Please be patient as the page loads
|
KCNH4_HUMAN
|
||||||
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
KCNH3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.980859 (rank : 3) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
KCNH3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.980755 (rank : 4) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
KCNH4_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 177 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
KCNH8_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.988115 (rank : 2) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96L42 | Gene names | KCNH8 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 8 (Voltage-gated potassium channel subunit Kv12.1) (Ether-a-go-go-like potassium channel 3) (ELK channel 3) (ELK3) (ELK1) (hElk1). | |||||
|
KCNH6_HUMAN
|
||||||
| θ value | 2.85966e-155 (rank : 5) | NC score | 0.965650 (rank : 5) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
KCNH1_HUMAN
|
||||||
| θ value | 1.09426e-130 (rank : 6) | NC score | 0.963922 (rank : 9) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O95259, O76035 | Gene names | KCNH1, EAG, EAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (hEAG1) (h-eag). | |||||
|
KCNH1_MOUSE
|
||||||
| θ value | 1.86652e-130 (rank : 7) | NC score | 0.964032 (rank : 8) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
KCNH5_HUMAN
|
||||||
| θ value | 3.1838e-130 (rank : 8) | NC score | 0.964158 (rank : 7) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
KCNH5_MOUSE
|
||||||
| θ value | 4.15819e-130 (rank : 9) | NC score | 0.964338 (rank : 6) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q920E3, Q8C035 | Gene names | Kcnh5, Eag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (Eag2). | |||||
|
KCNH2_MOUSE
|
||||||
| θ value | 4.60808e-121 (rank : 10) | NC score | 0.957133 (rank : 11) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
KCNH2_HUMAN
|
||||||
| θ value | 6.01832e-121 (rank : 11) | NC score | 0.957521 (rank : 10) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
KCNH7_MOUSE
|
||||||
| θ value | 2.61662e-116 (rank : 12) | NC score | 0.956178 (rank : 12) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
KCNH7_HUMAN
|
||||||
| θ value | 3.1986e-114 (rank : 13) | NC score | 0.954927 (rank : 13) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
HCN3_HUMAN
|
||||||
| θ value | 2.96089e-35 (rank : 14) | NC score | 0.794494 (rank : 21) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
CNGA1_MOUSE
|
||||||
| θ value | 1.12514e-34 (rank : 15) | NC score | 0.805134 (rank : 17) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA1_HUMAN
|
||||||
| θ value | 2.12192e-33 (rank : 16) | NC score | 0.809193 (rank : 15) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
HCN1_HUMAN
|
||||||
| θ value | 3.61944e-33 (rank : 17) | NC score | 0.798329 (rank : 19) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
|
HCN1_MOUSE
|
||||||
| θ value | 3.61944e-33 (rank : 18) | NC score | 0.794930 (rank : 20) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
HCN3_MOUSE
|
||||||
| θ value | 4.72714e-33 (rank : 19) | NC score | 0.794314 (rank : 22) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
CNGA3_HUMAN
|
||||||
| θ value | 6.17384e-33 (rank : 20) | NC score | 0.804260 (rank : 18) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16281, Q9UP64 | Gene names | CNGA3, CNCG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 6.17384e-33 (rank : 21) | NC score | 0.775166 (rank : 26) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
HCN4_HUMAN
|
||||||
| θ value | 1.0531e-32 (rank : 22) | NC score | 0.780310 (rank : 24) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
HCN2_MOUSE
|
||||||
| θ value | 2.34606e-32 (rank : 23) | NC score | 0.782878 (rank : 23) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 5.22648e-32 (rank : 24) | NC score | 0.777796 (rank : 25) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
CNGA3_MOUSE
|
||||||
| θ value | 5.77852e-31 (rank : 25) | NC score | 0.807657 (rank : 16) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9JJZ8, Q9WV01 | Gene names | Cnga3, Cng3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA2_MOUSE
|
||||||
| θ value | 1.28732e-30 (rank : 26) | NC score | 0.818220 (rank : 14) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q62398 | Gene names | Cnga2, Cncg2, Cncg4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated olfactory channel (Cyclic nucleotide-gated cation channel 2) (CNG channel 2) (CNG-2) (CNG2). | |||||
|
CNGB1_HUMAN
|
||||||
| θ value | 5.07402e-19 (rank : 27) | NC score | 0.728513 (rank : 29) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
CNGB3_MOUSE
|
||||||
| θ value | 8.10077e-17 (rank : 28) | NC score | 0.743110 (rank : 27) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JJZ9 | Gene names | Cngb3, Cng6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit) (Cyclic nucleotide-gated channel subunit CNG6). | |||||
|
CNGB3_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 29) | NC score | 0.730724 (rank : 28) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
KCNKH_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 30) | NC score | 0.243485 (rank : 30) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96T54, Q5TCF4, Q8TAW4, Q9BXD1, Q9H592 | Gene names | KCNK17, TALK2, TASK4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 17 (TWIK-related alkaline pH- activated K(+) channel 2) (2P domain potassium channel Talk-2) (TWIK- related acid-sensitive K(+) channel 4) (TASK-4). | |||||
|
KCNS2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 31) | NC score | 0.051069 (rank : 67) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 32) | NC score | 0.050616 (rank : 68) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
FOSL1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 33) | NC score | 0.038708 (rank : 77) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48755, O35285 | Gene names | Fosl1, Fra1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
GA2L1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 34) | NC score | 0.027219 (rank : 103) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99501, Q53EN7, Q92640, Q9BUY9 | Gene names | GAS2L1, GAR22 | |||
|
Domain Architecture |

|
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1) (GAS2-related protein on chromosome 22) (GAR22 protein). | |||||
|
PLIN_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 35) | NC score | 0.031239 (rank : 92) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CGN5 | Gene names | Plin, Peri | |||
|
Domain Architecture |

|
|||||
| Description | Perilipin (PERI) (Lipid droplet-associated protein). | |||||
|
KCND1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 36) | NC score | 0.080980 (rank : 51) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 37) | NC score | 0.027461 (rank : 102) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
KCNK3_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 38) | NC score | 0.172990 (rank : 32) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O14649 | Gene names | KCNK3, TASK, TASK1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Two pore potassium channel KT3.1). | |||||
|
KCNK4_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 39) | NC score | 0.159814 (rank : 38) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 40) | NC score | 0.032688 (rank : 90) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
AGGF1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 41) | NC score | 0.036441 (rank : 83) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TN31, Q8R2S6, Q9CQR9, Q9CU87, Q9D768 | Gene names | Aggf1, Vg5q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (mVG5Q). | |||||
|
KCND1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 42) | NC score | 0.075094 (rank : 54) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCND2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 43) | NC score | 0.073564 (rank : 56) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 44) | NC score | 0.073593 (rank : 55) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
TS101_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 45) | NC score | 0.026030 (rank : 107) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99816, Q9BUM5 | Gene names | TSG101 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
KCNKF_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 46) | NC score | 0.181314 (rank : 31) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H427, Q9HBC8 | Gene names | KCNK15, TASK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 15 (Acid-sensitive potassium channel protein TASK-5) (TWIK-related acid-sensitive K(+) channel 5) (Two pore potassium channel KT3.3). | |||||
|
CEBPA_MOUSE
|
||||||
| θ value | 0.125558 (rank : 47) | NC score | 0.047384 (rank : 70) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P53566, Q91XB6 | Gene names | Cebpa | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
CEBPD_MOUSE
|
||||||
| θ value | 0.125558 (rank : 48) | NC score | 0.045896 (rank : 71) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q00322 | Gene names | Cebpd, Crp3 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein delta (C/EBP delta) (C/EBP-related protein 3). | |||||
|
KCNK4_MOUSE
|
||||||
| θ value | 0.125558 (rank : 49) | NC score | 0.148131 (rank : 40) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O88454 | Gene names | Kcnk4, Traak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK). | |||||
|
KCNK6_HUMAN
|
||||||
| θ value | 0.125558 (rank : 50) | NC score | 0.166799 (rank : 37) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
KCND3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 51) | NC score | 0.071730 (rank : 61) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 52) | NC score | 0.071706 (rank : 62) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCNK3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 53) | NC score | 0.168974 (rank : 35) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O35111, O35163 | Gene names | Kcnk3, Ctbak, Task, Task1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Cardiac two-pore background K(+) channel) (cTBAK-1) (Two pore potassium channel KT3.1). | |||||
|
KCNKA_HUMAN
|
||||||
| θ value | 0.163984 (rank : 54) | NC score | 0.167517 (rank : 36) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P57789, Q8TDK7, Q8TDK8, Q9HB59 | Gene names | KCNK10, TREK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 10 (Outward rectifying potassium channel protein TREK-2) (TREK-2 K(+) channel subunit). | |||||
|
PHF1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 55) | NC score | 0.020159 (rank : 115) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43189, O60929, Q96KM7 | Gene names | PHF1 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 56) | NC score | 0.024975 (rank : 109) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
AGGF1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 57) | NC score | 0.034270 (rank : 87) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
CEBPA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 58) | NC score | 0.045409 (rank : 72) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P49715, P78319 | Gene names | CEBPA | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
NKX61_HUMAN
|
||||||
| θ value | 0.365318 (rank : 59) | NC score | 0.007632 (rank : 156) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P78426 | Gene names | NKX6-1, NKX6A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-6.1. | |||||
|
DDIT3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 60) | NC score | 0.041950 (rank : 74) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35639, Q91YW9 | Gene names | Ddit3, Chop, Gadd153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
KCNK9_HUMAN
|
||||||
| θ value | 0.47712 (rank : 61) | NC score | 0.145273 (rank : 41) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NPC2, Q2M290, Q540F2 | Gene names | KCNK9, TASK3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3) (Two pore potassium channel KT3.2). | |||||
|
KCNK9_MOUSE
|
||||||
| θ value | 0.47712 (rank : 62) | NC score | 0.145235 (rank : 42) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q3LS21 | Gene names | Kcnk9, Task3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 0.47712 (rank : 63) | NC score | 0.016314 (rank : 127) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 64) | NC score | 0.027067 (rank : 104) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
HNRPG_HUMAN
|
||||||
| θ value | 0.62314 (rank : 65) | NC score | 0.011981 (rank : 145) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P38159, Q5JQ67, Q969R3 | Gene names | RBMX, HNRPG, RBMXP1 | |||
|
Domain Architecture |
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein G (hnRNP G) (RNA-binding motif protein, X chromosome) (Glycoprotein p43). | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 66) | NC score | 0.003436 (rank : 171) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.813845 (rank : 67) | NC score | 0.017722 (rank : 121) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
IRK13_HUMAN
|
||||||
| θ value | 0.813845 (rank : 68) | NC score | 0.016836 (rank : 123) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60928, O76023, Q8N3Y4 | Gene names | KCNJ13 | |||
|
Domain Architecture |
|
|||||
| Description | Inward rectifier potassium channel 13 (Potassium channel, inwardly rectifying subfamily J member 13) (Inward rectifier K(+) channel Kir7.1). | |||||
|
KCMA1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 69) | NC score | 0.071900 (rank : 60) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q12791, Q12886, Q12917, Q12921, Q12960, Q13150, Q96LG8, Q9UBB0, Q9UQK6 | Gene names | KCNMA1, KCNMA | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (hSlo). | |||||
|
KCMA1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 70) | NC score | 0.072015 (rank : 59) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q08460, Q64703, Q8VHF1, Q9R196 | Gene names | Kcnma1, Kcnma | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (mSlo) (mSlo1). | |||||
|
KCNK2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 71) | NC score | 0.171811 (rank : 33) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O95069, Q9UNE3 | Gene names | KCNK2, TREK, TREK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (TREK-1 K(+) channel subunit) (Two-pore potassium channel TPKC1). | |||||
|
KCNK2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 72) | NC score | 0.171288 (rank : 34) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97438 | Gene names | Kcnk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (Two-pore potassium channel TPKC1) (TREK-1 K(+) channel subunit). | |||||
|
NOTC4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 73) | NC score | 0.001172 (rank : 179) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
CIC_MOUSE
|
||||||
| θ value | 1.06291 (rank : 74) | NC score | 0.015877 (rank : 132) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
EDN3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 75) | NC score | 0.018956 (rank : 118) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14138, Q03229 | Gene names | EDN3 | |||
|
Domain Architecture |
|
|||||
| Description | Endothelin-3 precursor (ET-3) (Preproendothelin-3) (PPET3). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 76) | NC score | 0.016520 (rank : 126) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 1.06291 (rank : 77) | NC score | 0.037860 (rank : 81) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NKX61_MOUSE
|
||||||
| θ value | 1.06291 (rank : 78) | NC score | 0.006263 (rank : 169) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99MA9, Q9ERQ7 | Gene names | Nkx6-1, Nkx6.1, Nkx6a | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-6.1. | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 79) | NC score | 0.019008 (rank : 117) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
ABL1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 80) | NC score | -0.002158 (rank : 187) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P00520, P97896, Q61252, Q61253, Q61254, Q61255, Q61256, Q61257, Q61258, Q61259, Q61260, Q61261, Q6PCM5, Q8C1X4 | Gene names | Abl1, Abl | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
CREB5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 81) | NC score | 0.017108 (rank : 122) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
DVL3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 82) | NC score | 0.010874 (rank : 148) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61062 | Gene names | Dvl3 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
H6ST3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 83) | NC score | 0.011854 (rank : 146) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZP7, Q5W0L0, Q68CW6 | Gene names | HS6ST3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 84) | NC score | 0.034860 (rank : 85) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | 0.007181 (rank : 161) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
FOSL1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.027008 (rank : 105) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P15407 | Gene names | FOSL1, FRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
HNF1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 87) | NC score | 0.012523 (rank : 143) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20823, Q99861 | Gene names | TCF1, HNF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1) (Transcription factor 1) (TCF-1). | |||||
|
KCNN2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 88) | NC score | 0.038828 (rank : 76) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H2S1 | Gene names | KCNN2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 89) | NC score | 0.038859 (rank : 75) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P58390 | Gene names | Kcnn2, Sk2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
MORN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 90) | NC score | 0.015850 (rank : 133) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5T089, Q8WW30, Q9H852 | Gene names | MORN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 1. | |||||
|
MYRIP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 91) | NC score | 0.016541 (rank : 124) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 92) | NC score | 0.022008 (rank : 112) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
PI5PA_MOUSE
|
||||||
| θ value | 1.81305 (rank : 93) | NC score | 0.016130 (rank : 130) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
RIN3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 94) | NC score | 0.014653 (rank : 138) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TB24, Q8NF30, Q8TEE8, Q8WYP4, Q9H6A5, Q9HAG1 | Gene names | RIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
CEBPB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 95) | NC score | 0.035978 (rank : 84) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P28033 | Gene names | Cebpb | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Interleukin-6- dependent-binding protein) (IL-6DBP) (Liver-enriched transcriptional activator) (LAP) (AGP/EBP). | |||||
|
CEBPD_HUMAN
|
||||||
| θ value | 2.36792 (rank : 96) | NC score | 0.037344 (rank : 82) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49716, Q14937 | Gene names | CEBPD | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein delta (C/EBP delta) (Nuclear factor NF- IL6-beta) (NF-IL6-beta). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 97) | NC score | -0.002103 (rank : 186) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 98) | NC score | -0.000411 (rank : 182) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 99) | NC score | 0.034742 (rank : 86) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
IRK9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 100) | NC score | 0.007358 (rank : 159) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92806, Q5JW75 | Gene names | KCNJ9, GIRK3 | |||
|
Domain Architecture |
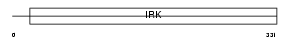
|
|||||
| Description | G protein-activated inward rectifier potassium channel 3 (GIRK3) (Potassium channel, inwardly rectifying subfamily J member 9) (Inwardly rectifier K(+) channel Kir3.3). | |||||
|
IRK9_MOUSE
|
||||||
| θ value | 2.36792 (rank : 101) | NC score | 0.007375 (rank : 158) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48543, Q9WUE1 | Gene names | Kcnj9, Girk3 | |||
|
Domain Architecture |
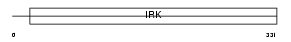
|
|||||
| Description | G protein-activated inward rectifier potassium channel 3 (GIRK3) (Potassium channel, inwardly rectifying subfamily J member 9) (Inwardly rectifier K(+) channel Kir3.3). | |||||
|
KGP1A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 102) | NC score | 0.038514 (rank : 78) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13976 | Gene names | PRKG1, PRKGR1A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, alpha isozyme (EC 2.7.11.12) (CGK 1 alpha) (cGKI-alpha). | |||||
|
KGP1B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 103) | NC score | 0.038497 (rank : 79) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 921 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P14619 | Gene names | PRKG1, PRKG1B, PRKGR1B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
MAVS_MOUSE
|
||||||
| θ value | 2.36792 (rank : 104) | NC score | 0.016173 (rank : 129) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VCF0 | Gene names | Mavs, Ips1, Visa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif). | |||||
|
PS1C2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 105) | NC score | 0.028551 (rank : 98) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80ZC9 | Gene names | Psors1c2, Spr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein homolog precursor (SPR1 protein). | |||||
|
SF3B4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 106) | NC score | 0.013061 (rank : 140) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
WNK4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 107) | NC score | -0.000001 (rank : 180) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
ANR25_HUMAN
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.006876 (rank : 164) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q63ZY3, Q3KQZ3, Q6GUF5, Q9H8S4, Q9NUP0, Q9P210 | Gene names | ANKRD25, KIAA1518, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25 (SRC-1-interacting protein) (SRC1-interacting protein). | |||||
|
CEBPE_HUMAN
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | 0.043139 (rank : 73) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
CENPA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 110) | NC score | 0.009920 (rank : 151) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49450, Q53T74, Q9BVW2 | Gene names | CENPA | |||
|
Domain Architecture |
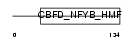
|
|||||
| Description | Histone H3-like centromeric protein A (Centromere protein A) (CENP-A) (Centromere autoantigen A). | |||||
|
CJ026_MOUSE
|
||||||
| θ value | 3.0926 (rank : 111) | NC score | 0.014037 (rank : 139) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BGW2, Q3TF48, Q3U3M2, Q6PD43, Q8R0W8 | Gene names | D19Wsu162e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C10orf26 homolog. | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 112) | NC score | 0.002879 (rank : 174) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
DDIT3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 113) | NC score | 0.030772 (rank : 94) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35638 | Gene names | DDIT3, CHOP, GADD153 | |||
|
Domain Architecture |

|
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
HNRPG_MOUSE
|
||||||
| θ value | 3.0926 (rank : 114) | NC score | 0.009832 (rank : 152) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35479 | Gene names | Rbmx, Hnrnpg, Hnrpg, Rbmxp1, Rbmxrt | |||
|
Domain Architecture |
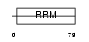
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein G (hnRNP G) (RNA-binding motif protein, X chromosome). | |||||
|
KCNKG_HUMAN
|
||||||
| θ value | 3.0926 (rank : 115) | NC score | 0.152470 (rank : 39) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96T55, Q5TCF3, Q9H591 | Gene names | KCNK16, TALK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 16 (TWIK-related alkaline pH- activated K(+) channel 1) (2P domain potassium channel Talk-1). | |||||
|
M3K11_HUMAN
|
||||||
| θ value | 3.0926 (rank : 116) | NC score | -0.002828 (rank : 189) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1136 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q16584, Q6P2G4 | Gene names | MAP3K11, MLK3, SPRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 11 (EC 2.7.11.25) (Mixed lineage kinase 3) (Src-homology 3 domain-containing proline- rich kinase). | |||||
|
NFAC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 117) | NC score | 0.008559 (rank : 155) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
PLXB1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 118) | NC score | 0.006770 (rank : 166) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
CEBPB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 119) | NC score | 0.034064 (rank : 89) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P17676, Q96IH2, Q9H4Z5 | Gene names | CEBPB, TCF5 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Nuclear factor NF- IL6) (Transcription factor 5). | |||||
|
DEK_MOUSE
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | 0.017857 (rank : 119) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
KCNK1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.118135 (rank : 44) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O00180, Q13307 | Gene names | KCNK1, HOHO1, KCNO1, TWIK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1) (Potassium channel KCNO1). | |||||
|
KCNN3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 122) | NC score | 0.034258 (rank : 88) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P58391 | Gene names | Kcnn3, Sk3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3). | |||||
|
NCOR2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 123) | NC score | 0.027868 (rank : 100) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
TMM51_HUMAN
|
||||||
| θ value | 4.03905 (rank : 124) | NC score | 0.016524 (rank : 125) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NW97 | Gene names | TMEM51, C1orf72 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 51. | |||||
|
VASP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 125) | NC score | 0.012654 (rank : 142) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P50552, Q6PIZ1, Q93035 | Gene names | VASP | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
VASP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 126) | NC score | 0.015611 (rank : 135) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P70460, Q3TAP0, Q3TCD2, Q3U0C2, Q3UDF1, Q91VD2, Q9R214 | Gene names | Vasp | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 127) | NC score | 0.016236 (rank : 128) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
BCAR3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 128) | NC score | 0.006094 (rank : 170) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QZK2, Q3TNC9, Q3UP10 | Gene names | Bcar3, And34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast cancer anti-estrogen resistance protein 3 (p130Cas-binding protein AND-34). | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.030855 (rank : 93) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
CS016_HUMAN
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.015323 (rank : 137) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.027802 (rank : 101) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
DPPA4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.009580 (rank : 153) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7L190, Q9H9N5, Q9NVI6 | Gene names | DPPA4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Developmental pluripotency-associated protein 4. | |||||
|
K1802_MOUSE
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.012196 (rank : 144) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
KCNK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.113652 (rank : 47) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O08581 | Gene names | Kcnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1). | |||||
|
KCNN3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 135) | NC score | 0.029572 (rank : 95) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UGI6, O43517 | Gene names | KCNN3, K3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3) (SKCa3). | |||||
|
KGP1B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 136) | NC score | 0.038059 (rank : 80) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Z0Z0 | Gene names | Prkg1, Prkg1b, Prkgr1b | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 137) | NC score | 0.028201 (rank : 99) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
NOTC4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 138) | NC score | -0.000029 (rank : 181) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 139) | NC score | 0.032609 (rank : 91) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 140) | NC score | 0.015760 (rank : 134) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
UBQL4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 141) | NC score | 0.006432 (rank : 168) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99NB8, Q8BP88 | Gene names | Ubqln4, Ubin | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-4 (Ataxin-1 ubiquitin-like-interacting protein A1U). | |||||
|
WASL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 142) | NC score | 0.015949 (rank : 131) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
ZN217_HUMAN
|
||||||
| θ value | 5.27518 (rank : 143) | NC score | -0.001778 (rank : 185) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75362 | Gene names | ZNF217, ZABC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 217. | |||||
|
6PGL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.011087 (rank : 147) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95336 | Gene names | PGLS | |||
|
Domain Architecture |
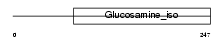
|
|||||
| Description | 6-phosphogluconolactonase (EC 3.1.1.31) (6PGL). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.022190 (rank : 111) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CT174_HUMAN
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.020171 (rank : 114) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5JPB2, Q5TDR4, Q8TCP0 | Gene names | C20orf174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf174. | |||||
|
DAZP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.006647 (rank : 167) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96EP5, Q96MJ3, Q9NRR9 | Gene names | DAZAP1 | |||
|
Domain Architecture |
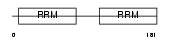
|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
DAZP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.007038 (rank : 162) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JII5 | Gene names | Dazap1 | |||
|
Domain Architecture |
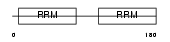
|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
DVL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.006775 (rank : 165) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92997, O14642, Q13531, Q8N5E9, Q92607 | Gene names | DVL3, KIAA0208 | |||
|
Domain Architecture |
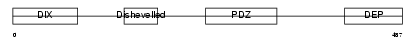
|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
ERRFI_HUMAN
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.009443 (rank : 154) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UJM3, Q9NTG9, Q9UD05 | Gene names | ERRFI1, MIG6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein) (Mig-6). | |||||
|
KCNN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.029320 (rank : 96) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q92952 | Gene names | KCNN1, SK | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCNN1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.029297 (rank : 97) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9EQR3, Q99JA9 | Gene names | Kcnn1, Sk1 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
MADCA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.010247 (rank : 149) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
RAI14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | -0.001459 (rank : 183) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
SI1L1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 155) | NC score | 0.003357 (rank : 172) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C0T5, Q6PDI8, Q80U02, Q8C026 | Gene names | Sipa1l1, Kiaa0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1. | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 156) | NC score | 0.019092 (rank : 116) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
TPO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 157) | NC score | 0.024360 (rank : 110) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
WASL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 158) | NC score | 0.015473 (rank : 136) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 159) | NC score | 0.020256 (rank : 113) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
ZN366_HUMAN
|
||||||
| θ value | 6.88961 (rank : 160) | NC score | -0.004807 (rank : 190) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N895, Q7RTV4 | Gene names | ZNF366 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 366. | |||||
|
ZN750_MOUSE
|
||||||
| θ value | 6.88961 (rank : 161) | NC score | 0.025923 (rank : 108) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
6PGL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 162) | NC score | 0.010046 (rank : 150) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CQ60, Q3TA62 | Gene names | Pgls | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 6-phosphogluconolactonase (EC 3.1.1.31) (6PGL). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | -0.002618 (rank : 188) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
BMP3B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.001678 (rank : 176) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55107, Q9UCX6 | Gene names | GDF10, BMP3B | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 3b precursor (BMP-3b) (Growth/differentiation factor 10) (GDF-10) (Bone-inducing protein) (BIP). | |||||
|
BORG3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | 0.007329 (rank : 160) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NZY7, Q8TB51 | Gene names | CDC42EP5, BORG3, CEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 5 (Binder of Rho GTPases 3). | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.002247 (rank : 175) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
GA2L2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.017828 (rank : 120) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.026966 (rank : 106) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
KCNK5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.117954 (rank : 45) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O95279 | Gene names | KCNK5, TASK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 5 (Acid-sensitive potassium channel protein TASK-2) (TWIK-related acid-sensitive K(+) channel 2). | |||||
|
KCNN4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.047432 (rank : 69) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15554 | Gene names | KCNN4, IK1, IKCA1, KCA4, SK4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1) (IKCa1) (Putative Gardos channel). | |||||
|
LZTS2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.001622 (rank : 177) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91YU6, Q569X6 | Gene names | Lzts2, Kiaa1813 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2. | |||||
|
NALP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.003196 (rank : 173) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
RFIP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.007020 (rank : 163) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D620, Q3UBC2, Q8BN24 | Gene names | Rab11fip1, Rcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
SLBP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.007529 (rank : 157) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14493 | Gene names | SLBP, HBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone RNA hairpin-binding protein (Histone stem-loop-binding protein). | |||||
|
TMM51_MOUSE
|
||||||
| θ value | 8.99809 (rank : 175) | NC score | 0.012702 (rank : 141) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99LG1, Q3TPZ3 | Gene names | Tmem51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 51. | |||||
|
TTBK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 176) | NC score | 0.001395 (rank : 178) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 177) | NC score | -0.001596 (rank : 184) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
CT152_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.059457 (rank : 66) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D5U8, Q8R083 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152 homolog. | |||||
|
KAP0_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.126785 (rank : 43) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P10644, Q567S7 | Gene names | PRKAR1A, PKR1, PRKAR1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue- specific extinguisher 1) (TSE1). | |||||
|
KAP0_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.114930 (rank : 46) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9DBC7, Q3UKU7, Q9JHR5, Q9JHR6 | Gene names | Prkar1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit. | |||||
|
KAP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.103839 (rank : 48) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.101371 (rank : 49) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.073289 (rank : 57) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P13861, Q16823 | Gene names | PRKAR2A, PKR2, PRKAR2 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.075415 (rank : 53) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12367 | Gene names | Prkar2a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.072715 (rank : 58) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P31323 | Gene names | PRKAR2B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.076071 (rank : 52) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P31324, Q3UTZ1, Q80ZM4, Q8BRZ7 | Gene names | Prkar2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KCNK7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.063607 (rank : 64) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z2T1, Q9QXY0, Q9QYE8, Q9R1V1, Q9R242 | Gene names | Kcnk7, Dpkch3, Kcnk6, Kcnk8, Knot1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7 (Putative potassium channel DP3) (Double-pore K(+) channel 3) (Neuromuscular two p domain potassium channel). | |||||
|
KCNKC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.059987 (rank : 65) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9HB15 | Gene names | KCNK12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 12 (Tandem pore domain halothane- inhibited potassium channel 2) (THIK-2). | |||||
|
KCNKD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.063794 (rank : 63) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9HB14, Q96E79 | Gene names | KCNK13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCNKD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.081180 (rank : 50) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8R1P5 | Gene names | Kcnk13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCNH4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 177 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
KCNH8_HUMAN
|
||||||
| NC score | 0.988115 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96L42 | Gene names | KCNH8 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 8 (Voltage-gated potassium channel subunit Kv12.1) (Ether-a-go-go-like potassium channel 3) (ELK channel 3) (ELK3) (ELK1) (hElk1). | |||||
|
KCNH3_HUMAN
|
||||||
| NC score | 0.980859 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9ULD8, Q9UQ06 | Gene names | KCNH3, KIAA1282 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (ELK2) (Brain-specific eag-like channel 1) (BEC1). | |||||
|
KCNH3_MOUSE
|
||||||
| NC score | 0.980755 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9WVJ0 | Gene names | Kcnh3, Elk2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 3 (Voltage-gated potassium channel subunit Kv12.2) (Ether-a-go-go-like potassium channel 2) (ELK channel 2) (mElk2). | |||||
|
KCNH6_HUMAN
|
||||||
| NC score | 0.965650 (rank : 5) | θ value | 2.85966e-155 (rank : 5) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
KCNH5_MOUSE
|
||||||
| NC score | 0.964338 (rank : 6) | θ value | 4.15819e-130 (rank : 9) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q920E3, Q8C035 | Gene names | Kcnh5, Eag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (Eag2). | |||||
|
KCNH5_HUMAN
|
||||||
| NC score | 0.964158 (rank : 7) | θ value | 3.1838e-130 (rank : 8) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
KCNH1_MOUSE
|
||||||
| NC score | 0.964032 (rank : 8) | θ value | 1.86652e-130 (rank : 7) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q60603 | Gene names | Kcnh1, Eag | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (EAG1) (m-eag). | |||||
|
KCNH1_HUMAN
|
||||||
| NC score | 0.963922 (rank : 9) | θ value | 1.09426e-130 (rank : 6) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O95259, O76035 | Gene names | KCNH1, EAG, EAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 1 (Voltage-gated potassium channel subunit Kv10.1) (Ether-a-go-go potassium channel 1) (hEAG1) (h-eag). | |||||
|
KCNH2_HUMAN
|
||||||
| NC score | 0.957521 (rank : 10) | θ value | 6.01832e-121 (rank : 11) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q12809, O75418, O75680, Q9BT72, Q9BUT7, Q9H3P0 | Gene names | KCNH2, ERG, ERG1, HERG, HERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (H-ERG) (Erg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1) (eag homolog). | |||||
|
KCNH2_MOUSE
|
||||||
| NC score | 0.957133 (rank : 11) | θ value | 4.60808e-121 (rank : 10) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O35219, O35220, O35221, O35989 | Gene names | Kcnh2, Erg, Merg1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 2 (Voltage-gated potassium channel subunit Kv11.1) (Ether-a-go-go-related gene potassium channel 1) (ERG1) (MERG) (Merg1) (Ether-a-go-go-related protein 1) (Eag-related protein 1). | |||||
|
KCNH7_MOUSE
|
||||||
| NC score | 0.956178 (rank : 12) | θ value | 2.61662e-116 (rank : 12) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9ER47 | Gene names | Kcnh7, Erg3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (Ether-a-go-go-related protein 3) (Eag-related protein 3). | |||||
|
KCNH7_HUMAN
|
||||||
| NC score | 0.954927 (rank : 13) | θ value | 3.1986e-114 (rank : 13) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NS40 | Gene names | KCNH7, ERG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 7 (Voltage-gated potassium channel subunit Kv11.3) (Ether-a-go-go-related gene potassium channel 3) (HERG-3) (Ether-a-go-go-related protein 3) (Eag- related protein 3). | |||||
|
CNGA2_MOUSE
|
||||||
| NC score | 0.818220 (rank : 14) | θ value | 1.28732e-30 (rank : 26) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q62398 | Gene names | Cnga2, Cncg2, Cncg4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated olfactory channel (Cyclic nucleotide-gated cation channel 2) (CNG channel 2) (CNG-2) (CNG2). | |||||
|
CNGA1_HUMAN
|
||||||
| NC score | 0.809193 (rank : 15) | θ value | 2.12192e-33 (rank : 16) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P29973, Q16279, Q16485 | Gene names | CNGA1, CNCG, CNCG1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA3_MOUSE
|
||||||
| NC score | 0.807657 (rank : 16) | θ value | 5.77852e-31 (rank : 25) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9JJZ8, Q9WV01 | Gene names | Cnga3, Cng3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA1_MOUSE
|
||||||
| NC score | 0.805134 (rank : 17) | θ value | 1.12514e-34 (rank : 15) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
CNGA3_HUMAN
|
||||||
| NC score | 0.804260 (rank : 18) | θ value | 6.17384e-33 (rank : 20) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q16281, Q9UP64 | Gene names | CNGA3, CNCG3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel alpha 3 (CNG channel alpha 3) (CNG-3) (CNG3) (Cyclic nucleotide-gated channel alpha 3) (Cone photoreceptor cGMP-gated channel subunit alpha). | |||||
|
HCN1_HUMAN
|
||||||
| NC score | 0.798329 (rank : 19) | θ value | 3.61944e-33 (rank : 17) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
|
HCN1_MOUSE
|
||||||
| NC score | 0.794930 (rank : 20) | θ value | 3.61944e-33 (rank : 18) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O88704, O54899, Q9D613 | Gene names | Hcn1, Bcng1, Hac2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1) (Hyperpolarization-activated cation channel 2) (HAC-2). | |||||
|
HCN3_HUMAN
|
||||||
| NC score | 0.794494 (rank : 21) | θ value | 2.96089e-35 (rank : 14) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9P1Z3, Q4VX12, Q8N6W6, Q9BWQ2 | Gene names | HCN3, KIAA1535 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3. | |||||
|
HCN3_MOUSE
|
||||||
| NC score | 0.794314 (rank : 22) | θ value | 4.72714e-33 (rank : 19) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
HCN2_MOUSE
|
||||||
| NC score | 0.782878 (rank : 23) | θ value | 2.34606e-32 (rank : 23) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.780310 (rank : 24) | θ value | 1.0531e-32 (rank : 22) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.777796 (rank : 25) | θ value | 5.22648e-32 (rank : 24) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.775166 (rank : 26) | θ value | 6.17384e-33 (rank : 21) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
CNGB3_MOUSE
|
||||||
| NC score | 0.743110 (rank : 27) | θ value | 8.10077e-17 (rank : 28) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JJZ9 | Gene names | Cngb3, Cng6 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit) (Cyclic nucleotide-gated channel subunit CNG6). | |||||
|
CNGB3_HUMAN
|
||||||
| NC score | 0.730724 (rank : 28) | θ value | 4.02038e-16 (rank : 29) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
CNGB1_HUMAN
|
||||||
| NC score | 0.728513 (rank : 29) | θ value | 5.07402e-19 (rank : 27) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14028, Q14029 | Gene names | CNGB1, CNCG4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel 4 (CNG channel 4) (CNG-4) (CNG4) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
KCNKH_HUMAN
|
||||||
| NC score | 0.243485 (rank : 30) | θ value | 0.00298849 (rank : 30) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96T54, Q5TCF4, Q8TAW4, Q9BXD1, Q9H592 | Gene names | KCNK17, TALK2, TASK4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 17 (TWIK-related alkaline pH- activated K(+) channel 2) (2P domain potassium channel Talk-2) (TWIK- related acid-sensitive K(+) channel 4) (TASK-4). | |||||
|
KCNKF_HUMAN
|
||||||
| NC score | 0.181314 (rank : 31) | θ value | 0.0961366 (rank : 46) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9H427, Q9HBC8 | Gene names | KCNK15, TASK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 15 (Acid-sensitive potassium channel protein TASK-5) (TWIK-related acid-sensitive K(+) channel 5) (Two pore potassium channel KT3.3). | |||||
|
KCNK3_HUMAN
|
||||||
| NC score | 0.172990 (rank : 32) | θ value | 0.0563607 (rank : 38) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O14649 | Gene names | KCNK3, TASK, TASK1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Two pore potassium channel KT3.1). | |||||
|
KCNK2_HUMAN
|
||||||
| NC score | 0.171811 (rank : 33) | θ value | 0.813845 (rank : 71) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O95069, Q9UNE3 | Gene names | KCNK2, TREK, TREK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (TREK-1 K(+) channel subunit) (Two-pore potassium channel TPKC1). | |||||
|
KCNK2_MOUSE
|
||||||
| NC score | 0.171288 (rank : 34) | θ value | 0.813845 (rank : 72) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97438 | Gene names | Kcnk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 2 (Outward rectifying potassium channel protein TREK-1) (Two-pore potassium channel TPKC1) (TREK-1 K(+) channel subunit). | |||||
|
KCNK3_MOUSE
|
||||||
| NC score | 0.168974 (rank : 35) | θ value | 0.163984 (rank : 53) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O35111, O35163 | Gene names | Kcnk3, Ctbak, Task, Task1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 3 (Acid-sensitive potassium channel protein TASK-1) (TWIK-related acid-sensitive K(+) channel 1) (Cardiac two-pore background K(+) channel) (cTBAK-1) (Two pore potassium channel KT3.1). | |||||
|
KCNKA_HUMAN
|
||||||
| NC score | 0.167517 (rank : 36) | θ value | 0.163984 (rank : 54) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P57789, Q8TDK7, Q8TDK8, Q9HB59 | Gene names | KCNK10, TREK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 10 (Outward rectifying potassium channel protein TREK-2) (TREK-2 K(+) channel subunit). | |||||
|
KCNK6_HUMAN
|
||||||
| NC score | 0.166799 (rank : 37) | θ value | 0.125558 (rank : 50) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y257, Q9HB47 | Gene names | KCNK6, TOSS, TWIK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 6 (Inward rectifying potassium channel protein TWIK-2) (TWIK-originated similarity sequence). | |||||
|
KCNK4_HUMAN
|
||||||
| NC score | 0.159814 (rank : 38) | θ value | 0.0563607 (rank : 39) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
KCNKG_HUMAN
|
||||||
| NC score | 0.152470 (rank : 39) | θ value | 3.0926 (rank : 115) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96T55, Q5TCF3, Q9H591 | Gene names | KCNK16, TALK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 16 (TWIK-related alkaline pH- activated K(+) channel 1) (2P domain potassium channel Talk-1). | |||||
|
KCNK4_MOUSE
|
||||||
| NC score | 0.148131 (rank : 40) | θ value | 0.125558 (rank : 49) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O88454 | Gene names | Kcnk4, Traak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK). | |||||
|
KCNK9_HUMAN
|
||||||
| NC score | 0.145273 (rank : 41) | θ value | 0.47712 (rank : 61) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NPC2, Q2M290, Q540F2 | Gene names | KCNK9, TASK3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3) (Two pore potassium channel KT3.2). | |||||
|
KCNK9_MOUSE
|
||||||
| NC score | 0.145235 (rank : 42) | θ value | 0.47712 (rank : 62) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q3LS21 | Gene names | Kcnk9, Task3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 9 (Acid-sensitive potassium channel protein TASK-3) (TWIK-related acid-sensitive K(+) channel 3). | |||||
|
KAP0_HUMAN
|
||||||
| NC score | 0.126785 (rank : 43) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P10644, Q567S7 | Gene names | PRKAR1A, PKR1, PRKAR1 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit (Tissue- specific extinguisher 1) (TSE1). | |||||
|
KCNK1_HUMAN
|
||||||
| NC score | 0.118135 (rank : 44) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O00180, Q13307 | Gene names | KCNK1, HOHO1, KCNO1, TWIK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1) (Potassium channel KCNO1). | |||||
|
KCNK5_HUMAN
|
||||||
| NC score | 0.117954 (rank : 45) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O95279 | Gene names | KCNK5, TASK2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 5 (Acid-sensitive potassium channel protein TASK-2) (TWIK-related acid-sensitive K(+) channel 2). | |||||
|
KAP0_MOUSE
|
||||||
| NC score | 0.114930 (rank : 46) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9DBC7, Q3UKU7, Q9JHR5, Q9JHR6 | Gene names | Prkar1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type I-alpha regulatory subunit. | |||||
|
KCNK1_MOUSE
|
||||||
| NC score | 0.113652 (rank : 47) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O08581 | Gene names | Kcnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 1 (Inward rectifying potassium channel protein TWIK-1). | |||||
|
KAP1_HUMAN
|
||||||
| NC score | 0.103839 (rank : 48) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P31321 | Gene names | PRKAR1B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KAP1_MOUSE
|
||||||
| NC score | 0.101371 (rank : 49) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P12849 | Gene names | Prkar1b | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type I-beta regulatory subunit. | |||||
|
KCNKD_MOUSE
|
||||||
| NC score | 0.081180 (rank : 50) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8R1P5 | Gene names | Kcnk13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCND1_MOUSE
|
||||||
| NC score | 0.080980 (rank : 51) | θ value | 0.0431538 (rank : 36) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KAP3_MOUSE
|
||||||
| NC score | 0.076071 (rank : 52) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P31324, Q3UTZ1, Q80ZM4, Q8BRZ7 | Gene names | Prkar2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KAP2_MOUSE
|
||||||
| NC score | 0.075415 (rank : 53) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12367 | Gene names | Prkar2a | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KCND1_HUMAN
|
||||||
| NC score | 0.075094 (rank : 54) | θ value | 0.0736092 (rank : 42) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCND2_MOUSE
|
||||||
| NC score | 0.073593 (rank : 55) | θ value | 0.0736092 (rank : 44) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_HUMAN
|
||||||
| NC score | 0.073564 (rank : 56) | θ value | 0.0736092 (rank : 43) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KAP2_HUMAN
|
||||||
| NC score | 0.073289 (rank : 57) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P13861, Q16823 | Gene names | PRKAR2A, PKR2, PRKAR2 | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-alpha regulatory subunit. | |||||
|
KAP3_HUMAN
|
||||||
| NC score | 0.072715 (rank : 58) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P31323 | Gene names | PRKAR2B | |||
|
Domain Architecture |

|
|||||
| Description | cAMP-dependent protein kinase type II-beta regulatory subunit. | |||||
|
KCMA1_MOUSE
|
||||||
| NC score | 0.072015 (rank : 59) | θ value | 0.813845 (rank : 70) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q08460, Q64703, Q8VHF1, Q9R196 | Gene names | Kcnma1, Kcnma | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (mSlo) (mSlo1). | |||||
|
KCMA1_HUMAN
|
||||||
| NC score | 0.071900 (rank : 60) | θ value | 0.813845 (rank : 69) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q12791, Q12886, Q12917, Q12921, Q12960, Q13150, Q96LG8, Q9UBB0, Q9UQK6 | Gene names | KCNMA1, KCNMA | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (hSlo). | |||||
|
KCND3_HUMAN
|
||||||
| NC score | 0.071730 (rank : 61) | θ value | 0.163984 (rank : 51) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_MOUSE
|
||||||
| NC score | 0.071706 (rank : 62) | θ value | 0.163984 (rank : 52) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCNKD_HUMAN
|
||||||
| NC score | 0.063794 (rank : 63) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9HB14, Q96E79 | Gene names | KCNK13 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 13 (Tandem pore domain halothane- inhibited potassium channel 1) (THIK-1). | |||||
|
KCNK7_MOUSE
|
||||||
| NC score | 0.063607 (rank : 64) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z2T1, Q9QXY0, Q9QYE8, Q9R1V1, Q9R242 | Gene names | Kcnk7, Dpkch3, Kcnk6, Kcnk8, Knot1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 7 (Putative potassium channel DP3) (Double-pore K(+) channel 3) (Neuromuscular two p domain potassium channel). | |||||
|
KCNKC_HUMAN
|
||||||
| NC score | 0.059987 (rank : 65) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9HB15 | Gene names | KCNK12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily K member 12 (Tandem pore domain halothane- inhibited potassium channel 2) (THIK-2). | |||||
|
CT152_MOUSE
|
||||||
| NC score | 0.059457 (rank : 66) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9D5U8, Q8R083 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf152 homolog. | |||||
|
KCNS2_HUMAN
|
||||||
| NC score | 0.051069 (rank : 67) | θ value | 0.0193708 (rank : 31) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_MOUSE
|
||||||
| NC score | 0.050616 (rank : 68) | θ value | 0.0193708 (rank : 32) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNN4_HUMAN
|
||||||
| NC score | 0.047432 (rank : 69) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O15554 | Gene names | KCNN4, IK1, IKCA1, KCA4, SK4 | |||
|
Domain Architecture |

|
|||||
| Description | Intermediate conductance calcium-activated potassium channel protein 4 (SK4) (KCa4) (IK1) (IKCa1) (Putative Gardos channel). | |||||
|
CEBPA_MOUSE
|
||||||
| NC score | 0.047384 (rank : 70) | θ value | 0.125558 (rank : 47) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P53566, Q91XB6 | Gene names | Cebpa | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
CEBPD_MOUSE
|
||||||
| NC score | 0.045896 (rank : 71) | θ value | 0.125558 (rank : 48) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q00322 | Gene names | Cebpd, Crp3 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein delta (C/EBP delta) (C/EBP-related protein 3). | |||||
|
CEBPA_HUMAN
|
||||||
| NC score | 0.045409 (rank : 72) | θ value | 0.279714 (rank : 58) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P49715, P78319 | Gene names | CEBPA | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein alpha (C/EBP alpha). | |||||
|
CEBPE_HUMAN
|
||||||
| NC score | 0.043139 (rank : 73) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
DDIT3_MOUSE
|
||||||
| NC score | 0.041950 (rank : 74) | θ value | 0.47712 (rank : 60) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35639, Q91YW9 | Gene names | Ddit3, Chop, Gadd153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
KCNN2_MOUSE
|
||||||
| NC score | 0.038859 (rank : 75) | θ value | 1.81305 (rank : 89) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P58390 | Gene names | Kcnn2, Sk2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
KCNN2_HUMAN
|
||||||
| NC score | 0.038828 (rank : 76) | θ value | 1.81305 (rank : 88) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H2S1 | Gene names | KCNN2 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 2 (SK2). | |||||
|
FOSL1_MOUSE
|
||||||
| NC score | 0.038708 (rank : 77) | θ value | 0.0252991 (rank : 33) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48755, O35285 | Gene names | Fosl1, Fra1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
KGP1A_HUMAN
|
||||||
| NC score | 0.038514 (rank : 78) | θ value | 2.36792 (rank : 102) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 901 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13976 | Gene names | PRKG1, PRKGR1A | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, alpha isozyme (EC 2.7.11.12) (CGK 1 alpha) (cGKI-alpha). | |||||
|
KGP1B_HUMAN
|
||||||
| NC score | 0.038497 (rank : 79) | θ value | 2.36792 (rank : 103) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 921 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P14619 | Gene names | PRKG1, PRKG1B, PRKGR1B | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
KGP1B_MOUSE
|
||||||
| NC score | 0.038059 (rank : 80) | θ value | 5.27518 (rank : 136) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Z0Z0 | Gene names | Prkg1, Prkg1b, Prkgr1b | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 1, beta isozyme (EC 2.7.11.12) (cGK 1 beta) (cGKI-beta). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.037860 (rank : 81) | θ value | 1.06291 (rank : 77) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
CEBPD_HUMAN
|
||||||
| NC score | 0.037344 (rank : 82) | θ value | 2.36792 (rank : 96) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P49716, Q14937 | Gene names | CEBPD | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein delta (C/EBP delta) (Nuclear factor NF- IL6-beta) (NF-IL6-beta). | |||||
|
AGGF1_MOUSE
|
||||||
| NC score | 0.036441 (rank : 83) | θ value | 0.0736092 (rank : 41) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TN31, Q8R2S6, Q9CQR9, Q9CU87, Q9D768 | Gene names | Aggf1, Vg5q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (mVG5Q). | |||||
|
CEBPB_MOUSE
|
||||||
| NC score | 0.035978 (rank : 84) | θ value | 2.36792 (rank : 95) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P28033 | Gene names | Cebpb | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Interleukin-6- dependent-binding protein) (IL-6DBP) (Liver-enriched transcriptional activator) (LAP) (AGP/EBP). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.034860 (rank : 85) | θ value | 1.38821 (rank : 84) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.034742 (rank : 86) | θ value | 2.36792 (rank : 99) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
AGGF1_HUMAN
|
||||||
| NC score | 0.034270 (rank : 87) | θ value | 0.279714 (rank : 57) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
KCNN3_MOUSE
|
||||||
| NC score | 0.034258 (rank : 88) | θ value | 4.03905 (rank : 122) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P58391 | Gene names | Kcnn3, Sk3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3). | |||||
|
CEBPB_HUMAN
|
||||||
| NC score | 0.034064 (rank : 89) | θ value | 4.03905 (rank : 119) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P17676, Q96IH2, Q9H4Z5 | Gene names | CEBPB, TCF5 | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein beta (C/EBP beta) (Nuclear factor NF- IL6) (Transcription factor 5). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.032688 (rank : 90) | θ value | 0.0563607 (rank : 40) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.032609 (rank : 91) | θ value | 5.27518 (rank : 139) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PLIN_MOUSE
|
||||||
| NC score | 0.031239 (rank : 92) | θ value | 0.0330416 (rank : 35) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CGN5 | Gene names | Plin, Peri | |||
|
Domain Architecture |

|
|||||
| Description | Perilipin (PERI) (Lipid droplet-associated protein). | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.030855 (rank : 93) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
DDIT3_HUMAN
|
||||||
| NC score | 0.030772 (rank : 94) | θ value | 3.0926 (rank : 113) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P35638 | Gene names | DDIT3, CHOP, GADD153 | |||
|
Domain Architecture |

|
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
KCNN3_HUMAN
|
||||||
| NC score | 0.029572 (rank : 95) | θ value | 5.27518 (rank : 135) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UGI6, O43517 | Gene names | KCNN3, K3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3) (SKCa3). | |||||
|
KCNN1_HUMAN
|
||||||
| NC score | 0.029320 (rank : 96) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q92952 | Gene names | KCNN1, SK | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
KCNN1_MOUSE
|
||||||
| NC score | 0.029297 (rank : 97) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9EQR3, Q99JA9 | Gene names | Kcnn1, Sk1 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
PS1C2_MOUSE
|
||||||
| NC score | 0.028551 (rank : 98) | θ value | 2.36792 (rank : 105) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80ZC9 | Gene names | Psors1c2, Spr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Psoriasis susceptibility 1 candidate gene 2 protein homolog precursor (SPR1 protein). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.028201 (rank : 99) | θ value | 5.27518 (rank : 137) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
NCOR2_MOUSE
|
||||||
| NC score | 0.027868 (rank : 100) | θ value | 4.03905 (rank : 123) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 790 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9WU42, Q9WU43, Q9WUC1 | Gene names | Ncor2, Smrt | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.027802 (rank : 101) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.027461 (rank : 102) | θ value | 0.0431538 (rank : 37) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
GA2L1_HUMAN
|
||||||
| NC score | 0.027219 (rank : 103) | θ value | 0.0252991 (rank : 34) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99501, Q53EN7, Q92640, Q9BUY9 | Gene names | GAS2L1, GAR22 | |||
|
Domain Architecture |

|
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1) (GAS2-related protein on chromosome 22) (GAR22 protein). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.027067 (rank : 104) | θ value | 0.47712 (rank : 64) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
FOSL1_HUMAN
|
||||||
| NC score | 0.027008 (rank : 105) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P15407 | Gene names | FOSL1, FRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Fos-related antigen 1 (FRA-1). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.026966 (rank : 106) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
TS101_HUMAN
|
||||||
| NC score | 0.026030 (rank : 107) | θ value | 0.0736092 (rank : 45) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99816, Q9BUM5 | Gene names | TSG101 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
ZN750_MOUSE
|
||||||
| NC score | 0.025923 (rank : 108) | θ value | 6.88961 (rank : 161) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BH05, Q66JP3, Q8C0L1 | Gene names | Znf750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.024975 (rank : 109) | θ value | 0.21417 (rank : 56) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
TPO_MOUSE
|
||||||
| NC score | 0.024360 (rank : 110) | θ value | 6.88961 (rank : 157) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.022190 (rank : 111) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.022008 (rank : 112) | θ value | 1.81305 (rank : 92) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.020256 (rank : 113) | θ value | 6.88961 (rank : 159) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
CT174_HUMAN
|
||||||
| NC score | 0.020171 (rank : 114) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5JPB2, Q5TDR4, Q8TCP0 | Gene names | C20orf174 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf174. | |||||
|
PHF1_HUMAN
|
||||||
| NC score | 0.020159 (rank : 115) | θ value | 0.163984 (rank : 55) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43189, O60929, Q96KM7 | Gene names | PHF1 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.019092 (rank : 116) | θ value | 6.88961 (rank : 156) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.019008 (rank : 117) | θ value | 1.06291 (rank : 79) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
EDN3_HUMAN
|
||||||
| NC score | 0.018956 (rank : 118) | θ value | 1.06291 (rank : 75) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14138, Q03229 | Gene names | EDN3 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-3 precursor (ET-3) (Preproendothelin-3) (PPET3). | |||||
|
DEK_MOUSE
|
||||||
| NC score | 0.017857 (rank : 119) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
GA2L2_HUMAN
|
||||||
| NC score | 0.017828 (rank : 120) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.017722 (rank : 121) | θ value | 0.813845 (rank : 67) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CREB5_HUMAN
|
||||||
| NC score | 0.017108 (rank : 122) | θ value | 1.38821 (rank : 81) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
IRK13_HUMAN
|
||||||
| NC score | 0.016836 (rank : 123) | θ value | 0.813845 (rank : 68) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60928, O76023, Q8N3Y4 | Gene names | KCNJ13 | |||
|
Domain Architecture |

|
|||||
| Description | Inward rectifier potassium channel 13 (Potassium channel, inwardly rectifying subfamily J member 13) (Inward rectifier K(+) channel Kir7.1). | |||||
|
MYRIP_HUMAN
|
||||||
| NC score | 0.016541 (rank : 124) | θ value | 1.81305 (rank : 91) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
TMM51_HUMAN
|
||||||
| NC score | 0.016524 (rank : 125) | θ value | 4.03905 (rank : 124) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NW97 | Gene names | TMEM51, C1orf72 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 51. | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.016520 (rank : 126) | θ value | 1.06291 (rank : 76) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.016314 (rank : 127) | θ value | 0.47712 (rank : 63) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.016236 (rank : 128) | θ value | 4.03905 (rank : 127) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
MAVS_MOUSE
|
||||||
| NC score | 0.016173 (rank : 129) | θ value | 2.36792 (rank : 104) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VCF0 | Gene names | Mavs, Ips1, Visa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif). | |||||
|
PI5PA_MOUSE
|
||||||
| NC score | 0.016130 (rank : 130) | θ value | 1.81305 (rank : 93) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
WASL_HUMAN
|
||||||
| NC score | 0.015949 (rank : 131) | θ value | 5.27518 (rank : 142) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.015877 (rank : 132) | θ value | 1.06291 (rank : 74) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
MORN1_HUMAN
|
||||||
| NC score | 0.015850 (rank : 133) | θ value | 1.81305 (rank : 90) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5T089, Q8WW30, Q9H852 | Gene names | MORN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORN repeat-containing protein 1. | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.015760 (rank : 134) | θ value | 5.27518 (rank : 140) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
VASP_MOUSE
|
||||||
| NC score | 0.015611 (rank : 135) | θ value | 4.03905 (rank : 126) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P70460, Q3TAP0, Q3TCD2, Q3U0C2, Q3UDF1, Q91VD2, Q9R214 | Gene names | Vasp | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.015473 (rank : 136) | θ value | 6.88961 (rank : 158) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.015323 (rank : 137) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
RIN3_HUMAN
|
||||||
| NC score | 0.014653 (rank : 138) | θ value | 1.81305 (rank : 94) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TB24, Q8NF30, Q8TEE8, Q8WYP4, Q9H6A5, Q9HAG1 | Gene names | RIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
CJ026_MOUSE
|
||||||
| NC score | 0.014037 (rank : 139) | θ value | 3.0926 (rank : 111) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BGW2, Q3TF48, Q3U3M2, Q6PD43, Q8R0W8 | Gene names | D19Wsu162e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C10orf26 homolog. | |||||
|
SF3B4_HUMAN
|
||||||
| NC score | 0.013061 (rank : 140) | θ value | 2.36792 (rank : 106) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
TMM51_MOUSE
|
||||||
| NC score | 0.012702 (rank : 141) | θ value | 8.99809 (rank : 175) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99LG1, Q3TPZ3 | Gene names | Tmem51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 51. | |||||
|
VASP_HUMAN
|
||||||
| NC score | 0.012654 (rank : 142) | θ value | 4.03905 (rank : 125) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P50552, Q6PIZ1, Q93035 | Gene names | VASP | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
HNF1A_HUMAN
|
||||||
| NC score | 0.012523 (rank : 143) | θ value | 1.81305 (rank : 87) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20823, Q99861 | Gene names | TCF1, HNF1A | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1) (Transcription factor 1) (TCF-1). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.012196 (rank : 144) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
HNRPG_HUMAN
|
||||||
| NC score | 0.011981 (rank : 145) | θ value | 0.62314 (rank : 65) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P38159, Q5JQ67, Q969R3 | Gene names | RBMX, HNRPG, RBMXP1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein G (hnRNP G) (RNA-binding motif protein, X chromosome) (Glycoprotein p43). | |||||
|
H6ST3_HUMAN
|
||||||
| NC score | 0.011854 (rank : 146) | θ value | 1.38821 (rank : 83) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZP7, Q5W0L0, Q68CW6 | Gene names | HS6ST3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3). | |||||
|
6PGL_HUMAN
|
||||||
| NC score | 0.011087 (rank : 147) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95336 | Gene names | PGLS | |||
|
Domain Architecture |

|
|||||
| Description | 6-phosphogluconolactonase (EC 3.1.1.31) (6PGL). | |||||
|
DVL3_MOUSE
|
||||||
| NC score | 0.010874 (rank : 148) | θ value | 1.38821 (rank : 82) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61062 | Gene names | Dvl3 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
MADCA_HUMAN
|
||||||
| NC score | 0.010247 (rank : 149) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13477, O60222, O75867 | Gene names | MADCAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (hMAdCAM-1). | |||||
|
6PGL_MOUSE
|
||||||
| NC score | 0.010046 (rank : 150) | θ value | 8.99809 (rank : 162) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CQ60, Q3TA62 | Gene names | Pgls | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 6-phosphogluconolactonase (EC 3.1.1.31) (6PGL). | |||||
|
CENPA_HUMAN
|
||||||
| NC score | 0.009920 (rank : 151) | θ value | 3.0926 (rank : 110) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49450, Q53T74, Q9BVW2 | Gene names | CENPA | |||
|
Domain Architecture |

|
|||||
| Description | Histone H3-like centromeric protein A (Centromere protein A) (CENP-A) (Centromere autoantigen A). | |||||
|
HNRPG_MOUSE
|
||||||
| NC score | 0.009832 (rank : 152) | θ value | 3.0926 (rank : 114) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35479 | Gene names | Rbmx, Hnrnpg, Hnrpg, Rbmxp1, Rbmxrt | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein G (hnRNP G) (RNA-binding motif protein, X chromosome). | |||||
|
DPPA4_HUMAN
|
||||||
| NC score | 0.009580 (rank : 153) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7L190, Q9H9N5, Q9NVI6 | Gene names | DPPA4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Developmental pluripotency-associated protein 4. | |||||
|
ERRFI_HUMAN
|
||||||
| NC score | 0.009443 (rank : 154) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UJM3, Q9NTG9, Q9UD05 | Gene names | ERRFI1, MIG6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein) (Mig-6). | |||||
|
NFAC1_HUMAN
|
||||||
| NC score | 0.008559 (rank : 155) | θ value | 3.0926 (rank : 117) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
NKX61_HUMAN
|
||||||
| NC score | 0.007632 (rank : 156) | θ value | 0.365318 (rank : 59) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P78426 | Gene names | NKX6-1, NKX6A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-6.1. | |||||
|
SLBP_HUMAN
|
||||||
| NC score | 0.007529 (rank : 157) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14493 | Gene names | SLBP, HBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone RNA hairpin-binding protein (Histone stem-loop-binding protein). | |||||
|
IRK9_MOUSE
|
||||||
| NC score | 0.007375 (rank : 158) | θ value | 2.36792 (rank : 101) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48543, Q9WUE1 | Gene names | Kcnj9, Girk3 | |||
|
Domain Architecture |

|
|||||
| Description | G protein-activated inward rectifier potassium channel 3 (GIRK3) (Potassium channel, inwardly rectifying subfamily J member 9) (Inwardly rectifier K(+) channel Kir3.3). | |||||
|
IRK9_HUMAN
|
||||||
| NC score | 0.007358 (rank : 159) | θ value | 2.36792 (rank : 100) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92806, Q5JW75 | Gene names | KCNJ9, GIRK3 | |||
|
Domain Architecture |

|
|||||
| Description | G protein-activated inward rectifier potassium channel 3 (GIRK3) (Potassium channel, inwardly rectifying subfamily J member 9) (Inwardly rectifier K(+) channel Kir3.3). | |||||
|
BORG3_HUMAN
|
||||||
| NC score | 0.007329 (rank : 160) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NZY7, Q8TB51 | Gene names | CDC42EP5, BORG3, CEP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cdc42 effector protein 5 (Binder of Rho GTPases 3). | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.007181 (rank : 161) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
DAZP1_MOUSE
|
||||||
| NC score | 0.007038 (rank : 162) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JII5 | Gene names | Dazap1 | |||
|
Domain Architecture |

|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
RFIP1_MOUSE
|
||||||
| NC score | 0.007020 (rank : 163) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D620, Q3UBC2, Q8BN24 | Gene names | Rab11fip1, Rcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
ANR25_HUMAN
|
||||||
| NC score | 0.006876 (rank : 164) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q63ZY3, Q3KQZ3, Q6GUF5, Q9H8S4, Q9NUP0, Q9P210 | Gene names | ANKRD25, KIAA1518, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25 (SRC-1-interacting protein) (SRC1-interacting protein). | |||||
|
DVL3_HUMAN
|
||||||
| NC score | 0.006775 (rank : 165) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92997, O14642, Q13531, Q8N5E9, Q92607 | Gene names | DVL3, KIAA0208 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
PLXB1_MOUSE
|
||||||
| NC score | 0.006770 (rank : 166) | θ value | 3.0926 (rank : 118) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
DAZP1_HUMAN
|
||||||
| NC score | 0.006647 (rank : 167) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96EP5, Q96MJ3, Q9NRR9 | Gene names | DAZAP1 | |||
|
Domain Architecture |

|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
UBQL4_MOUSE
|
||||||
| NC score | 0.006432 (rank : 168) | θ value | 5.27518 (rank : 141) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99NB8, Q8BP88 | Gene names | Ubqln4, Ubin | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-4 (Ataxin-1 ubiquitin-like-interacting protein A1U). | |||||
|
NKX61_MOUSE
|
||||||
| NC score | 0.006263 (rank : 169) | θ value | 1.06291 (rank : 78) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99MA9, Q9ERQ7 | Gene names | Nkx6-1, Nkx6.1, Nkx6a | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-6.1. | |||||
|
BCAR3_MOUSE
|
||||||
| NC score | 0.006094 (rank : 170) | θ value | 5.27518 (rank : 128) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QZK2, Q3TNC9, Q3UP10 | Gene names | Bcar3, And34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast cancer anti-estrogen resistance protein 3 (p130Cas-binding protein AND-34). | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.003436 (rank : 171) | θ value | 0.62314 (rank : 66) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
SI1L1_MOUSE
|
||||||
| NC score | 0.003357 (rank : 172) | θ value | 6.88961 (rank : 155) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C0T5, Q6PDI8, Q80U02, Q8C026 | Gene names | Sipa1l1, Kiaa0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1. | |||||
|
NALP1_HUMAN
|
||||||
| NC score | 0.003196 (rank : 173) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.002879 (rank : 174) | θ value | 3.0926 (rank : 112) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.002247 (rank : 175) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
BMP3B_HUMAN
|
||||||
| NC score | 0.001678 (rank : 176) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55107, Q9UCX6 | Gene names | GDF10, BMP3B | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 3b precursor (BMP-3b) (Growth/differentiation factor 10) (GDF-10) (Bone-inducing protein) (BIP). | |||||
|
LZTS2_MOUSE
|
||||||
| NC score | 0.001622 (rank : 177) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91YU6, Q569X6 | Gene names | Lzts2, Kiaa1813 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2. | |||||
|
TTBK1_HUMAN
|
||||||
| NC score | 0.001395 (rank : 178) | θ value | 8.99809 (rank : 176) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
NOTC4_HUMAN
|
||||||
| NC score | 0.001172 (rank : 179) | θ value | 0.813845 (rank : 73) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
WNK4_HUMAN
|
||||||
| NC score | -0.000001 (rank : 180) | θ value | 2.36792 (rank : 107) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
NOTC4_MOUSE
|
||||||
| NC score | -0.000029 (rank : 181) | θ value | 5.27518 (rank : 138) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | -0.000411 (rank : 182) | θ value | 2.36792 (rank : 98) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
RAI14_HUMAN
|
||||||
| NC score | -0.001459 (rank : 183) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
WNK1_MOUSE
|
||||||
| NC score | -0.001596 (rank : 184) | θ value | 8.99809 (rank : 177) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
ZN217_HUMAN
|
||||||
| NC score | -0.001778 (rank : 185) | θ value | 5.27518 (rank : 143) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75362 | Gene names | ZNF217, ZABC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 217. | |||||
|
GCC2_MOUSE
|
||||||
| NC score | -0.002103 (rank : 186) | θ value | 2.36792 (rank : 97) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
ABL1_MOUSE
|
||||||
| NC score | -0.002158 (rank : 187) | θ value | 1.38821 (rank : 80) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P00520, P97896, Q61252, Q61253, Q61254, Q61255, Q61256, Q61257, Q61258, Q61259, Q61260, Q61261, Q6PCM5, Q8C1X4 | Gene names | Abl1, Abl | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | -0.002618 (rank : 188) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
M3K11_HUMAN
|
||||||
| NC score | -0.002828 (rank : 189) | θ value | 3.0926 (rank : 116) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 1136 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q16584, Q6P2G4 | Gene names | MAP3K11, MLK3, SPRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 11 (EC 2.7.11.25) (Mixed lineage kinase 3) (Src-homology 3 domain-containing proline- rich kinase). | |||||
|
ZN366_HUMAN
|
||||||
| NC score | -0.004807 (rank : 190) | θ value | 6.88961 (rank : 160) | |||
| Query Neighborhood Hits | 177 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N895, Q7RTV4 | Gene names | ZNF366 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 366. | |||||