Please be patient as the page loads
|
CAC1C_HUMAN
|
||||||
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CAC1A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.943432 (rank : 13) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.952074 (rank : 11) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.953187 (rank : 10) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.947621 (rank : 12) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 158 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CAC1C_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.997417 (rank : 2) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1D_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.993159 (rank : 4) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1E_HUMAN
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.956906 (rank : 9) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | 0 (rank : 9) | NC score | 0.956957 (rank : 8) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1F_HUMAN
|
||||||
| θ value | 0 (rank : 10) | NC score | 0.991536 (rank : 7) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1F_MOUSE
|
||||||
| θ value | 0 (rank : 11) | NC score | 0.991921 (rank : 6) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1S_HUMAN
|
||||||
| θ value | 0 (rank : 12) | NC score | 0.993726 (rank : 3) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1S_MOUSE
|
||||||
| θ value | 0 (rank : 13) | NC score | 0.993102 (rank : 5) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
SCN7A_HUMAN
|
||||||
| θ value | 1.88829e-90 (rank : 14) | NC score | 0.848490 (rank : 19) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
SC11A_MOUSE
|
||||||
| θ value | 1.2268e-81 (rank : 15) | NC score | 0.854321 (rank : 17) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
CAC1I_HUMAN
|
||||||
| θ value | 7.71989e-68 (rank : 16) | NC score | 0.888878 (rank : 14) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
SC11A_HUMAN
|
||||||
| θ value | 1.90147e-66 (rank : 17) | NC score | 0.851551 (rank : 18) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
SCN3A_HUMAN
|
||||||
| θ value | 6.11685e-65 (rank : 18) | NC score | 0.836907 (rank : 23) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SCN2A_HUMAN
|
||||||
| θ value | 7.98882e-65 (rank : 19) | NC score | 0.834164 (rank : 25) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SCN9A_HUMAN
|
||||||
| θ value | 1.04338e-64 (rank : 20) | NC score | 0.833834 (rank : 26) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
SCN4A_HUMAN
|
||||||
| θ value | 1.77973e-64 (rank : 21) | NC score | 0.844145 (rank : 22) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
SCN1A_HUMAN
|
||||||
| θ value | 5.92465e-60 (rank : 22) | NC score | 0.831669 (rank : 28) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
SCN8A_HUMAN
|
||||||
| θ value | 6.55044e-59 (rank : 23) | NC score | 0.832798 (rank : 27) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN8A_MOUSE
|
||||||
| θ value | 1.45929e-58 (rank : 24) | NC score | 0.830152 (rank : 30) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 1.90589e-58 (rank : 25) | NC score | 0.859437 (rank : 16) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
SC10A_HUMAN
|
||||||
| θ value | 2.48917e-58 (rank : 26) | NC score | 0.830420 (rank : 29) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
SCN5A_HUMAN
|
||||||
| θ value | 2.48917e-58 (rank : 27) | NC score | 0.845448 (rank : 20) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
SC10A_MOUSE
|
||||||
| θ value | 3.25095e-58 (rank : 28) | NC score | 0.834721 (rank : 24) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 4.24587e-58 (rank : 29) | NC score | 0.844169 (rank : 21) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
CAC1H_MOUSE
|
||||||
| θ value | 7.24236e-58 (rank : 30) | NC score | 0.862526 (rank : 15) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
PK2L2_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 31) | NC score | 0.389723 (rank : 31) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PKD2_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 32) | NC score | 0.336289 (rank : 35) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
PK2L2_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 33) | NC score | 0.353315 (rank : 33) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L1_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 34) | NC score | 0.355677 (rank : 32) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
KCNF1_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 35) | NC score | 0.237582 (rank : 36) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
PKD2_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 36) | NC score | 0.341152 (rank : 34) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
KCNB2_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 37) | NC score | 0.206996 (rank : 38) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNB1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 38) | NC score | 0.194333 (rank : 42) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNB1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 39) | NC score | 0.194443 (rank : 41) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCNS3_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 40) | NC score | 0.173293 (rank : 45) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
KCND1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 41) | NC score | 0.160734 (rank : 48) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCNS2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 42) | NC score | 0.195064 (rank : 40) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 43) | NC score | 0.136162 (rank : 51) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCND3_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 44) | NC score | 0.176208 (rank : 44) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 45) | NC score | 0.176363 (rank : 43) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCNQ4_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 46) | NC score | 0.096294 (rank : 62) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
KCNQ2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 47) | NC score | 0.064952 (rank : 79) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNS2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 48) | NC score | 0.198476 (rank : 39) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNV2_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 49) | NC score | 0.223221 (rank : 37) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
KCND1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 50) | NC score | 0.156378 (rank : 49) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCND2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 51) | NC score | 0.169766 (rank : 47) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 52) | NC score | 0.169915 (rank : 46) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCNQ2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 53) | NC score | 0.068769 (rank : 75) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNS1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 54) | NC score | 0.146699 (rank : 50) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 55) | NC score | 0.014411 (rank : 112) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
RILP_HUMAN
|
||||||
| θ value | 0.125558 (rank : 56) | NC score | 0.034011 (rank : 91) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96NA2, Q71RE6, Q9BSL3, Q9BYS3 | Gene names | RILP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab-interacting lysosomal protein. | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.21417 (rank : 57) | NC score | 0.033805 (rank : 92) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 0.279714 (rank : 58) | NC score | 0.024042 (rank : 99) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 59) | NC score | 0.035299 (rank : 90) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
REL_MOUSE
|
||||||
| θ value | 0.47712 (rank : 60) | NC score | 0.010631 (rank : 120) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15307 | Gene names | Rel | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
SF3B1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 61) | NC score | 0.026677 (rank : 96) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 62) | NC score | 0.026715 (rank : 95) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
TPTE2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 63) | NC score | 0.130599 (rank : 52) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6XPS3, Q5VUH2, Q8WWL4, Q8WWL5 | Gene names | TPTE2, TPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase TPTE2 (EC 3.1.3.67) (TPTE and PTEN homologous inositol lipid phosphatase) (Lipid phosphatase TPIP). | |||||
|
ADDB_HUMAN
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.013852 (rank : 113) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
|
DJBP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.057734 (rank : 86) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P1E8, Q5DTV8, Q6PGK8, Q9D2D8 | Gene names | Djbp, Kiaa1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein. | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.019427 (rank : 102) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MB3L2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.017372 (rank : 104) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NHZ7 | Gene names | MBD3L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 3-like 2 (MBD3-like 2). | |||||
|
SEN34_MOUSE
|
||||||
| θ value | 1.06291 (rank : 68) | NC score | 0.021782 (rank : 100) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BMZ5, Q58EU2, Q99LV6, Q9CYW1 | Gene names | Tsen34, Leng5, Sen34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen34 (EC 3.1.27.9) (tRNA-intron endonuclease Sen34) (Leukocyte receptor cluster member 5 homolog). | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 1.38821 (rank : 69) | NC score | 0.012371 (rank : 119) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
APLP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 70) | NC score | 0.013382 (rank : 115) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q06481 | Gene names | APLP2, APPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 2 precursor (Amyloid protein homolog) (APPH) (CDEI box-binding protein) (CDEBP). | |||||
|
CXA12_MOUSE
|
||||||
| θ value | 1.38821 (rank : 71) | NC score | 0.004858 (rank : 149) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BQU6, Q6TLV2, Q8BQS2, Q9EPM1 | Gene names | Gja12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gap junction alpha-12 protein (Connexin-47) (Cx47). | |||||
|
FSCN1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.015424 (rank : 107) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16658, Q96IC5, Q9BRF1 | Gene names | FSCN1, FAN1, HSN, SNL | |||
|
Domain Architecture |
|
|||||
| Description | Fascin (Singed-like protein) (55 kDa actin-bundling protein) (p55). | |||||
|
FSCN1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.015542 (rank : 106) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61553, O09099, O09156 | Gene names | Fscn1, Fan1, Snl | |||
|
Domain Architecture |
|
|||||
| Description | Fascin (Singed-like protein). | |||||
|
ZFP13_MOUSE
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | -0.004531 (rank : 182) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P10754, Q80WS0, Q8CDD0 | Gene names | Zfp13, Krox-8, Zfp-13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 13 (Zfp-13) (Krox-8 protein). | |||||
|
ANX11_MOUSE
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.004932 (rank : 146) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97384 | Gene names | Anxa11, Anx11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50). | |||||
|
EHMT2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.001930 (rank : 167) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
KCNG2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 77) | NC score | 0.070328 (rank : 72) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNH4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 78) | NC score | 0.007181 (rank : 132) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
KCNQ1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 79) | NC score | 0.081093 (rank : 64) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
POT15_HUMAN
|
||||||
| θ value | 1.81305 (rank : 80) | NC score | 0.005558 (rank : 141) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
TENA_MOUSE
|
||||||
| θ value | 1.81305 (rank : 81) | NC score | 0.000427 (rank : 176) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80YX1, Q64706, Q80YX2 | Gene names | Tnc, Hxb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion). | |||||
|
TUR8_MOUSE
|
||||||
| θ value | 1.81305 (rank : 82) | NC score | 0.028404 (rank : 94) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19473 | Gene names | Trap1a, P1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor rejection antigen P815A. | |||||
|
ANR21_HUMAN
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.005354 (rank : 143) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86YR6 | Gene names | ANKRD21, POTE | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 21 (Protein POTE) (Prostate, ovary, testis-expressed protein). | |||||
|
AXN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.007143 (rank : 133) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15169, Q4TT26, Q4TT27, Q86YA7, Q8WVW6, Q96S28 | Gene names | AXIN1, AXIN | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (hAxin). | |||||
|
BSCL2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.024496 (rank : 98) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96G97, Q96SV1, Q9BSQ0 | Gene names | BSCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein). | |||||
|
NASP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | 0.018979 (rank : 103) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.014551 (rank : 111) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | -0.001550 (rank : 178) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CSTN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.004913 (rank : 147) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9EPL2 | Gene names | Clstn1, Cs1, Cstn1 | |||
|
Domain Architecture |
|
|||||
| Description | Calsyntenin-1 precursor. | |||||
|
FANCJ_HUMAN
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.007314 (rank : 131) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BX63, Q8NCI5 | Gene names | BRIP1, BACH1, FANCJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group J protein (EC 3.6.1.-) (ATP-dependent RNA helicase BRIP1) (Protein FACJ) (BRCA1-interacting protein C-terminal helicase 1) (BRCA1-interacting protein 1) (BRCA1-associated C-terminal helicase 1). | |||||
|
GATA4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.005308 (rank : 144) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08369, P97491, Q9QZK4 | Gene names | Gata4, Gata-4 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.015385 (rank : 108) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.015329 (rank : 109) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
APHC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.009186 (rank : 124) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NUN7 | Gene names | PHCA, APHC | |||
|
Domain Architecture |
|
|||||
| Description | Alkaline phytoceramidase (EC 3.5.1.-) (aPHC) (Alkaline ceramidase) (Alkaline dihydroceramidase SB89). | |||||
|
AT132_MOUSE
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.003475 (rank : 159) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CTG6, Q8CG98 | Gene names | Atp13a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable cation-transporting ATPase 13A2 (EC 3.6.3.-). | |||||
|
CD20_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.008252 (rank : 128) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11836, P08984, Q13963 | Gene names | MS4A1, CD20 | |||
|
Domain Architecture |
|
|||||
| Description | B-lymphocyte antigen CD20 (B-lymphocyte surface antigen B1) (Leu-16) (Bp35). | |||||
|
KCNA6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.076416 (rank : 68) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNC4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.120058 (rank : 55) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.013028 (rank : 116) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.004243 (rank : 151) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MYPN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.003952 (rank : 153) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5DTJ9, Q7TPW5, Q8BZ76 | Gene names | Mypn, Kiaa4170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myopalladin. | |||||
|
OR4B1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | -0.004195 (rank : 179) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 652 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NGF8, Q6IF75, Q96R64 | Gene names | OR4B1 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 4B1 (OST208) (Olfactory receptor OR11-106). | |||||
|
RNPC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.003128 (rank : 162) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62176, Q922Z2 | Gene names | Rnpc1, Seb4, Seb4l | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding region-containing protein 1 (ssDNA-binding protein SEB4). | |||||
|
SMC3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.003715 (rank : 155) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
SMC3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.003675 (rank : 157) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 106) | NC score | 0.001082 (rank : 173) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
TPTE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 107) | NC score | 0.091427 (rank : 63) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P56180, Q6XPS4, Q6XPS5, Q71JA8, Q8NCS8 | Gene names | TPTE | |||
|
Domain Architecture |

|
|||||
| Description | Putative tyrosine-protein phosphatase TPTE (EC 3.1.3.48) (Transmembrane phosphatase with tensin homology) (Protein BJ-HCC-5). | |||||
|
ANX11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.003668 (rank : 158) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P50995 | Gene names | ANXA11, ANX11 | |||
|
Domain Architecture |
|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50) (56 kDa autoantigen). | |||||
|
DNL3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.028612 (rank : 93) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P49916, Q16714 | Gene names | LIG3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
KCNQ1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.068252 (rank : 76) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.008738 (rank : 125) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
PCX3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.012770 (rank : 117) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VI59 | Gene names | Pcnxl3 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 3. | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.004047 (rank : 152) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
APLP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.006666 (rank : 135) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
BUB1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.012728 (rank : 118) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1S0 | Gene names | Bub1b, Mad3l | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta (EC 2.7.11.1) (MAD3/BUB1-related protein kinase) (Mitotic checkpoint kinase MAD3L). | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.002484 (rank : 164) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
CALX_MOUSE
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.005978 (rank : 140) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35564 | Gene names | Canx | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor. | |||||
|
CDA7L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.006077 (rank : 139) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96GN5, Q6PIL4, Q86YT0, Q8IXN5, Q96C70, Q9H9A2, Q9NPV2 | Gene names | CDCA7L, HR1, JPO2, R1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle-associated 7-like protein (Protein JPO2) (Transcription factor RAM2). | |||||
|
CDR2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.003250 (rank : 161) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q01850, Q13977 | Gene names | CDR2, PCD17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerebellar degeneration-related protein 2 (Paraneoplastic cerebellar degeneration-associated antigen) (Major Yo paraneoplastic antigen). | |||||
|
DNL3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.025473 (rank : 97) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97386, P97385 | Gene names | Lig3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
KCNC3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.107844 (rank : 57) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNC3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.106282 (rank : 58) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KLC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.004896 (rank : 148) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0B6, Q9H9C8, Q9HA20 | Gene names | KLC2 | |||
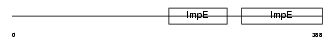
|
Domain Architecture |
|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
KLC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.004945 (rank : 145) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88448 | Gene names | Klc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
LDOC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.021643 (rank : 101) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TPY9 | Gene names | Ldoc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LDOC1. | |||||
|
MTMR5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.002473 (rank : 165) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.006498 (rank : 136) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
PKDRE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.101153 (rank : 59) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PROP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.001055 (rank : 174) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11680, Q3TB98, Q3U779 | Gene names | Cfp, Pfc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Properdin precursor (Factor P). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.016680 (rank : 105) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
SEN34_HUMAN
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.013556 (rank : 114) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BSV6, Q9BVT1, Q9H6H5 | Gene names | TSEN34, LENG5, SEN34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen34 (EC 3.1.27.9) (tRNA-intron endonuclease Sen34) (HsSen34) (Leukocyte receptor cluster member 5). | |||||
|
TXD13_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.008735 (rank : 126) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H1E5, Q8N4P7, Q8NCC1, Q9UJA1, Q9ULQ8 | Gene names | TXNDC13, KIAA1162 | |||
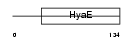
|
Domain Architecture |
|
|||||
| Description | Thioredoxin domain-containing protein 13 precursor. | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.006820 (rank : 134) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
APBA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.006119 (rank : 138) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q02410, O14914, O60570, Q5VYR8 | Gene names | APBA1, MINT1, X11 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 1 (Neuron- specific X11 protein) (Neuronal Munc18-1-interacting protein 1) (Mint- 1) (Adapter protein X11alpha). | |||||
|
B4GN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.003306 (rank : 160) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6L8S8 | Gene names | B4galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.001003 (rank : 175) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
BTC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.008500 (rank : 127) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35070, Q96F48 | Gene names | BTC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Betacellulin precursor (BTC). | |||||
|
CALX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.005486 (rank : 142) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P27824 | Gene names | CANX | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor (Major histocompatibility complex class I antigen- binding protein p88) (p90) (IP90). | |||||
|
COFA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.002041 (rank : 166) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35206, Q9EQD9 | Gene names | Col15a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV)]. | |||||
|
CRKL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.001619 (rank : 170) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P47941, Q3TQ18, Q8BGC5 | Gene names | Crkl, Crkol | |||
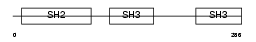
|
Domain Architecture |
|
|||||
| Description | Crk-like protein. | |||||
|
DGLA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.010314 (rank : 121) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y4D2, Q6WQJ0 | Gene names | NSDDR, C11orf11, KIAA0659 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sn1-specific diacylglycerol lipase alpha (EC 3.1.1.-) (DGL-alpha) (Neural stem cell-derived dendrite regulator). | |||||
|
DGLA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.009985 (rank : 122) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6WQJ1, Q6ZQ76 | Gene names | Nsddr, Kiaa0659 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sn1-specific diacylglycerol lipase alpha (EC 3.1.1.-) (DGL-alpha) (Neural stem cell-derived dendrite regulator). | |||||
|
DJBP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.015071 (rank : 110) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5THR3, Q5U5T6, Q9BY88, Q9H5C4, Q9NSF5 | Gene names | DJBP, KIAA1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein (CAP-binding protein complex-interacting protein 1). | |||||
|
ES8L3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.001893 (rank : 168) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TE67, Q5T8Q6, Q5T8Q7, Q5T8Q8, Q96E47, Q9H719 | Gene names | EPS8L3, EPS8R3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 3 (Epidermal growth factor receptor pathway substrate 8-related protein 3) (EPS8-like protein 3). | |||||
|
FRG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.007574 (rank : 130) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14331 | Gene names | FRG1 | |||
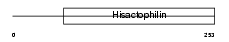
|
Domain Architecture |
|
|||||
| Description | Protein FRG1 (FSHD region gene 1 protein). | |||||
|
FRG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.007754 (rank : 129) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97376, Q99M42 | Gene names | Frg1 | |||
|
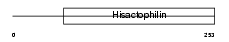
Domain Architecture |
|
|||||
| Description | Protein FRG1 (FSHD region gene 1 protein). | |||||
|
JHD1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.001861 (rank : 169) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P1G2, Q3V396, Q6PFD0, Q6ZPE8, Q9CSF7, Q9QZN6 | Gene names | Fbxl10, Fbl10, Jhdm1b, Kiaa3014 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10). | |||||
|
KI21A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | -0.000315 (rank : 177) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
KLC4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.003792 (rank : 154) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NSK0 | Gene names | KLC4, KNSL8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
MAST2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | -0.004313 (rank : 181) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MYB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.002967 (rank : 163) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06876, Q61929 | Gene names | Myb | |||
|
Domain Architecture |

|
|||||
| Description | Myb proto-oncogene protein (C-myb). | |||||
|
OR6Y1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | -0.004224 (rank : 180) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NGX8, Q6IFS0 | Gene names | OR6Y1, OR6Y2 | |||
|
Domain Architecture |
|
|||||
| Description | Olfactory receptor 6Y1 (Olfactory receptor OR1-11). | |||||
|
SH2D3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.006397 (rank : 137) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N5H7, Q5HYE5, Q6UY42, Q8N6X3, Q9Y2X5 | Gene names | SH2D3C, NSP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (Novel SH2-containing protein 3). | |||||
|
SKIL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.001442 (rank : 171) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P12757, O00464, P12756, Q07501 | Gene names | SKIL, SNO | |||
|
Domain Architecture |
|
|||||
| Description | Ski-like protein (Ski-related protein) (Ski-related oncogene). | |||||
|
T2R16_HUMAN
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.001294 (rank : 172) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYV7, Q502V3, Q549U8, Q645W1 | Gene names | TAS2R16 | |||
|
Domain Architecture |

|
|||||
| Description | Taste receptor type 2 member 16 (T2R16). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.004301 (rank : 150) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TXD13_MOUSE
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.009790 (rank : 123) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C0L0, Q3UHC6, Q6ZPW7, Q80X49 | Gene names | Txndc13, D2Bwg1356e, Kiaa1162 | |||
|
Domain Architecture |
|
|||||
| Description | Thioredoxin domain-containing protein 13 precursor. | |||||
|
ZC3H8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.003683 (rank : 156) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
KCNA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.078183 (rank : 66) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.080231 (rank : 65) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.076410 (rank : 69) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.077419 (rank : 67) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.065308 (rank : 78) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.064439 (rank : 80) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.055160 (rank : 88) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.055437 (rank : 87) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.062798 (rank : 81) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.067406 (rank : 77) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.069600 (rank : 74) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.124204 (rank : 53) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.122475 (rank : 54) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.111557 (rank : 56) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.070873 (rank : 71) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.101055 (rank : 60) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.099658 (rank : 61) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.062654 (rank : 82) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNQ3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.060717 (rank : 83) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.059645 (rank : 85) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.069916 (rank : 73) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
MCLN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.060453 (rank : 84) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K595, Q3UCG4, Q8K2T6, Q9CQD3 | Gene names | Mcoln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
PKDRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.072087 (rank : 70) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
TRPV3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.051267 (rank : 89) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 158 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CAC1C_MOUSE
|
||||||
| NC score | 0.997417 (rank : 2) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1S_HUMAN
|
||||||
| NC score | 0.993726 (rank : 3) | θ value | 0 (rank : 12) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1D_HUMAN
|
||||||
| NC score | 0.993159 (rank : 4) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1S_MOUSE
|
||||||
| NC score | 0.993102 (rank : 5) | θ value | 0 (rank : 13) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1F_MOUSE
|
||||||
| NC score | 0.991921 (rank : 6) | θ value | 0 (rank : 11) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1F_HUMAN
|
||||||
| NC score | 0.991536 (rank : 7) | θ value | 0 (rank : 10) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.956957 (rank : 8) | θ value | 0 (rank : 9) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_HUMAN
|
||||||
| NC score | 0.956906 (rank : 9) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.953187 (rank : 10) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.952074 (rank : 11) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.947621 (rank : 12) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1A_HUMAN
|
||||||
| NC score | 0.943432 (rank : 13) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1I_HUMAN
|
||||||
| NC score | 0.888878 (rank : 14) | θ value | 7.71989e-68 (rank : 16) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
CAC1H_MOUSE
|
||||||
| NC score | 0.862526 (rank : 15) | θ value | 7.24236e-58 (rank : 30) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.859437 (rank : 16) | θ value | 1.90589e-58 (rank : 25) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
SC11A_MOUSE
|
||||||
| NC score | 0.854321 (rank : 17) | θ value | 1.2268e-81 (rank : 15) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SC11A_HUMAN
|
||||||
| NC score | 0.851551 (rank : 18) | θ value | 1.90147e-66 (rank : 17) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
SCN7A_HUMAN
|
||||||
| NC score | 0.848490 (rank : 19) | θ value | 1.88829e-90 (rank : 14) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
SCN5A_HUMAN
|
||||||
| NC score | 0.845448 (rank : 20) | θ value | 2.48917e-58 (rank : 27) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.844169 (rank : 21) | θ value | 4.24587e-58 (rank : 29) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
SCN4A_HUMAN
|
||||||
| NC score | 0.844145 (rank : 22) | θ value | 1.77973e-64 (rank : 21) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
SCN3A_HUMAN
|
||||||
| NC score | 0.836907 (rank : 23) | θ value | 6.11685e-65 (rank : 18) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SC10A_MOUSE
|
||||||
| NC score | 0.834721 (rank : 24) | θ value | 3.25095e-58 (rank : 28) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
SCN2A_HUMAN
|
||||||
| NC score | 0.834164 (rank : 25) | θ value | 7.98882e-65 (rank : 19) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SCN9A_HUMAN
|
||||||
| NC score | 0.833834 (rank : 26) | θ value | 1.04338e-64 (rank : 20) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
SCN8A_HUMAN
|
||||||
| NC score | 0.832798 (rank : 27) | θ value | 6.55044e-59 (rank : 23) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN1A_HUMAN
|
||||||
| NC score | 0.831669 (rank : 28) | θ value | 5.92465e-60 (rank : 22) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
SC10A_HUMAN
|
||||||
| NC score | 0.830420 (rank : 29) | θ value | 2.48917e-58 (rank : 26) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
SCN8A_MOUSE
|
||||||
| NC score | 0.830152 (rank : 30) | θ value | 1.45929e-58 (rank : 24) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
PK2L2_HUMAN
|
||||||
| NC score | 0.389723 (rank : 31) | θ value | 2.36244e-08 (rank : 31) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L1_HUMAN
|
||||||
| NC score | 0.355677 (rank : 32) | θ value | 2.88788e-06 (rank : 34) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PK2L2_MOUSE
|
||||||
| NC score | 0.353315 (rank : 33) | θ value | 1.53129e-07 (rank : 33) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PKD2_MOUSE
|
||||||
| NC score | 0.341152 (rank : 34) | θ value | 2.44474e-05 (rank : 36) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
PKD2_HUMAN
|
||||||
| NC score | 0.336289 (rank : 35) | θ value | 8.97725e-08 (rank : 32) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
KCNF1_HUMAN
|
||||||
| NC score | 0.237582 (rank : 36) | θ value | 1.87187e-05 (rank : 35) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCNV2_HUMAN
|
||||||
| NC score | 0.223221 (rank : 37) | θ value | 0.0330416 (rank : 49) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
KCNB2_HUMAN
|
||||||
| NC score | 0.206996 (rank : 38) | θ value | 0.00102713 (rank : 37) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNS2_MOUSE
|
||||||
| NC score | 0.198476 (rank : 39) | θ value | 0.0330416 (rank : 48) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_HUMAN
|
||||||
| NC score | 0.195064 (rank : 40) | θ value | 0.0113563 (rank : 42) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNB1_MOUSE
|
||||||
| NC score | 0.194443 (rank : 41) | θ value | 0.00134147 (rank : 39) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCNB1_HUMAN
|
||||||
| NC score | 0.194333 (rank : 42) | θ value | 0.00134147 (rank : 38) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCND3_MOUSE
|
||||||
| NC score | 0.176363 (rank : 43) | θ value | 0.0252991 (rank : 45) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_HUMAN
|
||||||
| NC score | 0.176208 (rank : 44) | θ value | 0.0252991 (rank : 44) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCNS3_HUMAN
|
||||||
| NC score | 0.173293 (rank : 45) | θ value | 0.00134147 (rank : 40) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
KCND2_MOUSE
|
||||||
| NC score | 0.169915 (rank : 46) | θ value | 0.0431538 (rank : 52) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_HUMAN
|
||||||
| NC score | 0.169766 (rank : 47) | θ value | 0.0431538 (rank : 51) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND1_MOUSE
|
||||||
| NC score | 0.160734 (rank : 48) | θ value | 0.00509761 (rank : 41) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCND1_HUMAN
|
||||||
| NC score | 0.156378 (rank : 49) | θ value | 0.0431538 (rank : 50) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCNS1_HUMAN
|
||||||
| NC score | 0.146699 (rank : 50) | θ value | 0.0431538 (rank : 54) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNS1_MOUSE
|
||||||
| NC score | 0.136162 (rank : 51) | θ value | 0.0193708 (rank : 43) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
TPTE2_HUMAN
|
||||||
| NC score | 0.130599 (rank : 52) | θ value | 0.47712 (rank : 63) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6XPS3, Q5VUH2, Q8WWL4, Q8WWL5 | Gene names | TPTE2, TPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase TPTE2 (EC 3.1.3.67) (TPTE and PTEN homologous inositol lipid phosphatase) (Lipid phosphatase TPIP). | |||||
|
KCNC1_HUMAN
|
||||||
| NC score | 0.124204 (rank : 53) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC1_MOUSE
|
||||||
| NC score | 0.122475 (rank : 54) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC4_HUMAN
|
||||||
| NC score | 0.120058 (rank : 55) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNC4_MOUSE
|
||||||
| NC score | 0.111557 (rank : 56) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNC3_HUMAN
|
||||||
| NC score | 0.107844 (rank : 57) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNC3_MOUSE
|
||||||
| NC score | 0.106282 (rank : 58) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
PKDRE_HUMAN
|
||||||
| NC score | 0.101153 (rank : 59) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
KCNG3_HUMAN
|
||||||
| NC score | 0.101055 (rank : 60) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG3_MOUSE
|
||||||
| NC score | 0.099658 (rank : 61) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNQ4_HUMAN
|
||||||
| NC score | 0.096294 (rank : 62) | θ value | 0.0252991 (rank : 46) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
TPTE_HUMAN
|
||||||
| NC score | 0.091427 (rank : 63) | θ value | 4.03905 (rank : 107) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P56180, Q6XPS4, Q6XPS5, Q71JA8, Q8NCS8 | Gene names | TPTE | |||
|
Domain Architecture |

|
|||||
| Description | Putative tyrosine-protein phosphatase TPTE (EC 3.1.3.48) (Transmembrane phosphatase with tensin homology) (Protein BJ-HCC-5). | |||||
|
KCNQ1_MOUSE
|
||||||
| NC score | 0.081093 (rank : 64) | θ value | 1.81305 (rank : 79) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNA1_MOUSE
|
||||||
| NC score | 0.080231 (rank : 65) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA1_HUMAN
|
||||||
| NC score | 0.078183 (rank : 66) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA2_MOUSE
|
||||||
| NC score | 0.077419 (rank : 67) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNA6_MOUSE
|
||||||
| NC score | 0.076416 (rank : 68) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNA2_HUMAN
|
||||||
| NC score | 0.076410 (rank : 69) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
PKDRE_MOUSE
|
||||||
| NC score | 0.072087 (rank : 70) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
KCNG1_HUMAN
|
||||||
| NC score | 0.070873 (rank : 71) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNG2_HUMAN
|
||||||
| NC score | 0.070328 (rank : 72) | θ value | 1.81305 (rank : 77) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNQ5_HUMAN
|
||||||
| NC score | 0.069916 (rank : 73) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
KCNA6_HUMAN
|
||||||
| NC score | 0.069600 (rank : 74) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNQ2_HUMAN
|
||||||
| NC score | 0.068769 (rank : 75) | θ value | 0.0431538 (rank : 53) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ1_HUMAN
|
||||||
| NC score | 0.068252 (rank : 76) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNA5_MOUSE
|
||||||
| NC score | 0.067406 (rank : 77) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNA3_HUMAN
|
||||||
| NC score | 0.065308 (rank : 78) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNQ2_MOUSE
|
||||||
| NC score | 0.064952 (rank : 79) | θ value | 0.0330416 (rank : 47) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNA3_MOUSE
|
||||||
| NC score | 0.064439 (rank : 80) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNA5_HUMAN
|
||||||
| NC score | 0.062798 (rank : 81) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNG4_HUMAN
|
||||||
| NC score | 0.062654 (rank : 82) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNQ3_HUMAN
|
||||||
| NC score | 0.060717 (rank : 83) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
MCLN2_MOUSE
|
||||||
| NC score | 0.060453 (rank : 84) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K595, Q3UCG4, Q8K2T6, Q9CQD3 | Gene names | Mcoln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
KCNQ3_MOUSE
|
||||||
| NC score | 0.059645 (rank : 85) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
DJBP_MOUSE
|
||||||
| NC score | 0.057734 (rank : 86) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6P1E8, Q5DTV8, Q6PGK8, Q9D2D8 | Gene names | Djbp, Kiaa1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein. | |||||
|
KCNA4_MOUSE
|
||||||
| NC score | 0.055437 (rank : 87) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNA4_HUMAN
|
||||||
| NC score | 0.055160 (rank : 88) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
TRPV3_HUMAN
|
||||||
| NC score | 0.051267 (rank : 89) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.035299 (rank : 90) | θ value | 0.47712 (rank : 59) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RILP_HUMAN
|
||||||
| NC score | 0.034011 (rank : 91) | θ value | 0.125558 (rank : 56) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96NA2, Q71RE6, Q9BSL3, Q9BYS3 | Gene names | RILP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab-interacting lysosomal protein. | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.033805 (rank : 92) | θ value | 0.21417 (rank : 57) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
DNL3_HUMAN
|
||||||
| NC score | 0.028612 (rank : 93) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P49916, Q16714 | Gene names | LIG3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
TUR8_MOUSE
|
||||||
| NC score | 0.028404 (rank : 94) | θ value | 1.81305 (rank : 82) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19473 | Gene names | Trap1a, P1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor rejection antigen P815A. | |||||
|
SF3B1_MOUSE
|
||||||
| NC score | 0.026715 (rank : 95) | θ value | 0.47712 (rank : 62) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_HUMAN
|
||||||
| NC score | 0.026677 (rank : 96) | θ value | 0.47712 (rank : 61) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
DNL3_MOUSE
|
||||||
| NC score | 0.025473 (rank : 97) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97386, P97385 | Gene names | Lig3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
BSCL2_HUMAN
|
||||||
| NC score | 0.024496 (rank : 98) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96G97, Q96SV1, Q9BSQ0 | Gene names | BSCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.024042 (rank : 99) | θ value | 0.279714 (rank : 58) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
SEN34_MOUSE
|
||||||
| NC score | 0.021782 (rank : 100) | θ value | 1.06291 (rank : 68) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BMZ5, Q58EU2, Q99LV6, Q9CYW1 | Gene names | Tsen34, Leng5, Sen34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen34 (EC 3.1.27.9) (tRNA-intron endonuclease Sen34) (Leukocyte receptor cluster member 5 homolog). | |||||
|
LDOC1_MOUSE
|
||||||
| NC score | 0.021643 (rank : 101) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TPY9 | Gene names | Ldoc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LDOC1. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.019427 (rank : 102) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.018979 (rank : 103) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
MB3L2_HUMAN
|
||||||
| NC score | 0.017372 (rank : 104) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NHZ7 | Gene names | MBD3L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 3-like 2 (MBD3-like 2). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.016680 (rank : 105) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
FSCN1_MOUSE
|
||||||
| NC score | 0.015542 (rank : 106) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61553, O09099, O09156 | Gene names | Fscn1, Fan1, Snl | |||
|
Domain Architecture |

|
|||||
| Description | Fascin (Singed-like protein). | |||||
|
FSCN1_HUMAN
|
||||||
| NC score | 0.015424 (rank : 107) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16658, Q96IC5, Q9BRF1 | Gene names | FSCN1, FAN1, HSN, SNL | |||
|
Domain Architecture |

|
|||||
| Description | Fascin (Singed-like protein) (55 kDa actin-bundling protein) (p55). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.015385 (rank : 108) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.015329 (rank : 109) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
DJBP_HUMAN
|
||||||
| NC score | 0.015071 (rank : 110) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5THR3, Q5U5T6, Q9BY88, Q9H5C4, Q9NSF5 | Gene names | DJBP, KIAA1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein (CAP-binding protein complex-interacting protein 1). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.014551 (rank : 111) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.014411 (rank : 112) | θ value | 0.0961366 (rank : 55) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
ADDB_HUMAN
|
||||||
| NC score | 0.013852 (rank : 113) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
|
SEN34_HUMAN
|
||||||
| NC score | 0.013556 (rank : 114) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BSV6, Q9BVT1, Q9H6H5 | Gene names | TSEN34, LENG5, SEN34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | tRNA-splicing endonuclease subunit Sen34 (EC 3.1.27.9) (tRNA-intron endonuclease Sen34) (HsSen34) (Leukocyte receptor cluster member 5). | |||||
|
APLP2_HUMAN
|
||||||
| NC score | 0.013382 (rank : 115) | θ value | 1.38821 (rank : 70) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q06481 | Gene names | APLP2, APPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 2 precursor (Amyloid protein homolog) (APPH) (CDEI box-binding protein) (CDEBP). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.013028 (rank : 116) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
PCX3_MOUSE
|
||||||
| NC score | 0.012770 (rank : 117) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VI59 | Gene names | Pcnxl3 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 3. | |||||
|
BUB1B_MOUSE
|
||||||
| NC score | 0.012728 (rank : 118) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1S0 | Gene names | Bub1b, Mad3l | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta (EC 2.7.11.1) (MAD3/BUB1-related protein kinase) (Mitotic checkpoint kinase MAD3L). | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.012371 (rank : 119) | θ value | 1.38821 (rank : 69) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
REL_MOUSE
|
||||||
| NC score | 0.010631 (rank : 120) | θ value | 0.47712 (rank : 60) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15307 | Gene names | Rel | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
DGLA_HUMAN
|
||||||
| NC score | 0.010314 (rank : 121) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y4D2, Q6WQJ0 | Gene names | NSDDR, C11orf11, KIAA0659 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sn1-specific diacylglycerol lipase alpha (EC 3.1.1.-) (DGL-alpha) (Neural stem cell-derived dendrite regulator). | |||||
|
DGLA_MOUSE
|
||||||
| NC score | 0.009985 (rank : 122) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6WQJ1, Q6ZQ76 | Gene names | Nsddr, Kiaa0659 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sn1-specific diacylglycerol lipase alpha (EC 3.1.1.-) (DGL-alpha) (Neural stem cell-derived dendrite regulator). | |||||
|
TXD13_MOUSE
|
||||||
| NC score | 0.009790 (rank : 123) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C0L0, Q3UHC6, Q6ZPW7, Q80X49 | Gene names | Txndc13, D2Bwg1356e, Kiaa1162 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin domain-containing protein 13 precursor. | |||||
|
APHC_HUMAN
|
||||||
| NC score | 0.009186 (rank : 124) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NUN7 | Gene names | PHCA, APHC | |||
|
Domain Architecture |

|
|||||
| Description | Alkaline phytoceramidase (EC 3.5.1.-) (aPHC) (Alkaline ceramidase) (Alkaline dihydroceramidase SB89). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.008738 (rank : 125) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
TXD13_HUMAN
|
||||||
| NC score | 0.008735 (rank : 126) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H1E5, Q8N4P7, Q8NCC1, Q9UJA1, Q9ULQ8 | Gene names | TXNDC13, KIAA1162 | |||
|
Domain Architecture |

|
|||||
| Description | Thioredoxin domain-containing protein 13 precursor. | |||||
|
BTC_HUMAN
|
||||||
| NC score | 0.008500 (rank : 127) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35070, Q96F48 | Gene names | BTC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Betacellulin precursor (BTC). | |||||
|
CD20_HUMAN
|
||||||
| NC score | 0.008252 (rank : 128) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11836, P08984, Q13963 | Gene names | MS4A1, CD20 | |||
|
Domain Architecture |

|
|||||
| Description | B-lymphocyte antigen CD20 (B-lymphocyte surface antigen B1) (Leu-16) (Bp35). | |||||
|
FRG1_MOUSE
|
||||||
| NC score | 0.007754 (rank : 129) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97376, Q99M42 | Gene names | Frg1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein FRG1 (FSHD region gene 1 protein). | |||||
|
FRG1_HUMAN
|
||||||
| NC score | 0.007574 (rank : 130) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14331 | Gene names | FRG1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein FRG1 (FSHD region gene 1 protein). | |||||
|
FANCJ_HUMAN
|
||||||
| NC score | 0.007314 (rank : 131) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BX63, Q8NCI5 | Gene names | BRIP1, BACH1, FANCJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group J protein (EC 3.6.1.-) (ATP-dependent RNA helicase BRIP1) (Protein FACJ) (BRCA1-interacting protein C-terminal helicase 1) (BRCA1-interacting protein 1) (BRCA1-associated C-terminal helicase 1). | |||||
|
KCNH4_HUMAN
|
||||||
| NC score | 0.007181 (rank : 132) | θ value | 1.81305 (rank : 78) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UQ05 | Gene names | KCNH4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 4 (Voltage-gated potassium channel subunit Kv12.3) (Ether-a-go-go-like potassium channel 1) (ELK channel 1) (ELK1) (Brain-specific eag-like channel 2) (BEC2). | |||||
|
AXN1_HUMAN
|
||||||
| NC score | 0.007143 (rank : 133) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15169, Q4TT26, Q4TT27, Q86YA7, Q8WVW6, Q96S28 | Gene names | AXIN1, AXIN | |||
|
Domain Architecture |

|
|||||
| Description | Axin-1 (Axis inhibition protein 1) (hAxin). | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.006820 (rank : 134) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
APLP1_MOUSE
|
||||||
| NC score | 0.006666 (rank : 135) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03157, Q8VC38 | Gene names | Aplp1 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid-like protein 1 precursor (APLP) (APLP-1) [Contains: C30]. | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.006498 (rank : 136) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
SH2D3_HUMAN
|
||||||
| NC score | 0.006397 (rank : 137) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N5H7, Q5HYE5, Q6UY42, Q8N6X3, Q9Y2X5 | Gene names | SH2D3C, NSP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3C (Novel SH2-containing protein 3). | |||||
|
APBA1_HUMAN
|
||||||
| NC score | 0.006119 (rank : 138) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q02410, O14914, O60570, Q5VYR8 | Gene names | APBA1, MINT1, X11 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 1 (Neuron- specific X11 protein) (Neuronal Munc18-1-interacting protein 1) (Mint- 1) (Adapter protein X11alpha). | |||||
|
CDA7L_HUMAN
|
||||||
| NC score | 0.006077 (rank : 139) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96GN5, Q6PIL4, Q86YT0, Q8IXN5, Q96C70, Q9H9A2, Q9NPV2 | Gene names | CDCA7L, HR1, JPO2, R1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle-associated 7-like protein (Protein JPO2) (Transcription factor RAM2). | |||||
|
CALX_MOUSE
|
||||||
| NC score | 0.005978 (rank : 140) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35564 | Gene names | Canx | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor. | |||||
|
POT15_HUMAN
|
||||||
| NC score | 0.005558 (rank : 141) | θ value | 1.81305 (rank : 80) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
CALX_HUMAN
|
||||||
| NC score | 0.005486 (rank : 142) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P27824 | Gene names | CANX | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor (Major histocompatibility complex class I antigen- binding protein p88) (p90) (IP90). | |||||
|
ANR21_HUMAN
|
||||||
| NC score | 0.005354 (rank : 143) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86YR6 | Gene names | ANKRD21, POTE | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 21 (Protein POTE) (Prostate, ovary, testis-expressed protein). | |||||
|
GATA4_MOUSE
|
||||||
| NC score | 0.005308 (rank : 144) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08369, P97491, Q9QZK4 | Gene names | Gata4, Gata-4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor GATA-4 (GATA-binding factor 4). | |||||
|
KLC2_MOUSE
|
||||||
| NC score | 0.004945 (rank : 145) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88448 | Gene names | Klc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
ANX11_MOUSE
|
||||||
| NC score | 0.004932 (rank : 146) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97384 | Gene names | Anxa11, Anx11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50). | |||||
|
CSTN1_MOUSE
|
||||||
| NC score | 0.004913 (rank : 147) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9EPL2 | Gene names | Clstn1, Cs1, Cstn1 | |||
|
Domain Architecture |

|
|||||
| Description | Calsyntenin-1 precursor. | |||||
|
KLC2_HUMAN
|
||||||
| NC score | 0.004896 (rank : 148) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0B6, Q9H9C8, Q9HA20 | Gene names | KLC2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
CXA12_MOUSE
|
||||||
| NC score | 0.004858 (rank : 149) | θ value | 1.38821 (rank : 71) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BQU6, Q6TLV2, Q8BQS2, Q9EPM1 | Gene names | Gja12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gap junction alpha-12 protein (Connexin-47) (Cx47). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.004301 (rank : 150) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.004243 (rank : 151) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.004047 (rank : 152) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
MYPN_MOUSE
|
||||||
| NC score | 0.003952 (rank : 153) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5DTJ9, Q7TPW5, Q8BZ76 | Gene names | Mypn, Kiaa4170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myopalladin. | |||||
|
KLC4_HUMAN
|
||||||
| NC score | 0.003792 (rank : 154) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NSK0 | Gene names | KLC4, KNSL8 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 4 (KLC 4) (Kinesin-like protein 8). | |||||
|
SMC3_HUMAN
|
||||||
| NC score | 0.003715 (rank : 155) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
ZC3H8_MOUSE
|
||||||
| NC score | 0.003683 (rank : 156) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JJ48, Q80X92 | Gene names | Zc3h8, Fliz1, Zc3hdc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 8 (Fetal liver zinc finger protein 1). | |||||
|
SMC3_MOUSE
|
||||||
| NC score | 0.003675 (rank : 157) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
ANX11_HUMAN
|
||||||
| NC score | 0.003668 (rank : 158) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P50995 | Gene names | ANXA11, ANX11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50) (56 kDa autoantigen). | |||||
|
AT132_MOUSE
|
||||||
| NC score | 0.003475 (rank : 159) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CTG6, Q8CG98 | Gene names | Atp13a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable cation-transporting ATPase 13A2 (EC 3.6.3.-). | |||||
|
B4GN3_MOUSE
|
||||||
| NC score | 0.003306 (rank : 160) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6L8S8 | Gene names | B4galnt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N- acetylgalactosaminyltransferase 2 (EC 2.4.1.244) (NGalNAc-T2) (Beta- 1,4-N-acetylgalactosaminyltransferase III) (Beta4GalNAc-T3) (Beta4GalNAcT3). | |||||
|
CDR2_HUMAN
|
||||||
| NC score | 0.003250 (rank : 161) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q01850, Q13977 | Gene names | CDR2, PCD17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerebellar degeneration-related protein 2 (Paraneoplastic cerebellar degeneration-associated antigen) (Major Yo paraneoplastic antigen). | |||||
|
RNPC1_MOUSE
|
||||||
| NC score | 0.003128 (rank : 162) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62176, Q922Z2 | Gene names | Rnpc1, Seb4, Seb4l | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding region-containing protein 1 (ssDNA-binding protein SEB4). | |||||
|
MYB_MOUSE
|
||||||
| NC score | 0.002967 (rank : 163) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P06876, Q61929 | Gene names | Myb | |||
|
Domain Architecture |

|
|||||
| Description | Myb proto-oncogene protein (C-myb). | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.002484 (rank : 164) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
MTMR5_HUMAN
|
||||||
| NC score | 0.002473 (rank : 165) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95248, O60228, Q5JXD8, Q5PPM2, Q96GR9, Q9UGB8 | Gene names | SBF1, MTMR5 | |||
|
Domain Architecture |

|
|||||
| Description | SET-binding factor 1 (Sbf1) (Myotubularin-related protein 5). | |||||
|
COFA1_MOUSE
|
||||||
| NC score | 0.002041 (rank : 166) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35206, Q9EQD9 | Gene names | Col15a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV)]. | |||||
|
EHMT2_HUMAN
|
||||||
| NC score | 0.001930 (rank : 167) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
ES8L3_HUMAN
|
||||||
| NC score | 0.001893 (rank : 168) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TE67, Q5T8Q6, Q5T8Q7, Q5T8Q8, Q96E47, Q9H719 | Gene names | EPS8L3, EPS8R3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 3 (Epidermal growth factor receptor pathway substrate 8-related protein 3) (EPS8-like protein 3). | |||||
|
JHD1B_MOUSE
|
||||||
| NC score | 0.001861 (rank : 169) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6P1G2, Q3V396, Q6PFD0, Q6ZPE8, Q9CSF7, Q9QZN6 | Gene names | Fbxl10, Fbl10, Jhdm1b, Kiaa3014 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1B (EC 1.14.11.-) (F-box/LRR-repeat protein 10) (F-box and leucine-rich repeat protein 10) (F-box protein FBL10). | |||||
|
CRKL_MOUSE
|
||||||
| NC score | 0.001619 (rank : 170) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P47941, Q3TQ18, Q8BGC5 | Gene names | Crkl, Crkol | |||
|
Domain Architecture |

|
|||||
| Description | Crk-like protein. | |||||
|
SKIL_HUMAN
|
||||||
| NC score | 0.001442 (rank : 171) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P12757, O00464, P12756, Q07501 | Gene names | SKIL, SNO | |||
|
Domain Architecture |

|
|||||
| Description | Ski-like protein (Ski-related protein) (Ski-related oncogene). | |||||
|
T2R16_HUMAN
|
||||||
| NC score | 0.001294 (rank : 172) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYV7, Q502V3, Q549U8, Q645W1 | Gene names | TAS2R16 | |||
|
Domain Architecture |

|
|||||
| Description | Taste receptor type 2 member 16 (T2R16). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.001082 (rank : 173) | θ value | 4.03905 (rank : 106) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
PROP_MOUSE
|
||||||
| NC score | 0.001055 (rank : 174) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11680, Q3TB98, Q3U779 | Gene names | Cfp, Pfc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Properdin precursor (Factor P). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.001003 (rank : 175) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
TENA_MOUSE
|
||||||
| NC score | 0.000427 (rank : 176) | θ value | 1.81305 (rank : 81) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80YX1, Q64706, Q80YX2 | Gene names | Tnc, Hxb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion). | |||||
|
KI21A_MOUSE
|
||||||
| NC score | -0.000315 (rank : 177) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | -0.001550 (rank : 178) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
OR4B1_HUMAN
|
||||||
| NC score | -0.004195 (rank : 179) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 652 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NGF8, Q6IF75, Q96R64 | Gene names | OR4B1 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 4B1 (OST208) (Olfactory receptor OR11-106). | |||||
|
OR6Y1_HUMAN
|
||||||
| NC score | -0.004224 (rank : 180) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NGX8, Q6IFS0 | Gene names | OR6Y1, OR6Y2 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 6Y1 (Olfactory receptor OR1-11). | |||||
|
MAST2_MOUSE
|
||||||
| NC score | -0.004313 (rank : 181) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
ZFP13_MOUSE
|
||||||
| NC score | -0.004531 (rank : 182) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P10754, Q80WS0, Q8CDD0 | Gene names | Zfp13, Krox-8, Zfp-13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 13 (Zfp-13) (Krox-8 protein). | |||||