Please be patient as the page loads
|
PKDRE_HUMAN
|
||||||
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PKDRE_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PKDRE_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.984787 (rank : 2) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PK1L1_HUMAN
|
||||||
| θ value | 1.28732e-30 (rank : 3) | NC score | 0.604179 (rank : 5) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
PK2L1_HUMAN
|
||||||
| θ value | 2.12685e-25 (rank : 4) | NC score | 0.601621 (rank : 6) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PKD2_MOUSE
|
||||||
| θ value | 1.80048e-24 (rank : 5) | NC score | 0.607761 (rank : 3) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
PKD2_HUMAN
|
||||||
| θ value | 1.5242e-23 (rank : 6) | NC score | 0.600242 (rank : 7) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
PK2L2_MOUSE
|
||||||
| θ value | 9.24701e-21 (rank : 7) | NC score | 0.604381 (rank : 4) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L2_HUMAN
|
||||||
| θ value | 2.5182e-18 (rank : 8) | NC score | 0.587415 (rank : 8) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PKD1_HUMAN
|
||||||
| θ value | 1.63225e-17 (rank : 9) | NC score | 0.325663 (rank : 13) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
LOX5_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 10) | NC score | 0.189461 (rank : 16) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P09917 | Gene names | ALOX5, LOG5 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 5-lipoxygenase (EC 1.13.11.34) (5-lipoxygenase) (5-LO). | |||||
|
LOX5_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 11) | NC score | 0.178279 (rank : 17) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P48999 | Gene names | Alox5 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 5-lipoxygenase (EC 1.13.11.34) (5-lipoxygenase) (5-LO). | |||||
|
MCLN1_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 12) | NC score | 0.337938 (rank : 9) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9GZU1, Q7Z4F7, Q9H292, Q9H4B3, Q9H4B5 | Gene names | MCOLN1, ML4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-1 (Mucolipidin) (MG-2). | |||||
|
MCLN1_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 13) | NC score | 0.331424 (rank : 11) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99J21 | Gene names | Mcoln1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-1 (Mucolipidin). | |||||
|
MCLN3_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 14) | NC score | 0.333927 (rank : 10) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TDD5, Q5T4H5, Q5T4H6, Q9NV09 | Gene names | MCOLN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-3. | |||||
|
MCLN3_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 15) | NC score | 0.329507 (rank : 12) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R4F0, Q8BS73, Q8BSG1, Q8CDB2 | Gene names | Mcoln3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-3. | |||||
|
MCLN2_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 16) | NC score | 0.316608 (rank : 14) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IZK6, Q8N9R3 | Gene names | MCOLN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
MCLN2_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 17) | NC score | 0.309969 (rank : 15) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K595, Q3UCG4, Q8K2T6, Q9CQD3 | Gene names | Mcoln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
SCN5A_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 18) | NC score | 0.148035 (rank : 18) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
SC11A_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 19) | NC score | 0.125784 (rank : 22) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SC10A_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 20) | NC score | 0.132002 (rank : 19) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
RA6I1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 21) | NC score | 0.097460 (rank : 42) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6IQ26, Q96GN3, Q9H6U7, Q9UFV0, Q9UPR1 | Gene names | RAB6IP1, KIAA1091 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
RA6I1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 22) | NC score | 0.093160 (rank : 46) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PAL8, Q62146, Q8C829, Q8VDF6, Q9QYZ2 | Gene names | Rab6ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
CAC1C_MOUSE
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.107530 (rank : 32) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
SC10A_MOUSE
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.125789 (rank : 21) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
HPS6_HUMAN
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.046115 (rank : 62) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86YV9, Q9H685 | Gene names | HPS6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 6 protein (Ruby-eye protein homolog) (Ru). | |||||
|
SCN1A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 26) | NC score | 0.119512 (rank : 23) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
SCN2A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.119034 (rank : 24) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.020234 (rank : 69) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.101955 (rank : 36) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
LIPE_HUMAN
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.046177 (rank : 61) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y5X9, Q6UW82 | Gene names | LIPG | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial lipase precursor (EC 3.1.1.3) (Endothelial cell-derived lipase) (EDL) (EL). | |||||
|
LX12B_HUMAN
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.109577 (rank : 30) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75342 | Gene names | ALOX12B | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, 12R type (EC 1.13.11.-) (Epidermis-type lipoxygenase 12) (12R-lipoxygenase) (12R-LOX). | |||||
|
SCN3A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.118674 (rank : 25) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SC11A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.127851 (rank : 20) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
CAC1F_MOUSE
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.107784 (rank : 31) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
SNX10_HUMAN
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.041957 (rank : 64) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5X0 | Gene names | SNX10 | |||
|
Domain Architecture |
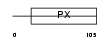
|
|||||
| Description | Sorting nexin-10. | |||||
|
SNX10_MOUSE
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.041871 (rank : 65) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CWT3, Q8C1E0 | Gene names | Snx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-10. | |||||
|
LX12B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.102062 (rank : 35) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70582 | Gene names | Alox12b, Aloxe2 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, 12R type (EC 1.13.11.-) (Epidermis-type lipoxygenase 12) (12R-lipoxygenase) (12R-LOX) (Epidermis-type lipoxygenase 2) (e-LOX 2). | |||||
|
LIPE_MOUSE
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.044901 (rank : 63) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WVG5, Q8VDU2 | Gene names | Lipg | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial lipase precursor (EC 3.1.1.3) (Endothelial cell-derived lipase) (EDL). | |||||
|
SCN8A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.111722 (rank : 29) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN8A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.111879 (rank : 28) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.090949 (rank : 48) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
DEK_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.031667 (rank : 66) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35659, Q5TGV4, Q5TGV5 | Gene names | DEK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
LAMA1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.006363 (rank : 77) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 804 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P25391 | Gene names | LAMA1, LAMA | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
LOXE3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.090142 (rank : 49) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WV07 | Gene names | Aloxe3 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermis-type lipoxygenase 3 (EC 1.13.11.-) (e-LOX-3). | |||||
|
SCN4A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.113899 (rank : 27) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
LRC27_MOUSE
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.021454 (rank : 68) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80YS5, Q9D6Z2 | Gene names | Lrrc27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 27. | |||||
|
TRY3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.003657 (rank : 82) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35030, P15951, Q15665, Q9UQV3 | Gene names | PRSS3, PRSS4, TRY3, TRY4 | |||
|
Domain Architecture |
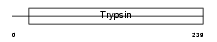
|
|||||
| Description | Trypsin-3 precursor (EC 3.4.21.4) (Trypsin III) (Brain trypsinogen) (Mesotrypsinogen) (Trypsin IV). | |||||
|
O12D2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | -0.000858 (rank : 83) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P58182 | Gene names | OR12D2 | |||
|
Domain Architecture |
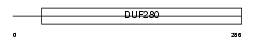
|
|||||
| Description | Olfactory receptor 12D2 (Hs6M1-20). | |||||
|
SCN9A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.118177 (rank : 26) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
AFAD_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.010332 (rank : 74) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
ARY3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.016843 (rank : 70) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50296 | Gene names | Nat3, Aac3 | |||
|
Domain Architecture |

|
|||||
| Description | Arylamine N-acetyltransferase 3 (EC 2.3.1.5) (Arylamide acetylase 3) (N-acetyltransferase type 3) (NAT-3). | |||||
|
CEND3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.024195 (rank : 67) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
HB2P_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.004764 (rank : 78) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04440, O19698, O19700, O19702 | Gene names | HLA-DPB1 | |||
|
Domain Architecture |

|
|||||
| Description | HLA class II histocompatibility antigen, DP(W4) beta chain precursor. | |||||
|
HB2Q_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.004705 (rank : 80) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13763 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HLA class II histocompatibility antigen, DP(W2) beta chain precursor (SB-2-beta). | |||||
|
LRC8B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.004756 (rank : 79) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5DU41 | Gene names | Lrrc8b, Kiaa0231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8B. | |||||
|
TCEA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.011417 (rank : 71) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15560, Q8TD37, Q8TD38 | Gene names | TCEA2 | |||
|
Domain Architecture |
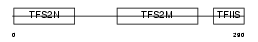
|
|||||
| Description | Transcription elongation factor A protein 2 (Transcription elongation factor S-II protein 2) (Testis-specific S-II) (Transcription elongation factor TFIIS.l). | |||||
|
ABCA4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.004078 (rank : 81) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P78363, O15112, O60438, O60915 | Gene names | ABCA4, ABCR | |||
|
Domain Architecture |

|
|||||
| Description | Retinal-specific ATP-binding cassette transporter (ATP-binding cassette sub-family A member 4) (RIM ABC transporter) (RIM protein) (RmP) (Stargardt disease protein). | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.101153 (rank : 37) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CAC1E_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.107096 (rank : 34) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.107262 (rank : 33) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CEND3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.009709 (rank : 75) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
GPR88_MOUSE
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.011177 (rank : 72) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EPB7 | Gene names | Gpr88, Strg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 88 (Striatum-specific G-protein coupled receptor). | |||||
|
ALMS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.008428 (rank : 76) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
COG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.010963 (rank : 73) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WTW3, Q9NPV9, Q9P2G6 | Gene names | COG1, KIAA1381 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Conserved oligomeric Golgi complex component 1. | |||||
|
CAC1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.088986 (rank : 51) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.089350 (rank : 50) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.098972 (rank : 39) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.094989 (rank : 44) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.097186 (rank : 43) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1F_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.100316 (rank : 38) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1H_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.083230 (rank : 52) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
CAC1I_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.098614 (rank : 40) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
CAC1S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.093217 (rank : 45) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1S_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.092659 (rank : 47) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
LOX12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.076807 (rank : 56) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P18054, O95569, Q6ISF8, Q9UQM4 | Gene names | ALOX12, LOG12 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, 12S-type (EC 1.13.11.31) (12-LOX) (Platelet-type lipoxygenase 12). | |||||
|
LOX12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.073961 (rank : 58) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P39655, Q8BHG4 | Gene names | Alox12, Alox12p | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, 12S-type (EC 1.13.11.31) (12-LOX) (Platelet-type lipoxygenase 12). | |||||
|
LOX15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.066564 (rank : 60) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P16050, Q8N6R7, Q99657 | Gene names | ALOX15, LOG15 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 15-lipoxygenase (EC 1.13.11.33) (Arachidonate omega-6 lipoxygenase) (15-LOX). | |||||
|
LOXE3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.082312 (rank : 53) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYJ1, Q9H4F2, Q9HC22 | Gene names | ALOXE3 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermis-type lipoxygenase 3 (EC 1.13.11.-) (e-LOX-3). | |||||
|
LX12E_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.070990 (rank : 59) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P55249, Q91YW6 | Gene names | Alox12e, Alox12-ps2, Aloxe | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, epidermal-type (EC 1.13.11.31) (12-LOX). | |||||
|
LX12L_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.076560 (rank : 57) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P39654, Q4FJY9, Q5F2E3, Q6PHB2 | Gene names | Alox12l, Alox15 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, leukocyte-type (EC 1.13.11.31) (12-LOX). | |||||
|
LX15B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.077278 (rank : 55) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15296, Q8IYQ2, Q8TEV3, Q8TEV4, Q8TEV5, Q8TEV6, Q9UKM4 | Gene names | ALOX15B | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 15-lipoxygenase type II (EC 1.13.11.33) (15-LOX-2) (15- lipoxygenase 2). | |||||
|
LX15B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.081033 (rank : 54) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35936 | Gene names | Alox15b, Alox8 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 15-lipoxygenase type II (EC 1.13.11.33) (15-LOX-2) (8S- lipoxygenase) (8S-LOX). | |||||
|
SCN7A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.097757 (rank : 41) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
PKDRE_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PKDRE_MOUSE
|
||||||
| NC score | 0.984787 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PKD2_MOUSE
|
||||||
| NC score | 0.607761 (rank : 3) | θ value | 1.80048e-24 (rank : 5) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
PK2L2_MOUSE
|
||||||
| NC score | 0.604381 (rank : 4) | θ value | 9.24701e-21 (rank : 7) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK1L1_HUMAN
|
||||||
| NC score | 0.604179 (rank : 5) | θ value | 1.28732e-30 (rank : 3) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
PK2L1_HUMAN
|
||||||
| NC score | 0.601621 (rank : 6) | θ value | 2.12685e-25 (rank : 4) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PKD2_HUMAN
|
||||||
| NC score | 0.600242 (rank : 7) | θ value | 1.5242e-23 (rank : 6) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
PK2L2_HUMAN
|
||||||
| NC score | 0.587415 (rank : 8) | θ value | 2.5182e-18 (rank : 8) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
MCLN1_HUMAN
|
||||||
| NC score | 0.337938 (rank : 9) | θ value | 1.17247e-07 (rank : 12) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9GZU1, Q7Z4F7, Q9H292, Q9H4B3, Q9H4B5 | Gene names | MCOLN1, ML4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-1 (Mucolipidin) (MG-2). | |||||
|
MCLN3_HUMAN
|
||||||
| NC score | 0.333927 (rank : 10) | θ value | 4.45536e-07 (rank : 14) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TDD5, Q5T4H5, Q5T4H6, Q9NV09 | Gene names | MCOLN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-3. | |||||
|
MCLN1_MOUSE
|
||||||
| NC score | 0.331424 (rank : 11) | θ value | 1.99992e-07 (rank : 13) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99J21 | Gene names | Mcoln1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-1 (Mucolipidin). | |||||
|
MCLN3_MOUSE
|
||||||
| NC score | 0.329507 (rank : 12) | θ value | 9.92553e-07 (rank : 15) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R4F0, Q8BS73, Q8BSG1, Q8CDB2 | Gene names | Mcoln3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-3. | |||||
|
PKD1_HUMAN
|
||||||
| NC score | 0.325663 (rank : 13) | θ value | 1.63225e-17 (rank : 9) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P98161, Q15140, Q15141 | Gene names | PKD1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-1 precursor (Autosomal dominant polycystic kidney disease protein 1). | |||||
|
MCLN2_HUMAN
|
||||||
| NC score | 0.316608 (rank : 14) | θ value | 0.00020696 (rank : 16) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IZK6, Q8N9R3 | Gene names | MCOLN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
MCLN2_MOUSE
|
||||||
| NC score | 0.309969 (rank : 15) | θ value | 0.000461057 (rank : 17) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K595, Q3UCG4, Q8K2T6, Q9CQD3 | Gene names | Mcoln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
LOX5_HUMAN
|
||||||
| NC score | 0.189461 (rank : 16) | θ value | 1.47974e-10 (rank : 10) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P09917 | Gene names | ALOX5, LOG5 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 5-lipoxygenase (EC 1.13.11.34) (5-lipoxygenase) (5-LO). | |||||
|
LOX5_MOUSE
|
||||||
| NC score | 0.178279 (rank : 17) | θ value | 6.21693e-09 (rank : 11) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P48999 | Gene names | Alox5 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 5-lipoxygenase (EC 1.13.11.34) (5-lipoxygenase) (5-LO). | |||||
|
SCN5A_HUMAN
|
||||||
| NC score | 0.148035 (rank : 18) | θ value | 0.000602161 (rank : 18) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
SC10A_HUMAN
|
||||||
| NC score | 0.132002 (rank : 19) | θ value | 0.0252991 (rank : 20) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
SC11A_HUMAN
|
||||||
| NC score | 0.127851 (rank : 20) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
SC10A_MOUSE
|
||||||
| NC score | 0.125789 (rank : 21) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
SC11A_MOUSE
|
||||||
| NC score | 0.125784 (rank : 22) | θ value | 0.00390308 (rank : 19) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SCN1A_HUMAN
|
||||||
| NC score | 0.119512 (rank : 23) | θ value | 0.279714 (rank : 26) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
SCN2A_HUMAN
|
||||||
| NC score | 0.119034 (rank : 24) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SCN3A_HUMAN
|
||||||
| NC score | 0.118674 (rank : 25) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SCN9A_HUMAN
|
||||||
| NC score | 0.118177 (rank : 26) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
SCN4A_HUMAN
|
||||||
| NC score | 0.113899 (rank : 27) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
SCN8A_MOUSE
|
||||||
| NC score | 0.111879 (rank : 28) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN8A_HUMAN
|
||||||
| NC score | 0.111722 (rank : 29) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
LX12B_HUMAN
|
||||||
| NC score | 0.109577 (rank : 30) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O75342 | Gene names | ALOX12B | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, 12R type (EC 1.13.11.-) (Epidermis-type lipoxygenase 12) (12R-lipoxygenase) (12R-LOX). | |||||
|
CAC1F_MOUSE
|
||||||
| NC score | 0.107784 (rank : 31) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1C_MOUSE
|
||||||
| NC score | 0.107530 (rank : 32) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.107262 (rank : 33) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_HUMAN
|
||||||
| NC score | 0.107096 (rank : 34) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
LX12B_MOUSE
|
||||||
| NC score | 0.102062 (rank : 35) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70582 | Gene names | Alox12b, Aloxe2 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, 12R type (EC 1.13.11.-) (Epidermis-type lipoxygenase 12) (12R-lipoxygenase) (12R-LOX) (Epidermis-type lipoxygenase 2) (e-LOX 2). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.101955 (rank : 36) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.101153 (rank : 37) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CAC1F_HUMAN
|
||||||
| NC score | 0.100316 (rank : 38) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.098972 (rank : 39) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1I_HUMAN
|
||||||
| NC score | 0.098614 (rank : 40) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
SCN7A_HUMAN
|
||||||
| NC score | 0.097757 (rank : 41) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
RA6I1_HUMAN
|
||||||
| NC score | 0.097460 (rank : 42) | θ value | 0.0736092 (rank : 21) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6IQ26, Q96GN3, Q9H6U7, Q9UFV0, Q9UPR1 | Gene names | RAB6IP1, KIAA1091 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
CAC1D_HUMAN
|
||||||
| NC score | 0.097186 (rank : 43) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.094989 (rank : 44) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1S_HUMAN
|
||||||
| NC score | 0.093217 (rank : 45) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
RA6I1_MOUSE
|
||||||
| NC score | 0.093160 (rank : 46) | θ value | 0.125558 (rank : 22) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PAL8, Q62146, Q8C829, Q8VDF6, Q9QYZ2 | Gene names | Rab6ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
CAC1S_MOUSE
|
||||||
| NC score | 0.092659 (rank : 47) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.090949 (rank : 48) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
LOXE3_MOUSE
|
||||||
| NC score | 0.090142 (rank : 49) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WV07 | Gene names | Aloxe3 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermis-type lipoxygenase 3 (EC 1.13.11.-) (e-LOX-3). | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.089350 (rank : 50) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1A_HUMAN
|
||||||
| NC score | 0.088986 (rank : 51) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1H_MOUSE
|
||||||
| NC score | 0.083230 (rank : 52) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
LOXE3_HUMAN
|
||||||
| NC score | 0.082312 (rank : 53) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYJ1, Q9H4F2, Q9HC22 | Gene names | ALOXE3 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermis-type lipoxygenase 3 (EC 1.13.11.-) (e-LOX-3). | |||||
|
LX15B_MOUSE
|
||||||
| NC score | 0.081033 (rank : 54) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35936 | Gene names | Alox15b, Alox8 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 15-lipoxygenase type II (EC 1.13.11.33) (15-LOX-2) (8S- lipoxygenase) (8S-LOX). | |||||
|
LX15B_HUMAN
|
||||||
| NC score | 0.077278 (rank : 55) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15296, Q8IYQ2, Q8TEV3, Q8TEV4, Q8TEV5, Q8TEV6, Q9UKM4 | Gene names | ALOX15B | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 15-lipoxygenase type II (EC 1.13.11.33) (15-LOX-2) (15- lipoxygenase 2). | |||||
|
LOX12_HUMAN
|
||||||
| NC score | 0.076807 (rank : 56) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P18054, O95569, Q6ISF8, Q9UQM4 | Gene names | ALOX12, LOG12 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, 12S-type (EC 1.13.11.31) (12-LOX) (Platelet-type lipoxygenase 12). | |||||
|
LX12L_MOUSE
|
||||||
| NC score | 0.076560 (rank : 57) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P39654, Q4FJY9, Q5F2E3, Q6PHB2 | Gene names | Alox12l, Alox15 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, leukocyte-type (EC 1.13.11.31) (12-LOX). | |||||
|
LOX12_MOUSE
|
||||||
| NC score | 0.073961 (rank : 58) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P39655, Q8BHG4 | Gene names | Alox12, Alox12p | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, 12S-type (EC 1.13.11.31) (12-LOX) (Platelet-type lipoxygenase 12). | |||||
|
LX12E_MOUSE
|
||||||
| NC score | 0.070990 (rank : 59) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P55249, Q91YW6 | Gene names | Alox12e, Alox12-ps2, Aloxe | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, epidermal-type (EC 1.13.11.31) (12-LOX). | |||||
|
LOX15_HUMAN
|
||||||
| NC score | 0.066564 (rank : 60) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P16050, Q8N6R7, Q99657 | Gene names | ALOX15, LOG15 | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 15-lipoxygenase (EC 1.13.11.33) (Arachidonate omega-6 lipoxygenase) (15-LOX). | |||||
|
LIPE_HUMAN
|
||||||
| NC score | 0.046177 (rank : 61) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y5X9, Q6UW82 | Gene names | LIPG | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial lipase precursor (EC 3.1.1.3) (Endothelial cell-derived lipase) (EDL) (EL). | |||||
|
HPS6_HUMAN
|
||||||
| NC score | 0.046115 (rank : 62) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86YV9, Q9H685 | Gene names | HPS6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 6 protein (Ruby-eye protein homolog) (Ru). | |||||
|
LIPE_MOUSE
|
||||||
| NC score | 0.044901 (rank : 63) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WVG5, Q8VDU2 | Gene names | Lipg | |||
|
Domain Architecture |

|
|||||
| Description | Endothelial lipase precursor (EC 3.1.1.3) (Endothelial cell-derived lipase) (EDL). | |||||
|
SNX10_HUMAN
|
||||||
| NC score | 0.041957 (rank : 64) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5X0 | Gene names | SNX10 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-10. | |||||
|
SNX10_MOUSE
|
||||||
| NC score | 0.041871 (rank : 65) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CWT3, Q8C1E0 | Gene names | Snx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-10. | |||||
|
DEK_HUMAN
|
||||||
| NC score | 0.031667 (rank : 66) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35659, Q5TGV4, Q5TGV5 | Gene names | DEK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
CEND3_HUMAN
|
||||||
| NC score | 0.024195 (rank : 67) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WWN8 | Gene names | CENTD3, ARAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3). | |||||
|
LRC27_MOUSE
|
||||||
| NC score | 0.021454 (rank : 68) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80YS5, Q9D6Z2 | Gene names | Lrrc27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 27. | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.020234 (rank : 69) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
ARY3_MOUSE
|
||||||
| NC score | 0.016843 (rank : 70) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50296 | Gene names | Nat3, Aac3 | |||
|
Domain Architecture |

|
|||||
| Description | Arylamine N-acetyltransferase 3 (EC 2.3.1.5) (Arylamide acetylase 3) (N-acetyltransferase type 3) (NAT-3). | |||||
|
TCEA2_HUMAN
|
||||||
| NC score | 0.011417 (rank : 71) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15560, Q8TD37, Q8TD38 | Gene names | TCEA2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor A protein 2 (Transcription elongation factor S-II protein 2) (Testis-specific S-II) (Transcription elongation factor TFIIS.l). | |||||
|
GPR88_MOUSE
|
||||||
| NC score | 0.011177 (rank : 72) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EPB7 | Gene names | Gpr88, Strg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 88 (Striatum-specific G-protein coupled receptor). | |||||
|
COG1_HUMAN
|
||||||
| NC score | 0.010963 (rank : 73) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WTW3, Q9NPV9, Q9P2G6 | Gene names | COG1, KIAA1381 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Conserved oligomeric Golgi complex component 1. | |||||
|
AFAD_HUMAN
|
||||||
| NC score | 0.010332 (rank : 74) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
CEND3_MOUSE
|
||||||
| NC score | 0.009709 (rank : 75) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
ALMS1_HUMAN
|
||||||
| NC score | 0.008428 (rank : 76) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
LAMA1_HUMAN
|
||||||
| NC score | 0.006363 (rank : 77) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 804 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P25391 | Gene names | LAMA1, LAMA | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
HB2P_HUMAN
|
||||||
| NC score | 0.004764 (rank : 78) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04440, O19698, O19700, O19702 | Gene names | HLA-DPB1 | |||
|
Domain Architecture |

|
|||||
| Description | HLA class II histocompatibility antigen, DP(W4) beta chain precursor. | |||||
|
LRC8B_MOUSE
|
||||||
| NC score | 0.004756 (rank : 79) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5DU41 | Gene names | Lrrc8b, Kiaa0231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 8B. | |||||
|
HB2Q_HUMAN
|
||||||
| NC score | 0.004705 (rank : 80) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13763 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | HLA class II histocompatibility antigen, DP(W2) beta chain precursor (SB-2-beta). | |||||
|
ABCA4_HUMAN
|
||||||
| NC score | 0.004078 (rank : 81) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P78363, O15112, O60438, O60915 | Gene names | ABCA4, ABCR | |||
|
Domain Architecture |

|
|||||
| Description | Retinal-specific ATP-binding cassette transporter (ATP-binding cassette sub-family A member 4) (RIM ABC transporter) (RIM protein) (RmP) (Stargardt disease protein). | |||||
|
TRY3_HUMAN
|
||||||
| NC score | 0.003657 (rank : 82) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35030, P15951, Q15665, Q9UQV3 | Gene names | PRSS3, PRSS4, TRY3, TRY4 | |||
|
Domain Architecture |

|
|||||
| Description | Trypsin-3 precursor (EC 3.4.21.4) (Trypsin III) (Brain trypsinogen) (Mesotrypsinogen) (Trypsin IV). | |||||
|
O12D2_HUMAN
|
||||||
| NC score | -0.000858 (rank : 83) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P58182 | Gene names | OR12D2 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 12D2 (Hs6M1-20). | |||||