Please be patient as the page loads
|
KCNN1_HUMAN
|
||||||
| SwissProt Accessions | Q92952 | Gene names | KCNN1, SK | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
KCNN1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | Q92952 | Gene names | KCNN1, SK | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCNN1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.991905 (rank : 2) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9EQR3, Q99JA9 | Gene names | Kcnn1, Sk1 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCNN2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.988177 (rank : 3) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9H2S1 | Gene names | KCNN2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.987565 (rank : 4) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P58390 | Gene names | Kcnn2, Sk2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.974281 (rank : 6) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9UGI6, O43517 | Gene names | KCNN3, K3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3) (SKCa3). | |||||
|
KCNN3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.979626 (rank : 5) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P58391 | Gene names | Kcnn3, Sk3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3). | |||||
|
KCNN4_MOUSE
|
||||||
| θ value | 8.77142e-88 (rank : 7) | NC score | 0.953244 (rank : 7) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O89109, Q3U1J8, Q9CY39 | Gene names | Kcnn4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1). | |||||
|
KCNN4_HUMAN
|
||||||
| θ value | 2.38869e-85 (rank : 8) | NC score | 0.951083 (rank : 8) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O15554 | Gene names | KCNN4, IK1, IKCA1, KCA4, SK4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1) (IKCa1) (Putative Gardos channel). | |||||
|
KCNQ4_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 9) | NC score | 0.369166 (rank : 9) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
KCNQ5_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 10) | NC score | 0.363219 (rank : 10) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
KCNQ2_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 11) | NC score | 0.346176 (rank : 11) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCND1_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 12) | NC score | 0.295518 (rank : 17) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCNQ1_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 13) | NC score | 0.346154 (rank : 12) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCND1_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 14) | NC score | 0.295450 (rank : 20) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCND2_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 15) | NC score | 0.295475 (rank : 19) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 16) | NC score | 0.295478 (rank : 18) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCNQ2_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 17) | NC score | 0.343651 (rank : 14) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ3_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 18) | NC score | 0.338869 (rank : 15) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ3_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 19) | NC score | 0.335799 (rank : 16) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ1_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 20) | NC score | 0.345308 (rank : 13) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCND3_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 21) | NC score | 0.291319 (rank : 22) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 22) | NC score | 0.291446 (rank : 21) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCNB2_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 23) | NC score | 0.279277 (rank : 28) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
COGA1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 24) | NC score | 0.029882 (rank : 90) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q07092 | Gene names | COL16A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVI) chain precursor. | |||||
|
KCNS2_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 25) | NC score | 0.290684 (rank : 23) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 26) | NC score | 0.289714 (rank : 24) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNA6_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 27) | NC score | 0.286983 (rank : 25) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNA6_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 28) | NC score | 0.284528 (rank : 26) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNB1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 29) | NC score | 0.271492 (rank : 36) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNB1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 30) | NC score | 0.271634 (rank : 35) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCNA4_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 31) | NC score | 0.275621 (rank : 32) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNG3_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 32) | NC score | 0.264549 (rank : 41) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 33) | NC score | 0.264315 (rank : 42) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
WWC1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 34) | NC score | 0.044840 (rank : 85) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IX03, O94946, Q6MZX4, Q6Y2F8, Q7Z4G8, Q8WVM4, Q9BT29 | Gene names | WWC1, KIAA0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA) (HBeAg-binding protein 3). | |||||
|
KCNA2_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 35) | NC score | 0.277696 (rank : 29) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNA2_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 36) | NC score | 0.275401 (rank : 33) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNA5_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 37) | NC score | 0.281410 (rank : 27) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
ANGL7_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 38) | NC score | 0.036027 (rank : 87) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R1Q3 | Gene names | Angptl7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 7 precursor (Angiopoietin-like 7). | |||||
|
KCNA3_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 39) | NC score | 0.275682 (rank : 31) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNA3_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 40) | NC score | 0.275062 (rank : 34) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNA4_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 41) | NC score | 0.270539 (rank : 38) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 42) | NC score | 0.021587 (rank : 96) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
JUND_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 43) | NC score | 0.026599 (rank : 93) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P15066 | Gene names | Jund, Jun-d, Jund1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
KCNA1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 44) | NC score | 0.270648 (rank : 37) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 45) | NC score | 0.270406 (rank : 39) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA5_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 46) | NC score | 0.276445 (rank : 30) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNG2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 47) | NC score | 0.262701 (rank : 43) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNK1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 48) | NC score | 0.136484 (rank : 58) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O00180, Q13307 | Gene names | KCNK1, HOHO1, KCNO1, TWIK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1) (Potassium channel KCNO1). | |||||
|
KCMA1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 49) | NC score | 0.214005 (rank : 57) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q12791, Q12886, Q12917, Q12921, Q12960, Q13150, Q96LG8, Q9UBB0, Q9UQK6 | Gene names | KCNMA1, KCNMA | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (hSlo). | |||||
|
KCMA1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 50) | NC score | 0.214386 (rank : 56) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q08460, Q64703, Q8VHF1, Q9R196 | Gene names | Kcnma1, Kcnma | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (mSlo) (mSlo1). | |||||
|
ROCK2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 51) | NC score | 0.011461 (rank : 114) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
WWC1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 52) | NC score | 0.040756 (rank : 86) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5SXA9, Q571D0, Q8K1Y3, Q8VD17, Q922W3 | Gene names | Wwc1, Kiaa0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA). | |||||
|
KCNC3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 53) | NC score | 0.247046 (rank : 52) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNG1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 54) | NC score | 0.255504 (rank : 44) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNS3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 55) | NC score | 0.270034 (rank : 40) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
KCNC1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 56) | NC score | 0.247293 (rank : 51) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 57) | NC score | 0.247356 (rank : 50) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 58) | NC score | 0.253955 (rank : 47) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNC4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 59) | NC score | 0.254600 (rank : 46) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNK1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 60) | NC score | 0.121598 (rank : 60) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O08581 | Gene names | Kcnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1). | |||||
|
KCNC3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 61) | NC score | 0.243924 (rank : 55) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNK6_HUMAN
|
||||||
| θ value | 0.47712 (rank : 62) | NC score | 0.134896 (rank : 59) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
ROCK2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 63) | NC score | 0.010221 (rank : 121) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
KCNG4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 64) | NC score | 0.247372 (rank : 49) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNS1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 65) | NC score | 0.255254 (rank : 45) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
CEP55_MOUSE
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.017243 (rank : 100) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BT07, Q8C2J0, Q8K2I8, Q8R2Y4, Q9DBZ8 | Gene names | Cep55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55. | |||||
|
FBX41_HUMAN
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.020591 (rank : 97) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TF61 | Gene names | FBXO41, FBX41, KIAA1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
KCNS1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 68) | NC score | 0.245383 (rank : 53) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 1.06291 (rank : 69) | NC score | 0.002693 (rank : 134) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
RPGF1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 70) | NC score | 0.017672 (rank : 99) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13905 | Gene names | RAPGEF1, GRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 1 (Guanine nucleotide-releasing factor 2) (C3G protein) (CRK SH3-binding GNRP). | |||||
|
COFA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.022613 (rank : 95) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P39059, Q5T6J4, Q9Y4W4 | Gene names | COL15A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV) (Restin) (Related to endostatin)]. | |||||
|
COIA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.018988 (rank : 98) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
CO3A1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.012441 (rank : 110) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
GPS2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.035011 (rank : 88) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13227 | Gene names | GPS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein pathway suppressor 2 (Protein GPS2). | |||||
|
HOME1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.027615 (rank : 92) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 630 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Z2Y3, Q8K3E1, Q8K4M8, Q9Z0E9, Q9Z216 | Gene names | Homer1, Vesl1 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1 (VASP/Ena-related gene up-regulated during seizure and LTP) (Vesl-1). | |||||
|
IFT81_MOUSE
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.013924 (rank : 106) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
KCNK4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.082353 (rank : 71) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O88454 | Gene names | Kcnk4, Traak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK). | |||||
|
KCNK9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.109845 (rank : 64) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9NPC2, Q2M290, Q540F2 | Gene names | KCNK9, TASK3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3) (Two pore potassium channel KT3.2). | |||||
|
KCNK9_MOUSE
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.109356 (rank : 65) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q3LS21 | Gene names | Kcnk9, Task3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3). | |||||
|
SF3B4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 80) | NC score | 0.010454 (rank : 120) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
TXLNG_HUMAN
|
||||||
| θ value | 1.81305 (rank : 81) | NC score | 0.014160 (rank : 105) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NUQ3, Q9P0X1 | Gene names | TXLNG, CXorf15, LSR5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-taxilin (Lipopolysaccharide-specific response protein 5). | |||||
|
ZN409_HUMAN
|
||||||
| θ value | 1.81305 (rank : 82) | NC score | 0.007451 (rank : 126) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
AAT1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.045813 (rank : 84) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z4T9, Q68DX2, Q8TD41, Q96A45, Q96JE8 | Gene names | AAT1, C3orf15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AMY-1-associating protein expressed in testis 1 (AAT-1). | |||||
|
CABL2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.014248 (rank : 103) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BTV7, Q5JWL0, Q9BYK0 | Gene names | CABLES2, C20orf150 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
KCNF1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.249705 (rank : 48) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCNV2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | 0.244163 (rank : 54) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
CO9A1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.015402 (rank : 101) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
DIAP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.011838 (rank : 111) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
TAXB1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.012934 (rank : 108) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
TENX_HUMAN
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.003494 (rank : 133) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
CO2A1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.012858 (rank : 109) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P02458, Q12985, Q14009, Q14044, Q14046, Q14056, Q14058, Q16672, Q6LBY1, Q6LBY2, Q6LBY3, Q99227, Q9UE38, Q9UE39, Q9UE40, Q9UE41, Q9UE42, Q9UE43 | Gene names | COL2A1 | |||
|
Domain Architecture |
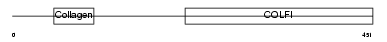
|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
HOME1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.024715 (rank : 94) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 564 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q86YM7, O96003, Q86YM5 | Gene names | HOMER1, SYN47 | |||
|
Domain Architecture |
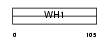
|
|||||
| Description | Homer protein homolog 1. | |||||
|
SMC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.011392 (rank : 115) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.010967 (rank : 117) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
INHBB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 95) | NC score | 0.004653 (rank : 131) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P09529, Q8N1D3 | Gene names | INHBB | |||
|
Domain Architecture |
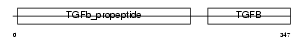
|
|||||
| Description | Inhibin beta B chain precursor (Activin beta-B chain). | |||||
|
KCNK3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.098080 (rank : 66) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O14649 | Gene names | KCNK3, TASK, TASK1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Two pore potassium channel KT3.1). | |||||
|
KCNK3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.098071 (rank : 67) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O35111, O35163 | Gene names | Kcnk3, Ctbak, Task, Task1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Cardiac two-pore background K(+) channel) (cTBAK-1) (Two pore potassium channel KT3.1). | |||||
|
KCNK5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.114114 (rank : 63) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O95279 | Gene names | KCNK5, TASK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 5 (Acid-sensitive potassium channel protein TASK-2) (TWIK-related acid-sensitive K(+) channel 2). | |||||
|
KCNKD_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.067150 (rank : 74) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HB14, Q96E79 | Gene names | KCNK13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
TEAD3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.006533 (rank : 129) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70210, O08516, O70623, P70209 | Gene names | Tead3, Tcf13r2, Tef5 | |||
|
Domain Architecture |
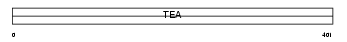
|
|||||
| Description | Transcriptional enhancer factor TEF-5 (TEA domain family member 3) (TEAD-3) (ETF-related factor 1) (ETFR-1) (DTEF-1). | |||||
|
CO9A1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.014650 (rank : 102) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.011702 (rank : 112) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
KCNH4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.029320 (rank : 91) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
KCNK2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.079493 (rank : 72) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P97438 | Gene names | Kcnk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (Two-pore potassium channel TPKC1) (TREK-1 K(+) channel subunit). | |||||
|
KCNKA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 105) | NC score | 0.114835 (rank : 62) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P57789, Q8TDK7, Q8TDK8, Q9HB59 | Gene names | KCNK10, TREK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 10 (Outward rectifying potassium channel protein TREK-2) (TREK-2 K(+) channel subunit). | |||||
|
KCNKD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 106) | NC score | 0.060270 (rank : 75) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8R1P5 | Gene names | Kcnk13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCNKG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 107) | NC score | 0.088057 (rank : 69) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q96T55, Q5TCF3, Q9H591 | Gene names | KCNK16, TALK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 16 (TWIK-related alkaline pH- activated K(+) channel 1) (2P domain potassium channel Talk-1). | |||||
|
LMNB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 108) | NC score | 0.013836 (rank : 107) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P20700, Q3SYN7, Q96EI6 | Gene names | LMNB1, LMN2, LMNB | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
MD1L1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 109) | NC score | 0.010775 (rank : 118) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.008262 (rank : 123) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.007828 (rank : 124) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.010657 (rank : 119) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
PTN23_HUMAN
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.003588 (rank : 132) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
ZN575_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | -0.003623 (rank : 136) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86XF7 | Gene names | ZNF575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 575. | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.007659 (rank : 125) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
COAA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.008534 (rank : 122) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q03692 | Gene names | COL10A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(X) chain precursor. | |||||
|
DIAP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.011485 (rank : 113) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.011152 (rank : 116) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
KCNKF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.115460 (rank : 61) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9H427, Q9HBC8 | Gene names | KCNK15, TASK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 15 (Acid-sensitive potassium channel protein TASK-5) (TWIK-related acid-sensitive K(+) channel 5) (Two pore potassium channel KT3.3). | |||||
|
LMNB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.014235 (rank : 104) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P14733, Q61791 | Gene names | Lmnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | -0.000827 (rank : 135) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.006935 (rank : 127) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
PEF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.005389 (rank : 130) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BFY6, Q8VCT5, Q9CYW8, Q9D934 | Gene names | Pef1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
RAB3I_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.032512 (rank : 89) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q68EF0 | Gene names | Rab3ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAB3A-interacting protein (Rabin-3) (SSX2-interacting protein). | |||||
|
WASP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.006867 (rank : 128) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P42768, Q9BU11, Q9UNJ9 | Gene names | WAS, IMD2 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein (WASp). | |||||
|
KCD15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.052958 (rank : 82) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96SI1, Q9BVI6 | Gene names | KCTD15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD15. | |||||
|
KCD15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.050683 (rank : 83) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K0E1 | Gene names | Kctd15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD15. | |||||
|
KCNK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.074722 (rank : 73) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O95069, Q9UNE3 | Gene names | KCNK2, TREK, TREK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (TREK-1 K(+) channel subunit) (Two-pore potassium channel TPKC1). | |||||
|
KCNK4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.060025 (rank : 76) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
KCNK7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.053925 (rank : 79) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Z2T1, Q9QXY0, Q9QYE8, Q9R1V1, Q9R242 | Gene names | Kcnk7, Dpkch3, Kcnk6, Kcnk8, Knot1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7 (Putative potassium channel DP3) (Double-pore K(+) channel 3) (Neuromuscular two p domain potassium channel). | |||||
|
KCNKH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.089168 (rank : 68) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q96T54, Q5TCF4, Q8TAW4, Q9BXD1, Q9H592 | Gene names | KCNK17, TALK2, TASK4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 17 (TWIK-related alkaline pH- activated K(+) channel 2) (2P domain potassium channel Talk-2) (TWIK- related acid-sensitive K(+) channel 4) (TASK-4). | |||||
|
KCNT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.084216 (rank : 70) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q5JUK3, Q9P2C5 | Gene names | KCNT1, KIAA1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1 (KCa4.1). | |||||
|
KCTD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.054134 (rank : 77) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q719H9 | Gene names | KCTD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD1. | |||||
|
KCTD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.054134 (rank : 78) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5M956, Q5EER9 | Gene names | Kctd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD1. | |||||
|
KCTD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.053412 (rank : 81) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8NC69, Q8NBS6, Q8TCA6 | Gene names | KCTD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD6. | |||||
|
KCTD6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.053768 (rank : 80) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BNL5, Q3TXX1 | Gene names | Kctd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD6. | |||||
|
KCNN1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 125 | |
| SwissProt Accessions | Q92952 | Gene names | KCNN1, SK | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCNN1_MOUSE
|
||||||
| NC score | 0.991905 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9EQR3, Q99JA9 | Gene names | Kcnn1, Sk1 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCNN2_HUMAN
|
||||||
| NC score | 0.988177 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9H2S1 | Gene names | KCNN2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN2_MOUSE
|
||||||
| NC score | 0.987565 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P58390 | Gene names | Kcnn2, Sk2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN3_MOUSE
|
||||||
| NC score | 0.979626 (rank : 5) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P58391 | Gene names | Kcnn3, Sk3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3). | |||||
|
KCNN3_HUMAN
|
||||||
| NC score | 0.974281 (rank : 6) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9UGI6, O43517 | Gene names | KCNN3, K3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3) (SKCa3). | |||||
|
KCNN4_MOUSE
|
||||||
| NC score | 0.953244 (rank : 7) | θ value | 8.77142e-88 (rank : 7) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O89109, Q3U1J8, Q9CY39 | Gene names | Kcnn4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1). | |||||
|
KCNN4_HUMAN
|
||||||
| NC score | 0.951083 (rank : 8) | θ value | 2.38869e-85 (rank : 8) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O15554 | Gene names | KCNN4, IK1, IKCA1, KCA4, SK4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1) (IKCa1) (Putative Gardos channel). | |||||
|
KCNQ4_HUMAN
|
||||||
| NC score | 0.369166 (rank : 9) | θ value | 1.09739e-05 (rank : 9) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
KCNQ5_HUMAN
|
||||||
| NC score | 0.363219 (rank : 10) | θ value | 1.43324e-05 (rank : 10) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
KCNQ2_MOUSE
|
||||||
| NC score | 0.346176 (rank : 11) | θ value | 0.000158464 (rank : 11) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ1_HUMAN
|
||||||
| NC score | 0.346154 (rank : 12) | θ value | 0.00035302 (rank : 13) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ1_MOUSE
|
||||||
| NC score | 0.345308 (rank : 13) | θ value | 0.000786445 (rank : 20) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNQ2_HUMAN
|
||||||
| NC score | 0.343651 (rank : 14) | θ value | 0.000602161 (rank : 17) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ3_HUMAN
|
||||||
| NC score | 0.338869 (rank : 15) | θ value | 0.000602161 (rank : 18) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ3_MOUSE
|
||||||
| NC score | 0.335799 (rank : 16) | θ value | 0.000602161 (rank : 19) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCND1_MOUSE
|
||||||
| NC score | 0.295518 (rank : 17) | θ value | 0.00035302 (rank : 12) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCND2_MOUSE
|
||||||
| NC score | 0.295478 (rank : 18) | θ value | 0.000461057 (rank : 16) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_HUMAN
|
||||||
| NC score | 0.295475 (rank : 19) | θ value | 0.000461057 (rank : 15) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND1_HUMAN
|
||||||
| NC score | 0.295450 (rank : 20) | θ value | 0.000461057 (rank : 14) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCND3_MOUSE
|
||||||
| NC score | 0.291446 (rank : 21) | θ value | 0.00102713 (rank : 22) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_HUMAN
|
||||||
| NC score | 0.291319 (rank : 22) | θ value | 0.00102713 (rank : 21) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCNS2_MOUSE
|
||||||
| NC score | 0.290684 (rank : 23) | θ value | 0.00665767 (rank : 25) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_HUMAN
|
||||||
| NC score | 0.289714 (rank : 24) | θ value | 0.00869519 (rank : 26) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNA6_HUMAN
|
||||||
| NC score | 0.286983 (rank : 25) | θ value | 0.0113563 (rank : 27) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNA6_MOUSE
|
||||||
| NC score | 0.284528 (rank : 26) | θ value | 0.0148317 (rank : 28) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNA5_MOUSE
|
||||||
| NC score | 0.281410 (rank : 27) | θ value | 0.0431538 (rank : 37) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNB2_HUMAN
|
||||||
| NC score | 0.279277 (rank : 28) | θ value | 0.00390308 (rank : 23) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNA2_HUMAN
|
||||||
| NC score | 0.277696 (rank : 29) | θ value | 0.0330416 (rank : 35) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNA5_HUMAN
|
||||||
| NC score | 0.276445 (rank : 30) | θ value | 0.0961366 (rank : 46) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNA3_HUMAN
|
||||||
| NC score | 0.275682 (rank : 31) | θ value | 0.0563607 (rank : 39) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNA4_HUMAN
|
||||||
| NC score | 0.275621 (rank : 32) | θ value | 0.0252991 (rank : 31) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNA2_MOUSE
|
||||||
| NC score | 0.275401 (rank : 33) | θ value | 0.0431538 (rank : 36) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNA3_MOUSE
|
||||||
| NC score | 0.275062 (rank : 34) | θ value | 0.0563607 (rank : 40) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNB1_MOUSE
|
||||||
| NC score | 0.271634 (rank : 35) | θ value | 0.0193708 (rank : 30) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCNB1_HUMAN
|
||||||
| NC score | 0.271492 (rank : 36) | θ value | 0.0193708 (rank : 29) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNA1_HUMAN
|
||||||
| NC score | 0.270648 (rank : 37) | θ value | 0.0961366 (rank : 44) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA4_MOUSE
|
||||||
| NC score | 0.270539 (rank : 38) | θ value | 0.0563607 (rank : 41) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNA1_MOUSE
|
||||||
| NC score | 0.270406 (rank : 39) | θ value | 0.0961366 (rank : 45) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNS3_HUMAN
|
||||||
| NC score | 0.270034 (rank : 40) | θ value | 0.279714 (rank : 55) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
KCNG3_HUMAN
|
||||||
| NC score | 0.264549 (rank : 41) | θ value | 0.0252991 (rank : 32) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG3_MOUSE
|
||||||
| NC score | 0.264315 (rank : 42) | θ value | 0.0252991 (rank : 33) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG2_HUMAN
|
||||||
| NC score | 0.262701 (rank : 43) | θ value | 0.125558 (rank : 47) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNG1_HUMAN
|
||||||
| NC score | 0.255504 (rank : 44) | θ value | 0.279714 (rank : 54) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNS1_HUMAN
|
||||||
| NC score | 0.255254 (rank : 45) | θ value | 0.813845 (rank : 65) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNC4_MOUSE
|
||||||
| NC score | 0.254600 (rank : 46) | θ value | 0.365318 (rank : 59) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNC4_HUMAN
|
||||||
| NC score | 0.253955 (rank : 47) | θ value | 0.365318 (rank : 58) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNF1_HUMAN
|
||||||
| NC score | 0.249705 (rank : 48) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCNG4_HUMAN
|
||||||
| NC score | 0.247372 (rank : 49) | θ value | 0.62314 (rank : 64) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNC1_MOUSE
|
||||||
| NC score | 0.247356 (rank : 50) | θ value | 0.365318 (rank : 57) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC1_HUMAN
|
||||||
| NC score | 0.247293 (rank : 51) | θ value | 0.365318 (rank : 56) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC3_MOUSE
|
||||||
| NC score | 0.247046 (rank : 52) | θ value | 0.279714 (rank : 53) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNS1_MOUSE
|
||||||
| NC score | 0.245383 (rank : 53) | θ value | 1.06291 (rank : 68) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNV2_HUMAN
|
||||||
| NC score | 0.244163 (rank : 54) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
KCNC3_HUMAN
|
||||||
| NC score | 0.243924 (rank : 55) | θ value | 0.47712 (rank : 61) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCMA1_MOUSE
|
||||||
| NC score | 0.214386 (rank : 56) | θ value | 0.21417 (rank : 50) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q08460, Q64703, Q8VHF1, Q9R196 | Gene names | Kcnma1, Kcnma | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (mSlo) (mSlo1). | |||||
|
KCMA1_HUMAN
|
||||||
| NC score | 0.214005 (rank : 57) | θ value | 0.21417 (rank : 49) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q12791, Q12886, Q12917, Q12921, Q12960, Q13150, Q96LG8, Q9UBB0, Q9UQK6 | Gene names | KCNMA1, KCNMA | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (hSlo). | |||||
|
KCNK1_HUMAN
|
||||||
| NC score | 0.136484 (rank : 58) | θ value | 0.125558 (rank : 48) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O00180, Q13307 | Gene names | KCNK1, HOHO1, KCNO1, TWIK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1) (Potassium channel KCNO1). | |||||
|
KCNK6_HUMAN
|
||||||
| NC score | 0.134896 (rank : 59) | θ value | 0.47712 (rank : 62) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
KCNK1_MOUSE
|
||||||
| NC score | 0.121598 (rank : 60) | θ value | 0.365318 (rank : 60) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O08581 | Gene names | Kcnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1). | |||||
|
KCNKF_HUMAN
|
||||||
| NC score | 0.115460 (rank : 61) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9H427, Q9HBC8 | Gene names | KCNK15, TASK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 15 (Acid-sensitive potassium channel protein TASK-5) (TWIK-related acid-sensitive K(+) channel 5) (Two pore potassium channel KT3.3). | |||||
|
KCNKA_HUMAN
|
||||||
| NC score | 0.114835 (rank : 62) | θ value | 6.88961 (rank : 105) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P57789, Q8TDK7, Q8TDK8, Q9HB59 | Gene names | KCNK10, TREK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 10 (Outward rectifying potassium channel protein TREK-2) (TREK-2 K(+) channel subunit). | |||||
|
KCNK5_HUMAN
|
||||||
| NC score | 0.114114 (rank : 63) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O95279 | Gene names | KCNK5, TASK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 5 (Acid-sensitive potassium channel protein TASK-2) (TWIK-related acid-sensitive K(+) channel 2). | |||||
|
KCNK9_HUMAN
|
||||||
| NC score | 0.109845 (rank : 64) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9NPC2, Q2M290, Q540F2 | Gene names | KCNK9, TASK3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3) (Two pore potassium channel KT3.2). | |||||
|
KCNK9_MOUSE
|
||||||
| NC score | 0.109356 (rank : 65) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q3LS21 | Gene names | Kcnk9, Task3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3). | |||||
|
KCNK3_HUMAN
|
||||||
| NC score | 0.098080 (rank : 66) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O14649 | Gene names | KCNK3, TASK, TASK1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Two pore potassium channel KT3.1). | |||||
|
KCNK3_MOUSE
|
||||||
| NC score | 0.098071 (rank : 67) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O35111, O35163 | Gene names | Kcnk3, Ctbak, Task, Task1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Cardiac two-pore background K(+) channel) (cTBAK-1) (Two pore potassium channel KT3.1). | |||||
|
KCNKH_HUMAN
|
||||||
| NC score | 0.089168 (rank : 68) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q96T54, Q5TCF4, Q8TAW4, Q9BXD1, Q9H592 | Gene names | KCNK17, TALK2, TASK4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 17 (TWIK-related alkaline pH- activated K(+) channel 2) (2P domain potassium channel Talk-2) (TWIK- related acid-sensitive K(+) channel 4) (TASK-4). | |||||
|
KCNKG_HUMAN
|
||||||
| NC score | 0.088057 (rank : 69) | θ value | 6.88961 (rank : 107) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q96T55, Q5TCF3, Q9H591 | Gene names | KCNK16, TALK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 16 (TWIK-related alkaline pH- activated K(+) channel 1) (2P domain potassium channel Talk-1). | |||||
|
KCNT1_HUMAN
|
||||||
| NC score | 0.084216 (rank : 70) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q5JUK3, Q9P2C5 | Gene names | KCNT1, KIAA1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1 (KCa4.1). | |||||
|
KCNK4_MOUSE
|
||||||
| NC score | 0.082353 (rank : 71) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O88454 | Gene names | Kcnk4, Traak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK). | |||||
|
KCNK2_MOUSE
|
||||||
| NC score | 0.079493 (rank : 72) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P97438 | Gene names | Kcnk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (Two-pore potassium channel TPKC1) (TREK-1 K(+) channel subunit). | |||||
|
KCNK2_HUMAN
|
||||||
| NC score | 0.074722 (rank : 73) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O95069, Q9UNE3 | Gene names | KCNK2, TREK, TREK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (TREK-1 K(+) channel subunit) (Two-pore potassium channel TPKC1). | |||||
|
KCNKD_HUMAN
|
||||||
| NC score | 0.067150 (rank : 74) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HB14, Q96E79 | Gene names | KCNK13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCNKD_MOUSE
|
||||||
| NC score | 0.060270 (rank : 75) | θ value | 6.88961 (rank : 106) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8R1P5 | Gene names | Kcnk13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCNK4_HUMAN
|
||||||
| NC score | 0.060025 (rank : 76) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
KCTD1_HUMAN
|
||||||
| NC score | 0.054134 (rank : 77) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q719H9 | Gene names | KCTD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD1. | |||||
|
KCTD1_MOUSE
|
||||||
| NC score | 0.054134 (rank : 78) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5M956, Q5EER9 | Gene names | Kctd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD1. | |||||
|
KCNK7_MOUSE
|
||||||
| NC score | 0.053925 (rank : 79) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Z2T1, Q9QXY0, Q9QYE8, Q9R1V1, Q9R242 | Gene names | Kcnk7, Dpkch3, Kcnk6, Kcnk8, Knot1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7 (Putative potassium channel DP3) (Double-pore K(+) channel 3) (Neuromuscular two p domain potassium channel). | |||||
|
KCTD6_MOUSE
|
||||||
| NC score | 0.053768 (rank : 80) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BNL5, Q3TXX1 | Gene names | Kctd6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD6. | |||||
|
KCTD6_HUMAN
|
||||||
| NC score | 0.053412 (rank : 81) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8NC69, Q8NBS6, Q8TCA6 | Gene names | KCTD6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD6. | |||||
|
KCD15_HUMAN
|
||||||
| NC score | 0.052958 (rank : 82) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96SI1, Q9BVI6 | Gene names | KCTD15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD15. | |||||
|
KCD15_MOUSE
|
||||||
| NC score | 0.050683 (rank : 83) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K0E1 | Gene names | Kctd15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein KCTD15. | |||||
|
AAT1_HUMAN
|
||||||
| NC score | 0.045813 (rank : 84) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7Z4T9, Q68DX2, Q8TD41, Q96A45, Q96JE8 | Gene names | AAT1, C3orf15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AMY-1-associating protein expressed in testis 1 (AAT-1). | |||||
|
WWC1_HUMAN
|
||||||
| NC score | 0.044840 (rank : 85) | θ value | 0.0252991 (rank : 34) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IX03, O94946, Q6MZX4, Q6Y2F8, Q7Z4G8, Q8WVM4, Q9BT29 | Gene names | WWC1, KIAA0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA) (HBeAg-binding protein 3). | |||||
|
WWC1_MOUSE
|
||||||
| NC score | 0.040756 (rank : 86) | θ value | 0.21417 (rank : 52) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5SXA9, Q571D0, Q8K1Y3, Q8VD17, Q922W3 | Gene names | Wwc1, Kiaa0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA). | |||||
|
ANGL7_MOUSE
|
||||||
| NC score | 0.036027 (rank : 87) | θ value | 0.0563607 (rank : 38) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R1Q3 | Gene names | Angptl7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiopoietin-related protein 7 precursor (Angiopoietin-like 7). | |||||
|
GPS2_HUMAN
|
||||||
| NC score | 0.035011 (rank : 88) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13227 | Gene names | GPS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein pathway suppressor 2 (Protein GPS2). | |||||
|
RAB3I_MOUSE
|
||||||
| NC score | 0.032512 (rank : 89) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q68EF0 | Gene names | Rab3ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAB3A-interacting protein (Rabin-3) (SSX2-interacting protein). | |||||
|
COGA1_HUMAN
|
||||||
| NC score | 0.029882 (rank : 90) | θ value | 0.00665767 (rank : 24) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q07092 | Gene names | COL16A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVI) chain precursor. | |||||
|
KCNH4_HUMAN
|
||||||
| NC score | 0.029320 (rank : 91) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
HOME1_MOUSE
|
||||||
| NC score | 0.027615 (rank : 92) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 630 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Z2Y3, Q8K3E1, Q8K4M8, Q9Z0E9, Q9Z216 | Gene names | Homer1, Vesl1 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1 (VASP/Ena-related gene up-regulated during seizure and LTP) (Vesl-1). | |||||
|
JUND_MOUSE
|
||||||
| NC score | 0.026599 (rank : 93) | θ value | 0.0961366 (rank : 43) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P15066 | Gene names | Jund, Jun-d, Jund1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor jun-D. | |||||
|
HOME1_HUMAN
|
||||||
| NC score | 0.024715 (rank : 94) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 564 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q86YM7, O96003, Q86YM5 | Gene names | HOMER1, SYN47 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1. | |||||
|
COFA1_HUMAN
|
||||||
| NC score | 0.022613 (rank : 95) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P39059, Q5T6J4, Q9Y4W4 | Gene names | COL15A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV) (Restin) (Related to endostatin)]. | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.021587 (rank : 96) | θ value | 0.0736092 (rank : 42) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
FBX41_HUMAN
|
||||||
| NC score | 0.020591 (rank : 97) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TF61 | Gene names | FBXO41, FBX41, KIAA1940 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 41. | |||||
|
COIA1_HUMAN
|
||||||
| NC score | 0.018988 (rank : 98) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
RPGF1_HUMAN
|
||||||
| NC score | 0.017672 (rank : 99) | θ value | 1.06291 (rank : 70) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13905 | Gene names | RAPGEF1, GRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 1 (Guanine nucleotide-releasing factor 2) (C3G protein) (CRK SH3-binding GNRP). | |||||
|
CEP55_MOUSE
|
||||||
| NC score | 0.017243 (rank : 100) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BT07, Q8C2J0, Q8K2I8, Q8R2Y4, Q9DBZ8 | Gene names | Cep55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55. | |||||
|
CO9A1_MOUSE
|
||||||
| NC score | 0.015402 (rank : 101) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CO9A1_HUMAN
|
||||||
| NC score | 0.014650 (rank : 102) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CABL2_HUMAN
|
||||||
| NC score | 0.014248 (rank : 103) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BTV7, Q5JWL0, Q9BYK0 | Gene names | CABLES2, C20orf150 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
LMNB1_MOUSE
|
||||||
| NC score | 0.014235 (rank : 104) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P14733, Q61791 | Gene names | Lmnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
TXLNG_HUMAN
|
||||||
| NC score | 0.014160 (rank : 105) | θ value | 1.81305 (rank : 81) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NUQ3, Q9P0X1 | Gene names | TXLNG, CXorf15, LSR5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gamma-taxilin (Lipopolysaccharide-specific response protein 5). | |||||
|
IFT81_MOUSE
|
||||||
| NC score | 0.013924 (rank : 106) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
LMNB1_HUMAN
|
||||||
| NC score | 0.013836 (rank : 107) | θ value | 6.88961 (rank : 108) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P20700, Q3SYN7, Q96EI6 | Gene names | LMNB1, LMN2, LMNB | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
TAXB1_HUMAN
|
||||||
| NC score | 0.012934 (rank : 108) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
CO2A1_HUMAN
|
||||||
| NC score | 0.012858 (rank : 109) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P02458, Q12985, Q14009, Q14044, Q14046, Q14056, Q14058, Q16672, Q6LBY1, Q6LBY2, Q6LBY3, Q99227, Q9UE38, Q9UE39, Q9UE40, Q9UE41, Q9UE42, Q9UE43 | Gene names | COL2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
CO3A1_MOUSE
|
||||||
| NC score | 0.012441 (rank : 110) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
DIAP1_HUMAN
|
||||||
| NC score | 0.011838 (rank : 111) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60610, Q9UC76 | Gene names | DIAPH1, DIAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1). | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.011702 (rank : 112) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
DIAP1_MOUSE
|
||||||
| NC score | 0.011485 (rank : 113) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 839 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O08808 | Gene names | Diaph1, Diap1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 1 (Diaphanous-related formin-1) (DRF1) (mDIA1) (p140mDIA). | |||||
|
ROCK2_HUMAN
|
||||||
| NC score | 0.011461 (rank : 114) | θ value | 0.21417 (rank : 51) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
SMC2_MOUSE
|
||||||
| NC score | 0.011392 (rank : 115) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.011152 (rank : 116) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.010967 (rank : 117) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
MD1L1_MOUSE
|
||||||
| NC score | 0.010775 (rank : 118) | θ value | 6.88961 (rank : 109) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.010657 (rank : 119) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
SF3B4_HUMAN
|
||||||
| NC score | 0.010454 (rank : 120) | θ value | 1.81305 (rank : 80) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
ROCK2_MOUSE
|
||||||
| NC score | 0.010221 (rank : 121) | θ value | 0.47712 (rank : 63) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
COAA1_HUMAN
|
||||||
| NC score | 0.008534 (rank : 122) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q03692 | Gene names | COL10A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(X) chain precursor. | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.008262 (rank : 123) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.007828 (rank : 124) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.007659 (rank : 125) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
ZN409_HUMAN
|
||||||
| NC score | 0.007451 (rank : 126) | θ value | 1.81305 (rank : 82) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.006935 (rank : 127) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
WASP_HUMAN
|
||||||
| NC score | 0.006867 (rank : 128) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P42768, Q9BU11, Q9UNJ9 | Gene names | WAS, IMD2 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein (WASp). | |||||
|
TEAD3_MOUSE
|
||||||
| NC score | 0.006533 (rank : 129) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70210, O08516, O70623, P70209 | Gene names | Tead3, Tcf13r2, Tef5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional enhancer factor TEF-5 (TEA domain family member 3) (TEAD-3) (ETF-related factor 1) (ETFR-1) (DTEF-1). | |||||
|
PEF1_MOUSE
|
||||||
| NC score | 0.005389 (rank : 130) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BFY6, Q8VCT5, Q9CYW8, Q9D934 | Gene names | Pef1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
INHBB_HUMAN
|
||||||
| NC score | 0.004653 (rank : 131) | θ value | 5.27518 (rank : 95) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P09529, Q8N1D3 | Gene names | INHBB | |||
|
Domain Architecture |

|
|||||
| Description | Inhibin beta B chain precursor (Activin beta-B chain). | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.003588 (rank : 132) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
TENX_HUMAN
|
||||||
| NC score | 0.003494 (rank : 133) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.002693 (rank : 134) | θ value | 1.06291 (rank : 69) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | -0.000827 (rank : 135) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
ZN575_HUMAN
|
||||||
| NC score | -0.003623 (rank : 136) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 125 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86XF7 | Gene names | ZNF575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 575. | |||||