Please be patient as the page loads
|
ODPX_MOUSE
|
||||||
| SwissProt Accessions | Q8BKZ9 | Gene names | Pdhx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ODPX_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.992209 (rank : 2) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00330, O60221, Q96FV8, Q99783 | Gene names | PDHX, PDX1 | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X) (E3-binding protein) (E3BP) (proX). | |||||
|
ODPX_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BKZ9 | Gene names | Pdhx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X). | |||||
|
ODP2_HUMAN
|
||||||
| θ value | 1.31376e-75 (rank : 3) | NC score | 0.925731 (rank : 3) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P10515, Q16783 | Gene names | DLAT, DLTA | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.12) (Pyruvate dehydrogenase complex E2 subunit) (PDCE2) (E2) (Dihydrolipoamide S- acetyltransferase component of pyruvate dehydrogenase complex) (PDC- E2) (70 kDa mitochondrial autoantigen of primary biliary cirrhosis) (PBC) (M2 antigen complex 70 kDa subunit). | |||||
|
ODB2_HUMAN
|
||||||
| θ value | 8.06329e-33 (rank : 4) | NC score | 0.808743 (rank : 4) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11182 | Gene names | DBT, BCATE2 | |||
|
Domain Architecture |
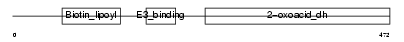
|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODB2_MOUSE
|
||||||
| θ value | 5.22648e-32 (rank : 5) | NC score | 0.802154 (rank : 5) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P53395 | Gene names | Dbt | |||
|
Domain Architecture |
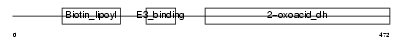
|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODO2_MOUSE
|
||||||
| θ value | 1.99067e-23 (rank : 6) | NC score | 0.739085 (rank : 7) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D2G2 | Gene names | Dlst | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
ODO2_HUMAN
|
||||||
| θ value | 1.6852e-22 (rank : 7) | NC score | 0.745265 (rank : 6) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P36957, Q9BQ32 | Gene names | DLST, DLTS | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
RFIP1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 8) | NC score | 0.099022 (rank : 8) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D620, Q3UBC2, Q8BN24 | Gene names | Rab11fip1, Rcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
MCCA_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 9) | NC score | 0.091044 (rank : 9) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99MR8, Q3TGU0, Q9D8R2 | Gene names | Mccc1, Mcca | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 10) | NC score | 0.019307 (rank : 18) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 11) | NC score | 0.028239 (rank : 15) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
MCCA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.090070 (rank : 10) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96RQ3, Q59ES4, Q9H959, Q9NS97 | Gene names | MCCC1, MCCA | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
ARRB2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 13) | NC score | 0.023551 (rank : 17) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P32121, Q0Z8D3, Q2PP19, Q6ICT3, Q8N7Y2, Q9UEQ6 | Gene names | ARRB2, ARB2, ARR2 | |||
|
Domain Architecture |
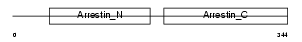
|
|||||
| Description | Beta-arrestin-2 (Arrestin beta 2). | |||||
|
DNMT1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.028882 (rank : 14) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P26358, Q9UHG5, Q9ULA2, Q9UMZ6 | Gene names | DNMT1, AIM, DNMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI). | |||||
|
RUNX2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.036490 (rank : 12) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08775, O35183, Q08776, Q9JLN0, Q9QUQ6, Q9QY29, Q9R0U4, Q9Z2J7 | Gene names | Runx2, Aml3, Cbfa1, Osf2, Pebp2a | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
COA1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.053320 (rank : 11) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13085 | Gene names | ACACA, ACAC, ACC1, ACCA | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 1 (EC 6.4.1.2) (ACC-alpha) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
RUNX2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.035371 (rank : 13) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13950, O14614, O14615, O95181 | Gene names | RUNX2, AML3, CBFA1, OSF2, PEBP2A | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
OPT_HUMAN
|
||||||
| θ value | 1.38821 (rank : 18) | NC score | 0.013698 (rank : 22) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UBM4 | Gene names | OPTC, OPT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Opticin precursor (Oculoglycan). | |||||
|
EDN3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 19) | NC score | 0.027630 (rank : 16) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48299, Q543L0 | Gene names | Edn3 | |||
|
Domain Architecture |
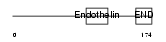
|
|||||
| Description | Endothelin-3 precursor (ET-3) (Preproendothelin-3) (PPET3). | |||||
|
EFS_MOUSE
|
||||||
| θ value | 2.36792 (rank : 20) | NC score | 0.019218 (rank : 19) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64355 | Gene names | Efs, Sin | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (SRC-interacting protein) (Signal- integrating protein). | |||||
|
KIF2C_HUMAN
|
||||||
| θ value | 2.36792 (rank : 21) | NC score | 0.008807 (rank : 27) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99661, Q96C18, Q96HB8, Q9BWV8 | Gene names | KIF2C, KNSL6 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF2C (Mitotic centromere-associated kinesin) (MCAK) (Kinesin-like protein 6). | |||||
|
CAC1S_MOUSE
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.006866 (rank : 28) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
FBLN2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.006286 (rank : 29) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.014139 (rank : 21) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
UBP43_MOUSE
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.008937 (rank : 26) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
UBP53_MOUSE
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.013431 (rank : 23) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15975, Q8CB37, Q8R251 | Gene names | Usp53, Phxr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53) (Per-hexamer repeat protein 3). | |||||
|
ZN496_HUMAN
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.000369 (rank : 32) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96IT1 | Gene names | ZNF496 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 496. | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.013143 (rank : 24) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
FOG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.001994 (rank : 31) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
K1383_HUMAN
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.015854 (rank : 20) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P2G4, Q58EZ9, Q5VV83 | Gene names | KIAA1383 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1383. | |||||
|
LRC17_HUMAN
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.004602 (rank : 30) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N6Y2, Q13288, Q6UWA7, Q75MG5 | Gene names | LRRC17, P37NB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 17 precursor (p37NB). | |||||
|
PI5PA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.011111 (rank : 25) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
ODPX_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BKZ9 | Gene names | Pdhx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X). | |||||
|
ODPX_HUMAN
|
||||||
| NC score | 0.992209 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00330, O60221, Q96FV8, Q99783 | Gene names | PDHX, PDX1 | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X) (E3-binding protein) (E3BP) (proX). | |||||
|
ODP2_HUMAN
|
||||||
| NC score | 0.925731 (rank : 3) | θ value | 1.31376e-75 (rank : 3) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P10515, Q16783 | Gene names | DLAT, DLTA | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.12) (Pyruvate dehydrogenase complex E2 subunit) (PDCE2) (E2) (Dihydrolipoamide S- acetyltransferase component of pyruvate dehydrogenase complex) (PDC- E2) (70 kDa mitochondrial autoantigen of primary biliary cirrhosis) (PBC) (M2 antigen complex 70 kDa subunit). | |||||
|
ODB2_HUMAN
|
||||||
| NC score | 0.808743 (rank : 4) | θ value | 8.06329e-33 (rank : 4) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11182 | Gene names | DBT, BCATE2 | |||
|
Domain Architecture |

|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODB2_MOUSE
|
||||||
| NC score | 0.802154 (rank : 5) | θ value | 5.22648e-32 (rank : 5) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P53395 | Gene names | Dbt | |||
|
Domain Architecture |

|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODO2_HUMAN
|
||||||
| NC score | 0.745265 (rank : 6) | θ value | 1.6852e-22 (rank : 7) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P36957, Q9BQ32 | Gene names | DLST, DLTS | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
ODO2_MOUSE
|
||||||
| NC score | 0.739085 (rank : 7) | θ value | 1.99067e-23 (rank : 6) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D2G2 | Gene names | Dlst | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
RFIP1_MOUSE
|
||||||
| NC score | 0.099022 (rank : 8) | θ value | 0.00509761 (rank : 8) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D620, Q3UBC2, Q8BN24 | Gene names | Rab11fip1, Rcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
MCCA_MOUSE
|
||||||
| NC score | 0.091044 (rank : 9) | θ value | 0.0961366 (rank : 9) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99MR8, Q3TGU0, Q9D8R2 | Gene names | Mccc1, Mcca | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
MCCA_HUMAN
|
||||||
| NC score | 0.090070 (rank : 10) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96RQ3, Q59ES4, Q9H959, Q9NS97 | Gene names | MCCC1, MCCA | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
COA1_HUMAN
|
||||||
| NC score | 0.053320 (rank : 11) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13085 | Gene names | ACACA, ACAC, ACC1, ACCA | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 1 (EC 6.4.1.2) (ACC-alpha) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
RUNX2_MOUSE
|
||||||
| NC score | 0.036490 (rank : 12) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08775, O35183, Q08776, Q9JLN0, Q9QUQ6, Q9QY29, Q9R0U4, Q9Z2J7 | Gene names | Runx2, Aml3, Cbfa1, Osf2, Pebp2a | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
RUNX2_HUMAN
|
||||||
| NC score | 0.035371 (rank : 13) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13950, O14614, O14615, O95181 | Gene names | RUNX2, AML3, CBFA1, OSF2, PEBP2A | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
DNMT1_HUMAN
|
||||||
| NC score | 0.028882 (rank : 14) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P26358, Q9UHG5, Q9ULA2, Q9UMZ6 | Gene names | DNMT1, AIM, DNMT | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase HsaI) (DNA MTase HsaI) (MCMT) (M.HsaI). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.028239 (rank : 15) | θ value | 0.21417 (rank : 11) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
EDN3_MOUSE
|
||||||
| NC score | 0.027630 (rank : 16) | θ value | 2.36792 (rank : 19) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48299, Q543L0 | Gene names | Edn3 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-3 precursor (ET-3) (Preproendothelin-3) (PPET3). | |||||
|
ARRB2_HUMAN
|
||||||
| NC score | 0.023551 (rank : 17) | θ value | 0.62314 (rank : 13) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P32121, Q0Z8D3, Q2PP19, Q6ICT3, Q8N7Y2, Q9UEQ6 | Gene names | ARRB2, ARB2, ARR2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-arrestin-2 (Arrestin beta 2). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.019307 (rank : 18) | θ value | 0.21417 (rank : 10) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
EFS_MOUSE
|
||||||
| NC score | 0.019218 (rank : 19) | θ value | 2.36792 (rank : 20) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64355 | Gene names | Efs, Sin | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (SRC-interacting protein) (Signal- integrating protein). | |||||
|
K1383_HUMAN
|
||||||
| NC score | 0.015854 (rank : 20) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P2G4, Q58EZ9, Q5VV83 | Gene names | KIAA1383 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1383. | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.014139 (rank : 21) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
OPT_HUMAN
|
||||||
| NC score | 0.013698 (rank : 22) | θ value | 1.38821 (rank : 18) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UBM4 | Gene names | OPTC, OPT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Opticin precursor (Oculoglycan). | |||||
|
UBP53_MOUSE
|
||||||
| NC score | 0.013431 (rank : 23) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15975, Q8CB37, Q8R251 | Gene names | Usp53, Phxr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53) (Per-hexamer repeat protein 3). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.013143 (rank : 24) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
PI5PA_MOUSE
|
||||||
| NC score | 0.011111 (rank : 25) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
UBP43_MOUSE
|
||||||
| NC score | 0.008937 (rank : 26) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BUM9, Q8VDP5 | Gene names | Usp43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 43 (EC 3.1.2.15) (Ubiquitin thioesterase 43) (Ubiquitin-specific-processing protease 43) (Deubiquitinating enzyme 43). | |||||
|
KIF2C_HUMAN
|
||||||
| NC score | 0.008807 (rank : 27) | θ value | 2.36792 (rank : 21) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99661, Q96C18, Q96HB8, Q9BWV8 | Gene names | KIF2C, KNSL6 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF2C (Mitotic centromere-associated kinesin) (MCAK) (Kinesin-like protein 6). | |||||
|
CAC1S_MOUSE
|
||||||
| NC score | 0.006866 (rank : 28) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q02789, Q99240 | Gene names | Cacna1s, Cach1, Cach1b, Cacn1, Cacnl1a3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1S (Voltage- gated calcium channel subunit alpha Cav1.1) (Calcium channel, L type, alpha-1 polypeptide, isoform 3, skeletal muscle). | |||||
|
FBLN2_MOUSE
|
||||||
| NC score | 0.006286 (rank : 29) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
LRC17_HUMAN
|
||||||
| NC score | 0.004602 (rank : 30) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N6Y2, Q13288, Q6UWA7, Q75MG5 | Gene names | LRRC17, P37NB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 17 precursor (p37NB). | |||||
|
FOG1_HUMAN
|
||||||
| NC score | 0.001994 (rank : 31) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
ZN496_HUMAN
|
||||||
| NC score | 0.000369 (rank : 32) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 743 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96IT1 | Gene names | ZNF496 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 496. | |||||