Please be patient as the page loads
|
CAC1S_MOUSE
|
||||||
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CAC1A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.948259 (rank : 13) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.957024 (rank : 11) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.957304 (rank : 10) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.953077 (rank : 12) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.993102 (rank : 7) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CAC1C_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.993423 (rank : 5) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1D_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.994651 (rank : 3) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1E_HUMAN
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.961896 (rank : 9) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_MOUSE
|
||||||
| θ value | 0 (rank : 9) | NC score | 0.962186 (rank : 8) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1F_HUMAN
|
||||||
| θ value | 0 (rank : 10) | NC score | 0.993419 (rank : 6) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1F_MOUSE
|
||||||
| θ value | 0 (rank : 11) | NC score | 0.993470 (rank : 4) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1S_HUMAN
|
||||||
| θ value | 0 (rank : 12) | NC score | 0.998457 (rank : 2) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1S_MOUSE
|
||||||
| θ value | 0 (rank : 13) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1I_HUMAN
|
||||||
| θ value | 1.89707e-74 (rank : 14) | NC score | 0.901279 (rank : 14) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
SC11A_HUMAN
|
||||||
| θ value | 4.22625e-74 (rank : 15) | NC score | 0.863594 (rank : 18) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
SCN2A_HUMAN
|
||||||
| θ value | 4.22625e-74 (rank : 16) | NC score | 0.846875 (rank : 26) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SCN3A_HUMAN
|
||||||
| θ value | 6.10265e-73 (rank : 17) | NC score | 0.849771 (rank : 23) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SCN4A_HUMAN
|
||||||
| θ value | 2.31901e-72 (rank : 18) | NC score | 0.856406 (rank : 22) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
SC10A_MOUSE
|
||||||
| θ value | 5.1662e-72 (rank : 19) | NC score | 0.848735 (rank : 24) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 8.81223e-72 (rank : 20) | NC score | 0.873680 (rank : 16) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
SCN9A_HUMAN
|
||||||
| θ value | 8.81223e-72 (rank : 21) | NC score | 0.846393 (rank : 27) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 2.17053e-70 (rank : 22) | NC score | 0.858005 (rank : 20) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
SCN8A_HUMAN
|
||||||
| θ value | 2.83479e-70 (rank : 23) | NC score | 0.847105 (rank : 25) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SC11A_MOUSE
|
||||||
| θ value | 3.70236e-70 (rank : 24) | NC score | 0.865257 (rank : 17) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SCN8A_MOUSE
|
||||||
| θ value | 6.31527e-70 (rank : 25) | NC score | 0.844506 (rank : 28) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
CAC1H_MOUSE
|
||||||
| θ value | 8.24801e-70 (rank : 26) | NC score | 0.877533 (rank : 15) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
SC10A_HUMAN
|
||||||
| θ value | 1.40689e-69 (rank : 27) | NC score | 0.843991 (rank : 30) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
SCN1A_HUMAN
|
||||||
| θ value | 9.11921e-69 (rank : 28) | NC score | 0.844435 (rank : 29) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
SCN5A_HUMAN
|
||||||
| θ value | 4.52582e-68 (rank : 29) | NC score | 0.858206 (rank : 19) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
SCN7A_HUMAN
|
||||||
| θ value | 3.36421e-55 (rank : 30) | NC score | 0.856526 (rank : 21) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
PK2L2_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 31) | NC score | 0.371891 (rank : 31) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
KCNV2_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 32) | NC score | 0.229680 (rank : 37) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
PK2L1_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 33) | NC score | 0.340281 (rank : 32) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PKD2_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 34) | NC score | 0.326012 (rank : 34) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
PKD2_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 35) | NC score | 0.319275 (rank : 35) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
PK2L2_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 36) | NC score | 0.335092 (rank : 33) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
KCNF1_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 37) | NC score | 0.240059 (rank : 36) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCNB2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 38) | NC score | 0.209465 (rank : 38) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCND1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 39) | NC score | 0.164309 (rank : 48) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCND1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 40) | NC score | 0.160242 (rank : 49) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCNS2_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 41) | NC score | 0.202351 (rank : 39) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNB1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 42) | NC score | 0.196531 (rank : 42) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCNB1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 43) | NC score | 0.196621 (rank : 41) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCND3_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 44) | NC score | 0.180375 (rank : 44) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 45) | NC score | 0.180503 (rank : 43) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCNS2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 46) | NC score | 0.198602 (rank : 40) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCND2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 47) | NC score | 0.174172 (rank : 47) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 48) | NC score | 0.174333 (rank : 46) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCNQ1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 49) | NC score | 0.079663 (rank : 67) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNS3_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 50) | NC score | 0.175241 (rank : 45) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
BRCA1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 51) | NC score | 0.018439 (rank : 90) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
KCNS1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 52) | NC score | 0.137332 (rank : 51) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNC3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 53) | NC score | 0.114884 (rank : 57) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNC3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 54) | NC score | 0.113366 (rank : 58) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
PO6F2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 55) | NC score | 0.007378 (rank : 108) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
KCNS1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 56) | NC score | 0.148182 (rank : 50) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.012389 (rank : 96) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
KCNQ1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.065992 (rank : 76) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
PSME2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 59) | NC score | 0.017440 (rank : 91) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UL46, Q15129 | Gene names | PSME2 | |||
|
Domain Architecture |
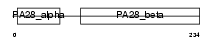
|
|||||
| Description | Proteasome activator complex subunit 2 (Proteasome activator 28- subunit beta) (PA28beta) (PA28b) (Activator of multicatalytic protease subunit 2) (11S regulator complex subunit beta) (REG-beta). | |||||
|
ACK1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | -0.000289 (rank : 122) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1080 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O54967, Q8C2U0, Q8K0K4 | Gene names | Tnk2, Ack1 | |||
|
Domain Architecture |
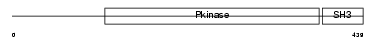
|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Non-receptor protein tyrosine kinase Ack) (Tyrosine kinase non-receptor protein 2). | |||||
|
KCNC4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.125569 (rank : 55) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNC4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.117360 (rank : 56) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
PO6F2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.015538 (rank : 94) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
PSME2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.016142 (rank : 93) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97372, O35562 | Gene names | Psme2, Pa28b1 | |||
|
Domain Architecture |
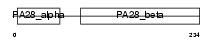
|
|||||
| Description | Proteasome activator complex subunit 2 (Proteasome activator 28- subunit beta) (PA28beta) (PA28b) (Activator of multicatalytic protease subunit 2) (11S regulator complex subunit beta) (REG-beta). | |||||
|
ALMS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.013081 (rank : 95) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
AVR2B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | -0.000137 (rank : 121) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13705, Q4VAV0 | Gene names | ACVR2B | |||
|
Domain Architecture |

|
|||||
| Description | Activin receptor type 2B precursor (EC 2.7.11.30) (Activin receptor type IIB) (ACTR-IIB). | |||||
|
AVR2B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | -0.000111 (rank : 120) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27040 | Gene names | Acvr2b | |||
|
Domain Architecture |

|
|||||
| Description | Activin receptor type 2B precursor (EC 2.7.11.30) (Activin receptor type IIB) (ACTR-IIB). | |||||
|
SCG2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.012215 (rank : 97) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13521, Q8TBH3 | Gene names | SCG2, CHGC | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-2 precursor (Secretogranin II) (SgII) (Chromogranin C) [Contains: Secretoneurin (SN)]. | |||||
|
PCQAP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.010833 (rank : 98) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
PDYN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.016517 (rank : 92) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01213 | Gene names | PDYN | |||
|
Domain Architecture |
|
|||||
| Description | Beta-neoendorphin-dynorphin precursor (Proenkephalin B) (Preprodynorphin) [Contains: Beta-neoendorphin; Dynorphin; Leu- enkephalin; Rimorphin; Leumorphin]. | |||||
|
CABP4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.008413 (rank : 104) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
KCNC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.129233 (rank : 53) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNG1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.072965 (rank : 70) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.003519 (rank : 116) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TFEB_MOUSE
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.010164 (rank : 99) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
ACTN2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.009088 (rank : 100) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35609, Q86TF4, Q86TI8 | Gene names | ACTN2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
ERCC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.008997 (rank : 101) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18074, Q8N721 | Gene names | ERCC2, XPD, XPDC | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex helicase subunit (EC 3.6.1.-) (DNA-repair protein complementing XP-D cells) (Xeroderma pigmentosum group D-complementing protein) (CXPD) (DNA excision repair protein ERCC-2). | |||||
|
KCNA2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.079886 (rank : 66) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.130929 (rank : 52) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
ODPX_MOUSE
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.006866 (rank : 109) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BKZ9 | Gene names | Pdhx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X). | |||||
|
ACTN2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.008020 (rank : 105) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JI91 | Gene names | Actn2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.008934 (rank : 102) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.001676 (rank : 119) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
KCNA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.078854 (rank : 68) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
ABTAP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.003598 (rank : 115) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H501, Q86X92, Q9H9Q5, Q9HA35, Q9NX93, Q9P1S6 | Gene names | ABTAP, C20orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
ACTN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.006189 (rank : 110) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08043 | Gene names | ACTN3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
ACTN3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.006145 (rank : 111) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88990 | Gene names | Actn3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
DJBP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.051497 (rank : 88) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P1E8, Q5DTV8, Q6PGK8, Q9D2D8 | Gene names | Djbp, Kiaa1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein. | |||||
|
ERCC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.007912 (rank : 106) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08811, Q9DC01 | Gene names | Ercc2, Xpd | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex helicase subunit (EC 3.6.1.-) (DNA-repair protein complementing XP-D cells) (Xeroderma pigmentosum group D-complementing protein) (CXPD) (DNA excision repair protein ERCC-2). | |||||
|
GA2L2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.003900 (rank : 114) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
ILF3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.007441 (rank : 107) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12906, O43409, Q6P1X1, Q86XY7, Q99544, Q99545, Q9BZH4, Q9BZH5, Q9NQ95, Q9NQ96, Q9NQ97, Q9NQ98, Q9NQ99, Q9NQA0, Q9NQA1, Q9NQA2, Q9NRN2, Q9NRN3, Q9NRN4, Q9UMZ9, Q9UN00, Q9UN84, Q9UNA2 | Gene names | ILF3, DRBF, MPHOSPH4, NF90 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3 (Nuclear factor of activated T- cells 90 kDa) (NF-AT-90) (Double-stranded RNA-binding protein 76) (DRBP76) (Translational control protein 80) (TCP80) (Nuclear factor associated with dsRNA) (NFAR) (M-phase phosphoprotein 4) (MPP4). | |||||
|
MCL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.005689 (rank : 112) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97287, Q3TUS0, Q792P0, Q9CRI4 | Gene names | Mcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Induced myeloid leukemia cell differentiation protein Mcl-1 homolog (Bcl-2-related protein EAT/mcl1). | |||||
|
TFR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.003465 (rank : 117) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P02786, Q59G55, Q9UCN0, Q9UCU5, Q9UDF9, Q9UK21 | Gene names | TFRC | |||
|
Domain Architecture |

|
|||||
| Description | Transferrin receptor protein 1 (TfR1) (TR) (TfR) (Trfr) (CD71 antigen) (T9) (p90). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.002488 (rank : 118) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
KCNA6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.078443 (rank : 69) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
PHF3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.004011 (rank : 113) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
SYP2L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.008719 (rank : 103) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H987 | Gene names | SYNPO2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
TPTE2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.126259 (rank : 54) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6XPS3, Q5VUH2, Q8WWL4, Q8WWL5 | Gene names | TPTE2, TPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase TPTE2 (EC 3.1.3.67) (TPTE and PTEN homologous inositol lipid phosphatase) (Lipid phosphatase TPIP). | |||||
|
KCNA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.080215 (rank : 65) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.082273 (rank : 64) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.067190 (rank : 74) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.066236 (rank : 75) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.057236 (rank : 82) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.057434 (rank : 81) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.064205 (rank : 78) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.069114 (rank : 72) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.071840 (rank : 71) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.069082 (rank : 73) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.102737 (rank : 59) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.101392 (rank : 60) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.063720 (rank : 79) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNQ2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.057110 (rank : 83) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.053679 (rank : 85) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.052945 (rank : 86) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.051943 (rank : 87) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.084484 (rank : 63) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
KCNQ5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.061499 (rank : 80) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
MCLN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.056680 (rank : 84) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K595, Q3UCG4, Q8K2T6, Q9CQD3 | Gene names | Mcoln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
PKDRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.092659 (rank : 61) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
PKDRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.065056 (rank : 77) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
TPTE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.086121 (rank : 62) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P56180, Q6XPS4, Q6XPS5, Q71JA8, Q8NCS8 | Gene names | TPTE | |||
|
Domain Architecture |

|
|||||
| Description | Putative tyrosine-protein phosphatase TPTE (EC 3.1.3.48) (Transmembrane phosphatase with tensin homology) (Protein BJ-HCC-5). | |||||
|
TRPV3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.050838 (rank : 89) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
CAC1S_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 13) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1S_HUMAN
|
||||||
| NC score | 0.998457 (rank : 2) | θ value | 0 (rank : 12) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q13698, Q12896, Q13934 | Gene names | CACNA1S, CACH1, CACN1, CACNL1A3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
CAC1D_HUMAN
|
||||||
| NC score | 0.994651 (rank : 3) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CAC1F_MOUSE
|
||||||
| NC score | 0.993470 (rank : 4) | θ value | 0 (rank : 11) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9JIS7 | Gene names | Cacna1f | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1C_MOUSE
|
||||||
| NC score | 0.993423 (rank : 5) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q01815, Q04476, Q61242, Q99242 | Gene names | Cacna1c, Cach2, Cacn2, Cacnl1a1, Cchl1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle) (Mouse brain class C) (MBC) (MELC-CC). | |||||
|
CAC1F_HUMAN
|
||||||
| NC score | 0.993419 (rank : 6) | θ value | 0 (rank : 10) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.993102 (rank : 7) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
CAC1E_MOUSE
|
||||||
| NC score | 0.962186 (rank : 8) | θ value | 0 (rank : 9) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q61290 | Gene names | Cacna1e, Cach6, Cacnl1a6, Cchra1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1E_HUMAN
|
||||||
| NC score | 0.961896 (rank : 9) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.957304 (rank : 10) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1A_MOUSE
|
||||||
| NC score | 0.957024 (rank : 11) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P97445 | Gene names | Cacna1a, Cach4, Cacn3, Cacnl1a4, Ccha1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.953077 (rank : 12) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CAC1A_HUMAN
|
||||||
| NC score | 0.948259 (rank : 13) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O00555, P78510, P78511, Q16290, Q92690, Q99790, Q99791, Q99792, Q99793 | Gene names | CACNA1A, CACH4, CACN3, CACNL1A4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent P/Q-type calcium channel subunit alpha-1A (Voltage- gated calcium channel subunit alpha Cav2.1) (Calcium channel, L type, alpha-1 polypeptide isoform 4) (Brain calcium channel I) (BI). | |||||
|
CAC1I_HUMAN
|
||||||
| NC score | 0.901279 (rank : 14) | θ value | 1.89707e-74 (rank : 14) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
CAC1H_MOUSE
|
||||||
| NC score | 0.877533 (rank : 15) | θ value | 8.24801e-70 (rank : 26) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.873680 (rank : 16) | θ value | 8.81223e-72 (rank : 20) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
SC11A_MOUSE
|
||||||
| NC score | 0.865257 (rank : 17) | θ value | 3.70236e-70 (rank : 24) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SC11A_HUMAN
|
||||||
| NC score | 0.863594 (rank : 18) | θ value | 4.22625e-74 (rank : 15) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UI33, Q68K15, Q8NDX3, Q9UHE0, Q9UHM0 | Gene names | SCN11A, PN5, SCN12A, SNS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (Peripheral nerve sodium channel 5) (hNaN). | |||||
|
SCN5A_HUMAN
|
||||||
| NC score | 0.858206 (rank : 19) | θ value | 4.52582e-68 (rank : 29) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q14524 | Gene names | SCN5A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 5 subunit alpha (Sodium channel protein type V subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.5) (Sodium channel protein, cardiac muscle alpha-subunit) (HH1). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.858005 (rank : 20) | θ value | 2.17053e-70 (rank : 22) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
SCN7A_HUMAN
|
||||||
| NC score | 0.856526 (rank : 21) | θ value | 3.36421e-55 (rank : 30) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q01118 | Gene names | SCN7A, SCN6A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 7 subunit alpha (Sodium channel protein type VII subunit alpha) (Putative voltage-gated sodium channel alpha subunit Nax) (Sodium channel protein, cardiac and skeletal muscle alpha-subunit). | |||||
|
SCN4A_HUMAN
|
||||||
| NC score | 0.856406 (rank : 22) | θ value | 2.31901e-72 (rank : 18) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P35499, Q15478, Q16447, Q7Z6B1 | Gene names | SCN4A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 4 subunit alpha (Sodium channel protein type IV subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.4) (Sodium channel protein, skeletal muscle subunit alpha) (SkM1). | |||||
|
SCN3A_HUMAN
|
||||||
| NC score | 0.849771 (rank : 23) | θ value | 6.10265e-73 (rank : 17) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SC10A_MOUSE
|
||||||
| NC score | 0.848735 (rank : 24) | θ value | 5.1662e-72 (rank : 19) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6QIY3, Q62243, Q6EWG7, Q6KCH7, Q703F9 | Gene names | Scn10a, Pn3, Sns | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (Sensory neuron sodium channel). | |||||
|
SCN8A_HUMAN
|
||||||
| NC score | 0.847105 (rank : 25) | θ value | 2.83479e-70 (rank : 23) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UQD0, O95788, Q9NYX2, Q9UPB2 | Gene names | SCN8A | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN2A_HUMAN
|
||||||
| NC score | 0.846875 (rank : 26) | θ value | 4.22625e-74 (rank : 16) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SCN9A_HUMAN
|
||||||
| NC score | 0.846393 (rank : 27) | θ value | 8.81223e-72 (rank : 21) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q15858, Q6B4R9, Q6B4S0, Q6B4S1, Q70HX1, Q70HX2, Q8WTU1, Q8WWN4 | Gene names | SCN9A, NENA, PN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 9 subunit alpha (Sodium channel protein type IX subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.7) (Neuroendocrine sodium channel) (hNE-Na) (Peripheral sodium channel 1). | |||||
|
SCN8A_MOUSE
|
||||||
| NC score | 0.844506 (rank : 28) | θ value | 6.31527e-70 (rank : 25) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9WTU3, Q60828, Q60858, Q62449 | Gene names | Scn8a, Nbna1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 8 subunit alpha (Sodium channel protein type VIII subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.6). | |||||
|
SCN1A_HUMAN
|
||||||
| NC score | 0.844435 (rank : 29) | θ value | 9.11921e-69 (rank : 28) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P35498, Q16172, Q96LA3, Q9C008 | Gene names | SCN1A, NAC1, SCN1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 1 subunit alpha (Sodium channel protein type I subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.1) (Sodium channel protein, brain I subunit alpha). | |||||
|
SC10A_HUMAN
|
||||||
| NC score | 0.843991 (rank : 30) | θ value | 1.40689e-69 (rank : 27) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
PK2L2_HUMAN
|
||||||
| NC score | 0.371891 (rank : 31) | θ value | 1.87187e-05 (rank : 31) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NZM6, Q9UNJ0 | Gene names | PKD2L2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PK2L1_HUMAN
|
||||||
| NC score | 0.340281 (rank : 32) | θ value | 3.19293e-05 (rank : 33) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9P0L9, O75972, Q9UP35, Q9UPA2 | Gene names | PKD2L1, PKD2L, PKDL | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 1 protein (Polycystin-L) (Polycystin- 2 homolog). | |||||
|
PK2L2_MOUSE
|
||||||
| NC score | 0.335092 (rank : 33) | θ value | 0.000602161 (rank : 36) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JLG4 | Gene names | Pkd2l2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 2-like 2 protein (Polycystin-L2). | |||||
|
PKD2_MOUSE
|
||||||
| NC score | 0.326012 (rank : 34) | θ value | 5.44631e-05 (rank : 34) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O35245, Q9QWP0, Q9Z193, Q9Z194 | Gene names | Pkd2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog). | |||||
|
PKD2_HUMAN
|
||||||
| NC score | 0.319275 (rank : 35) | θ value | 0.000158464 (rank : 35) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13563, O60441, Q15764 | Gene names | PKD2 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystin-2 (Polycystic kidney disease 2 protein homolog) (Autosomal dominant polycystic kidney disease type II protein) (Polycystwin) (R48321). | |||||
|
KCNF1_HUMAN
|
||||||
| NC score | 0.240059 (rank : 36) | θ value | 0.00175202 (rank : 37) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9H3M0, O43527 | Gene names | KCNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily F member 1 (Voltage-gated potassium channel subunit Kv5.1) (kH1). | |||||
|
KCNV2_HUMAN
|
||||||
| NC score | 0.229680 (rank : 37) | θ value | 2.44474e-05 (rank : 32) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8TDN2 | Gene names | KCNV2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily V member 2 (Voltage-gated potassium channel subunit Kv8.2). | |||||
|
KCNB2_HUMAN
|
||||||
| NC score | 0.209465 (rank : 38) | θ value | 0.00298849 (rank : 38) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q92953, Q9BXD3 | Gene names | KCNB2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 2 (Voltage-gated potassium channel subunit Kv2.2). | |||||
|
KCNS2_MOUSE
|
||||||
| NC score | 0.202351 (rank : 39) | θ value | 0.00869519 (rank : 41) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O35174, Q543P3 | Gene names | Kcns2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNS2_HUMAN
|
||||||
| NC score | 0.198602 (rank : 40) | θ value | 0.0193708 (rank : 46) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9ULS6 | Gene names | KCNS2, KIAA1144 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 2 (Voltage-gated potassium channel subunit Kv9.2) (Delayed-rectifier K(+) channel alpha subunit 2). | |||||
|
KCNB1_MOUSE
|
||||||
| NC score | 0.196621 (rank : 41) | θ value | 0.0113563 (rank : 43) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q03717 | Gene names | Kcnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (mShab). | |||||
|
KCNB1_HUMAN
|
||||||
| NC score | 0.196531 (rank : 42) | θ value | 0.0113563 (rank : 42) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q14721, Q14193 | Gene names | KCNB1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily B member 1 (Voltage-gated potassium channel subunit Kv2.1) (h-DRK1). | |||||
|
KCND3_MOUSE
|
||||||
| NC score | 0.180503 (rank : 43) | θ value | 0.0148317 (rank : 45) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Z0V1, Q8CC44, Q9Z0V0 | Gene names | Kcnd3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCND3_HUMAN
|
||||||
| NC score | 0.180375 (rank : 44) | θ value | 0.0148317 (rank : 44) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UK17, O60576, O60577, Q5T0M0, Q9UH85, Q9UH86, Q9UK16 | Gene names | KCND3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 3 (Voltage-gated potassium channel subunit Kv4.3). | |||||
|
KCNS3_HUMAN
|
||||||
| NC score | 0.175241 (rank : 45) | θ value | 0.0563607 (rank : 50) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9BQ31, O43651, Q96B56 | Gene names | KCNS3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 3 (Voltage-gated potassium channel subunit Kv9.3) (Delayed-rectifier K(+) channel alpha subunit 3). | |||||
|
KCND2_MOUSE
|
||||||
| NC score | 0.174333 (rank : 46) | θ value | 0.0252991 (rank : 48) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Z0V2, Q8BSK3, Q8CHB7, Q9JJ60 | Gene names | Kcnd2, Kiaa1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND2_HUMAN
|
||||||
| NC score | 0.174172 (rank : 47) | θ value | 0.0252991 (rank : 47) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NZV8, O95012, O95021, Q9UBY7, Q9UN98, Q9UNH9 | Gene names | KCND2, KIAA1044 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 2 (Voltage-gated potassium channel subunit Kv4.2). | |||||
|
KCND1_MOUSE
|
||||||
| NC score | 0.164309 (rank : 48) | θ value | 0.00509761 (rank : 39) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q03719, Q8CC68 | Gene names | Kcnd1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1) (mShal). | |||||
|
KCND1_HUMAN
|
||||||
| NC score | 0.160242 (rank : 49) | θ value | 0.00665767 (rank : 40) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NSA2, O75671 | Gene names | KCND1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily D member 1 (Voltage-gated potassium channel subunit Kv4.1). | |||||
|
KCNS1_HUMAN
|
||||||
| NC score | 0.148182 (rank : 50) | θ value | 0.62314 (rank : 56) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q96KK3, O43652, Q6DJU6 | Gene names | KCNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNS1_MOUSE
|
||||||
| NC score | 0.137332 (rank : 51) | θ value | 0.0961366 (rank : 52) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O35173 | Gene names | Kcns1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily S member 1 (Voltage-gated potassium channel subunit Kv9.1) (Delayed-rectifier K(+) channel alpha subunit 1). | |||||
|
KCNC1_HUMAN
|
||||||
| NC score | 0.130929 (rank : 52) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P48547 | Gene names | KCNC1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
KCNC1_MOUSE
|
||||||
| NC score | 0.129233 (rank : 53) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P15388 | Gene names | Kcnc1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 1 (Voltage-gated potassium channel subunit Kv3.1) (Kv4) (NGK2). | |||||
|
TPTE2_HUMAN
|
||||||
| NC score | 0.126259 (rank : 54) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6XPS3, Q5VUH2, Q8WWL4, Q8WWL5 | Gene names | TPTE2, TPIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase TPTE2 (EC 3.1.3.67) (TPTE and PTEN homologous inositol lipid phosphatase) (Lipid phosphatase TPIP). | |||||
|
KCNC4_HUMAN
|
||||||
| NC score | 0.125569 (rank : 55) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q03721, Q3MIM4, Q5TBI6 | Gene names | KCNC4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4) (KSHIIIC). | |||||
|
KCNC4_MOUSE
|
||||||
| NC score | 0.117360 (rank : 56) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8R1C0 | Gene names | Kcnc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily C member 4 (Voltage-gated potassium channel subunit Kv3.4). | |||||
|
KCNC3_HUMAN
|
||||||
| NC score | 0.114884 (rank : 57) | θ value | 0.365318 (rank : 53) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNC3_MOUSE
|
||||||
| NC score | 0.113366 (rank : 58) | θ value | 0.365318 (rank : 54) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q63959, Q62088 | Gene names | Kcnc3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
KCNG3_HUMAN
|
||||||
| NC score | 0.102737 (rank : 59) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TAE7 | Gene names | KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
KCNG3_MOUSE
|
||||||
| NC score | 0.101392 (rank : 60) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P59053 | Gene names | Kcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 3 (Voltage-gated potassium channel subunit Kv6.3) (Kv10.1). | |||||
|
PKDRE_HUMAN
|
||||||
| NC score | 0.092659 (rank : 61) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NTG1, O95850 | Gene names | PKDREJ | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
TPTE_HUMAN
|
||||||
| NC score | 0.086121 (rank : 62) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P56180, Q6XPS4, Q6XPS5, Q71JA8, Q8NCS8 | Gene names | TPTE | |||
|
Domain Architecture |

|
|||||
| Description | Putative tyrosine-protein phosphatase TPTE (EC 3.1.3.48) (Transmembrane phosphatase with tensin homology) (Protein BJ-HCC-5). | |||||
|
KCNQ4_HUMAN
|
||||||
| NC score | 0.084484 (rank : 63) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P56696, O96025 | Gene names | KCNQ4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 4 (Voltage-gated potassium channel subunit Kv7.4) (Potassium channel subunit alpha KvLQT4) (KQT-like 4). | |||||
|
KCNA1_MOUSE
|
||||||
| NC score | 0.082273 (rank : 64) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P16388 | Gene names | Kcna1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (MKI) (MBK1). | |||||
|
KCNA1_HUMAN
|
||||||
| NC score | 0.080215 (rank : 65) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q09470 | Gene names | KCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 1 (Voltage-gated potassium channel subunit Kv1.1) (HUKI) (HBK1). | |||||
|
KCNA2_MOUSE
|
||||||
| NC score | 0.079886 (rank : 66) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P63141, P15386, Q02010, Q8C8W4 | Gene names | Kcna2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (MK2). | |||||
|
KCNQ1_MOUSE
|
||||||
| NC score | 0.079663 (rank : 67) | θ value | 0.0252991 (rank : 49) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KCNA2_HUMAN
|
||||||
| NC score | 0.078854 (rank : 68) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P16389 | Gene names | KCNA2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 2 (Voltage-gated potassium channel subunit Kv1.2) (HBK5) (NGK1) (HUKIV). | |||||
|
KCNA6_MOUSE
|
||||||
| NC score | 0.078443 (rank : 69) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q61923 | Gene names | Kcna6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (MK1.6). | |||||
|
KCNG1_HUMAN
|
||||||
| NC score | 0.072965 (rank : 70) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UIX4, O43528, Q9BRC1 | Gene names | KCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 1 (Voltage-gated potassium channel subunit Kv6.1) (kH2). | |||||
|
KCNA6_HUMAN
|
||||||
| NC score | 0.071840 (rank : 71) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P17658 | Gene names | KCNA6 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 6 (Voltage-gated potassium channel subunit Kv1.6) (HBK2). | |||||
|
KCNA5_MOUSE
|
||||||
| NC score | 0.069114 (rank : 72) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61762 | Gene names | Kcna5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (KV1-5). | |||||
|
KCNG2_HUMAN
|
||||||
| NC score | 0.069082 (rank : 73) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UJ96 | Gene names | KCNG2, KCNF2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 2 (Voltage-gated potassium channel subunit Kv6.2) (Cardiac potassium channel subunit). | |||||
|
KCNA3_HUMAN
|
||||||
| NC score | 0.067190 (rank : 74) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P22001 | Gene names | KCNA3, HGK5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (HPCN3) (HGK5) (HuKIII) (HLK3). | |||||
|
KCNA3_MOUSE
|
||||||
| NC score | 0.066236 (rank : 75) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P16390 | Gene names | Kcna3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 3 (Voltage-gated potassium channel subunit Kv1.3) (MK3). | |||||
|
KCNQ1_HUMAN
|
||||||
| NC score | 0.065992 (rank : 76) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P51787, O00347, O60607, O94787, Q7Z6G9, Q92960, Q9UMN8, Q9UMN9 | Gene names | KCNQ1, KCNA8, KCNA9, KVLQT1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
PKDRE_MOUSE
|
||||||
| NC score | 0.065056 (rank : 77) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Z0T6 | Gene names | Pkdrej | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease and receptor for egg jelly-related protein precursor (PKD and REJ homolog). | |||||
|
KCNA5_HUMAN
|
||||||
| NC score | 0.064205 (rank : 78) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P22460 | Gene names | KCNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 5 (Voltage-gated potassium channel subunit Kv1.5) (HK2) (HPCN1). | |||||
|
KCNG4_HUMAN
|
||||||
| NC score | 0.063720 (rank : 79) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8TDN1, Q96H24 | Gene names | KCNG4, KCNG3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily G member 4 (Voltage-gated potassium channel subunit Kv6.4). | |||||
|
KCNQ5_HUMAN
|
||||||
| NC score | 0.061499 (rank : 80) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NR82, Q9NRN0, Q9NYA6 | Gene names | KCNQ5 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 5 (Voltage-gated potassium channel subunit Kv7.5) (Potassium channel subunit alpha KvLQT5) (KQT-like 5). | |||||
|
KCNA4_MOUSE
|
||||||
| NC score | 0.057434 (rank : 81) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61423 | Gene names | Kcna4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4). | |||||
|
KCNA4_HUMAN
|
||||||
| NC score | 0.057236 (rank : 82) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P22459 | Gene names | KCNA4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily A member 4 (Voltage-gated potassium channel subunit Kv1.4) (HK1) (HPCN2) (HBK4) (HUKII). | |||||
|
KCNQ2_HUMAN
|
||||||
| NC score | 0.057110 (rank : 83) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43526, O43796, O75580, O95845, Q5VYT8, Q96J59, Q99454 | Gene names | KCNQ2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Neuroblastoma-specific potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
MCLN2_MOUSE
|
||||||
| NC score | 0.056680 (rank : 84) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K595, Q3UCG4, Q8K2T6, Q9CQD3 | Gene names | Mcoln2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucolipin-2. | |||||
|
KCNQ2_MOUSE
|
||||||
| NC score | 0.053679 (rank : 85) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z351, Q8R498, Q9QWN9, Q9Z342, Q9Z343, Q9Z344, Q9Z345, Q9Z346, Q9Z347, Q9Z348, Q9Z349, Q9Z350 | Gene names | Kcnq2, Kqt2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 2 (Voltage-gated potassium channel subunit Kv7.2) (Potassium channel subunit alpha KvLQT2) (KQT-like 2). | |||||
|
KCNQ3_HUMAN
|
||||||
| NC score | 0.052945 (rank : 86) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O43525 | Gene names | KCNQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
KCNQ3_MOUSE
|
||||||
| NC score | 0.051943 (rank : 87) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K3F6 | Gene names | Kcnq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 3 (Voltage-gated potassium channel subunit Kv7.3) (Potassium channel subunit alpha KvLQT3) (KQT-like 3). | |||||
|
DJBP_MOUSE
|
||||||
| NC score | 0.051497 (rank : 88) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P1E8, Q5DTV8, Q6PGK8, Q9D2D8 | Gene names | Djbp, Kiaa1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein. | |||||
|
TRPV3_HUMAN
|
||||||
| NC score | 0.050838 (rank : 89) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NET8, Q8NDW7, Q8NET9, Q8NFH2 | Gene names | TRPV3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 3 (TrpV3) (Vanilloid receptor-like 3) (VRL-3). | |||||
|
BRCA1_MOUSE
|
||||||
| NC score | 0.018439 (rank : 90) | θ value | 0.0736092 (rank : 51) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
PSME2_HUMAN
|
||||||
| NC score | 0.017440 (rank : 91) | θ value | 0.813845 (rank : 59) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UL46, Q15129 | Gene names | PSME2 | |||
|
Domain Architecture |

|
|||||
| Description | Proteasome activator complex subunit 2 (Proteasome activator 28- subunit beta) (PA28beta) (PA28b) (Activator of multicatalytic protease subunit 2) (11S regulator complex subunit beta) (REG-beta). | |||||
|
PDYN_HUMAN
|
||||||
| NC score | 0.016517 (rank : 92) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01213 | Gene names | PDYN | |||
|
Domain Architecture |

|
|||||
| Description | Beta-neoendorphin-dynorphin precursor (Proenkephalin B) (Preprodynorphin) [Contains: Beta-neoendorphin; Dynorphin; Leu- enkephalin; Rimorphin; Leumorphin]. | |||||
|
PSME2_MOUSE
|
||||||
| NC score | 0.016142 (rank : 93) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97372, O35562 | Gene names | Psme2, Pa28b1 | |||
|
Domain Architecture |

|
|||||
| Description | Proteasome activator complex subunit 2 (Proteasome activator 28- subunit beta) (PA28beta) (PA28b) (Activator of multicatalytic protease subunit 2) (11S regulator complex subunit beta) (REG-beta). | |||||
|
PO6F2_HUMAN
|
||||||
| NC score | 0.015538 (rank : 94) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
ALMS1_HUMAN
|
||||||
| NC score | 0.013081 (rank : 95) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.012389 (rank : 96) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
SCG2_HUMAN
|
||||||
| NC score | 0.012215 (rank : 97) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13521, Q8TBH3 | Gene names | SCG2, CHGC | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-2 precursor (Secretogranin II) (SgII) (Chromogranin C) [Contains: Secretoneurin (SN)]. | |||||
|
PCQAP_MOUSE
|
||||||
| NC score | 0.010833 (rank : 98) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
TFEB_MOUSE
|
||||||
| NC score | 0.010164 (rank : 99) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R210 | Gene names | Tfeb, Tcfeb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor EB. | |||||
|
ACTN2_HUMAN
|
||||||
| NC score | 0.009088 (rank : 100) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35609, Q86TF4, Q86TI8 | Gene names | ACTN2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
ERCC2_HUMAN
|
||||||
| NC score | 0.008997 (rank : 101) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18074, Q8N721 | Gene names | ERCC2, XPD, XPDC | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex helicase subunit (EC 3.6.1.-) (DNA-repair protein complementing XP-D cells) (Xeroderma pigmentosum group D-complementing protein) (CXPD) (DNA excision repair protein ERCC-2). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.008934 (rank : 102) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
SYP2L_HUMAN
|
||||||
| NC score | 0.008719 (rank : 103) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H987 | Gene names | SYNPO2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
CABP4_HUMAN
|
||||||
| NC score | 0.008413 (rank : 104) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P57796, Q8N4Z2, Q8WWY5 | Gene names | CABP4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-binding protein 4 (CaBP4). | |||||
|
ACTN2_MOUSE
|
||||||
| NC score | 0.008020 (rank : 105) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JI91 | Gene names | Actn2 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-2 (Alpha actinin skeletal muscle isoform 2) (F-actin cross-linking protein). | |||||
|
ERCC2_MOUSE
|
||||||
| NC score | 0.007912 (rank : 106) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08811, Q9DC01 | Gene names | Ercc2, Xpd | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex helicase subunit (EC 3.6.1.-) (DNA-repair protein complementing XP-D cells) (Xeroderma pigmentosum group D-complementing protein) (CXPD) (DNA excision repair protein ERCC-2). | |||||
|
ILF3_HUMAN
|
||||||
| NC score | 0.007441 (rank : 107) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12906, O43409, Q6P1X1, Q86XY7, Q99544, Q99545, Q9BZH4, Q9BZH5, Q9NQ95, Q9NQ96, Q9NQ97, Q9NQ98, Q9NQ99, Q9NQA0, Q9NQA1, Q9NQA2, Q9NRN2, Q9NRN3, Q9NRN4, Q9UMZ9, Q9UN00, Q9UN84, Q9UNA2 | Gene names | ILF3, DRBF, MPHOSPH4, NF90 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin enhancer-binding factor 3 (Nuclear factor of activated T- cells 90 kDa) (NF-AT-90) (Double-stranded RNA-binding protein 76) (DRBP76) (Translational control protein 80) (TCP80) (Nuclear factor associated with dsRNA) (NFAR) (M-phase phosphoprotein 4) (MPP4). | |||||
|
PO6F2_MOUSE
|
||||||
| NC score | 0.007378 (rank : 108) | θ value | 0.365318 (rank : 55) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
ODPX_MOUSE
|
||||||
| NC score | 0.006866 (rank : 109) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BKZ9 | Gene names | Pdhx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X). | |||||
|
ACTN3_HUMAN
|
||||||
| NC score | 0.006189 (rank : 110) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08043 | Gene names | ACTN3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
ACTN3_MOUSE
|
||||||
| NC score | 0.006145 (rank : 111) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88990 | Gene names | Actn3 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-actinin-3 (Alpha actinin skeletal muscle isoform 3) (F-actin cross-linking protein). | |||||
|
MCL1_MOUSE
|
||||||
| NC score | 0.005689 (rank : 112) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97287, Q3TUS0, Q792P0, Q9CRI4 | Gene names | Mcl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Induced myeloid leukemia cell differentiation protein Mcl-1 homolog (Bcl-2-related protein EAT/mcl1). | |||||
|
PHF3_HUMAN
|
||||||
| NC score | 0.004011 (rank : 113) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
GA2L2_HUMAN
|
||||||
| NC score | 0.003900 (rank : 114) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
ABTAP_HUMAN
|
||||||
| NC score | 0.003598 (rank : 115) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H501, Q86X92, Q9H9Q5, Q9HA35, Q9NX93, Q9P1S6 | Gene names | ABTAP, C20orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.003519 (rank : 116) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TFR1_HUMAN
|
||||||
| NC score | 0.003465 (rank : 117) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P02786, Q59G55, Q9UCN0, Q9UCU5, Q9UDF9, Q9UK21 | Gene names | TFRC | |||
|
Domain Architecture |

|
|||||
| Description | Transferrin receptor protein 1 (TfR1) (TR) (TfR) (Trfr) (CD71 antigen) (T9) (p90). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.002488 (rank : 118) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.001676 (rank : 119) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
AVR2B_MOUSE
|
||||||
| NC score | -0.000111 (rank : 120) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27040 | Gene names | Acvr2b | |||
|
Domain Architecture |

|
|||||
| Description | Activin receptor type 2B precursor (EC 2.7.11.30) (Activin receptor type IIB) (ACTR-IIB). | |||||
|
AVR2B_HUMAN
|
||||||
| NC score | -0.000137 (rank : 121) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13705, Q4VAV0 | Gene names | ACVR2B | |||
|
Domain Architecture |

|
|||||
| Description | Activin receptor type 2B precursor (EC 2.7.11.30) (Activin receptor type IIB) (ACTR-IIB). | |||||
|
ACK1_MOUSE
|
||||||
| NC score | -0.000289 (rank : 122) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 98 | Target Neighborhood Hits | 1080 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O54967, Q8C2U0, Q8K0K4 | Gene names | Tnk2, Ack1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Non-receptor protein tyrosine kinase Ack) (Tyrosine kinase non-receptor protein 2). | |||||