950, O14614, O14615, O95181 | Gene names | RUNX2, AML3, CBFA1, OSF2, PEBP2A | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
TAF6L_MOUSE
|
||||||
| NC score | 0.009417 (rank : 139) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R2K4, Q8C5T2 | Gene names | Taf6l, Paf65a | |||
|
Domain Architecture |

|
|||||
| Description | TAF6-like RNA polymerase II p300/CBP-associated factor-associated factor 65 kDa subunit 6L (PCAF-associated factor 65 alpha) (PAF65- alpha). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.009140 (rank : 140) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.009026 (rank : 141) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.008849 (rank : 142) | θ value | 0.47712 (rank : 50) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.007521 (rank : 143) | θ value | 1.06291 (rank : 57) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ZFHX2_HUMAN
|
||||||
| NC score | 0.005710 (rank : 144) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
SIA7A_HUMAN
|
||||||
| NC score | 0.005688 (rank : 145) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NSC7, Q6UW90, Q9NSC6 | Gene names | ST6GALNAC1, SIAT7A | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (EC 2.4.99.3) (GalNAc alpha-2,6-sialyltransferase I) (ST6GalNAc I) (Sialyltransferase 7A). | |||||
|
PCD12_MOUSE
|
||||||
| NC score | 0.005103 (rank : 146) | θ value | 0.279714 (rank : 45) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O55134 | Gene names | Pcdh12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-12 precursor (Vascular cadherin-2) (Vascular endothelial cadherin-2) (VE-cadherin-2) (VE-cad-2). | |||||
|
SCYL2_MOUSE
|
||||||
| NC score | 0.004530 (rank : 147) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CFE4, Q3UT57, Q3UWU9, Q8K0M4 | Gene names | Scyl2, Cvak104, D10Ertd802e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCY1-like protein 2 (Coated vesicle-associated kinase of 104 kDa). | |||||
|
FREA_HUMAN
|
||||||
| NC score | 0.004442 (rank : 148) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43638, Q96D28 | Gene names | FKHL18, FREAC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead-related transcription factor 10 (FREAC-10) (Forkhead-like 18 protein). | |||||
|
GLT12_MOUSE
|
||||||
| NC score | 0.004147 (rank : 149) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BGT9 | Gene names | Galnt12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 12 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 12) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 12) (Polypeptide GalNAc transferase 12) (GalNAc-T12) (pp-GaNTase 12). | |||||
|
R4RL2_MOUSE
|
||||||
| NC score | 0.002874 (rank : 150) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7M6Z0, Q3UQ62 | Gene names | Rtn4rl2, Ngrl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-4 receptor-like 2 precursor (Nogo-66 receptor homolog 1) (Nogo-66 receptor-related protein 2) (NgR2) (Nogo receptor-like 3). | |||||
|
LRTM3_MOUSE
|
||||||
| NC score | 0.001552 (rank : 151) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BZ81, Q3TQA3, Q8BGJ7 | Gene names | Lrrtm3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat transmembrane neuronal protein 3 precursor. | |||||
|
DLX5_MOUSE
|
||||||
| NC score | 0.000891 (rank : 152) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70396, O54876, O54877, O54878, Q9JJ45 | Gene names | Dlx5 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein DLX-5. | |||||
|
MAST4_HUMAN
|
||||||
| NC score | -0.000287 (rank : 153) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
ERBB2_HUMAN
|
||||||
| NC score | -0.003128 (rank : 154) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 109 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||