Please be patient as the page loads
|
ODO2_HUMAN
|
||||||
| SwissProt Accessions | P36957, Q9BQ32 | Gene names | DLST, DLTS | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ODO2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P36957, Q9BQ32 | Gene names | DLST, DLTS | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
ODO2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.978854 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D2G2 | Gene names | Dlst | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
ODB2_HUMAN
|
||||||
| θ value | 1.92365e-26 (rank : 3) | NC score | 0.764564 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11182 | Gene names | DBT, BCATE2 | |||
|
Domain Architecture |
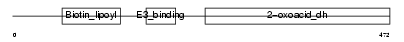
|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODPX_HUMAN
|
||||||
| θ value | 5.59698e-26 (rank : 4) | NC score | 0.757004 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00330, O60221, Q96FV8, Q99783 | Gene names | PDHX, PDX1 | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X) (E3-binding protein) (E3BP) (proX). | |||||
|
ODB2_MOUSE
|
||||||
| θ value | 2.12685e-25 (rank : 5) | NC score | 0.762066 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P53395 | Gene names | Dbt | |||
|
Domain Architecture |
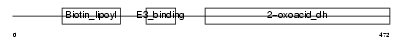
|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODP2_HUMAN
|
||||||
| θ value | 1.37858e-24 (rank : 6) | NC score | 0.765455 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P10515, Q16783 | Gene names | DLAT, DLTA | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.12) (Pyruvate dehydrogenase complex E2 subunit) (PDCE2) (E2) (Dihydrolipoamide S- acetyltransferase component of pyruvate dehydrogenase complex) (PDC- E2) (70 kDa mitochondrial autoantigen of primary biliary cirrhosis) (PBC) (M2 antigen complex 70 kDa subunit). | |||||
|
ODPX_MOUSE
|
||||||
| θ value | 1.6852e-22 (rank : 7) | NC score | 0.745265 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BKZ9 | Gene names | Pdhx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 8) | NC score | 0.070947 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 0.163984 (rank : 9) | NC score | 0.080761 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.279714 (rank : 10) | NC score | 0.037805 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
CIC_MOUSE
|
||||||
| θ value | 0.365318 (rank : 11) | NC score | 0.068687 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.049740 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.034249 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
ARMX2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.045801 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
MAVS_HUMAN
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.055347 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.053417 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.018528 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
LAMC1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 18) | NC score | 0.010704 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11047 | Gene names | LAMC1, LAMB2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.043743 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
NOTC2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.009438 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
SON_HUMAN
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.040826 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.039128 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
TACC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.020124 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.057066 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.028948 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
DEAF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.022464 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z1T5 | Gene names | Deaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.032922 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PO6F1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.029159 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q07916, Q91XK3 | Gene names | Pou6f1, Emb | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 1 (Octamer-binding transcription factor EMB) (Transcription regulatory protein MCP-1). | |||||
|
TM108_MOUSE
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.044479 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
CHMP6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.025202 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96FZ7, Q53FU4, Q9HAE8 | Gene names | CHMP6, VPS20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 6 (Chromatin-modifying protein 6) (Vacuolar protein sorting-associated protein 20) (hVps20). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.022127 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
HGS_HUMAN
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.016673 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
ZBT16_HUMAN
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.000301 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.021434 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.041547 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.042695 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CC28B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.021588 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BUN5, Q8TBV8 | Gene names | CCDC28B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 28B. | |||||
|
COIA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.016947 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
CS021_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.023999 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IVT2 | Gene names | C19orf21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf21. | |||||
|
G3BP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.012636 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97855 | Gene names | G3bp, G3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 1 (EC 3.6.1.-) (ATP- dependent DNA helicase VIII) (GAP SH3-domain-binding protein 1) (G3BP- 1) (HDH-VIII). | |||||
|
GORS2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.025849 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99JX3, Q3U9D2, Q8CCD0, Q91W68 | Gene names | Gorasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.007407 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.032068 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.022524 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.025961 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ALMS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.022614 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.042332 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
FREA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.006280 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43638, Q96D28 | Gene names | FKHL18, FREAC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead-related transcription factor 10 (FREAC-10) (Forkhead-like 18 protein). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.021506 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.029906 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
REL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.011423 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P15307 | Gene names | Rel | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.010139 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
DYNA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.006238 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.016687 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
MYLK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.005697 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
PHLB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.014488 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86UU1, O75133, Q4KMF8, Q8TEQ2 | Gene names | PHLDB1, DLNB07, KIAA0638, LL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
UBP10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.009814 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14694, Q9BWG7, Q9NSL7 | Gene names | USP10, KIAA0190 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.010453 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
ODO2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P36957, Q9BQ32 | Gene names | DLST, DLTS | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
ODO2_MOUSE
|
||||||
| NC score | 0.978854 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D2G2 | Gene names | Dlst | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
ODP2_HUMAN
|
||||||
| NC score | 0.765455 (rank : 3) | θ value | 1.37858e-24 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P10515, Q16783 | Gene names | DLAT, DLTA | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.12) (Pyruvate dehydrogenase complex E2 subunit) (PDCE2) (E2) (Dihydrolipoamide S- acetyltransferase component of pyruvate dehydrogenase complex) (PDC- E2) (70 kDa mitochondrial autoantigen of primary biliary cirrhosis) (PBC) (M2 antigen complex 70 kDa subunit). | |||||
|
ODB2_HUMAN
|
||||||
| NC score | 0.764564 (rank : 4) | θ value | 1.92365e-26 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11182 | Gene names | DBT, BCATE2 | |||
|
Domain Architecture |

|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODB2_MOUSE
|
||||||
| NC score | 0.762066 (rank : 5) | θ value | 2.12685e-25 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P53395 | Gene names | Dbt | |||
|
Domain Architecture |

|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODPX_HUMAN
|
||||||
| NC score | 0.757004 (rank : 6) | θ value | 5.59698e-26 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00330, O60221, Q96FV8, Q99783 | Gene names | PDHX, PDX1 | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X) (E3-binding protein) (E3BP) (proX). | |||||
|
ODPX_MOUSE
|
||||||
| NC score | 0.745265 (rank : 7) | θ value | 1.6852e-22 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BKZ9 | Gene names | Pdhx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.080761 (rank : 8) | θ value | 0.163984 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.070947 (rank : 9) | θ value | 0.0148317 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.068687 (rank : 10) | θ value | 0.365318 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.057066 (rank : 11) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
MAVS_HUMAN
|
||||||
| NC score | 0.055347 (rank : 12) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.053417 (rank : 13) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.049740 (rank : 14) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
ARMX2_MOUSE
|
||||||
| NC score | 0.045801 (rank : 15) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
TM108_MOUSE
|
||||||
| NC score | 0.044479 (rank : 16) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.043743 (rank : 17) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.042695 (rank : 18) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.042332 (rank : 19) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.041547 (rank : 20) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.040826 (rank : 21) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.039128 (rank : 22) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.037805 (rank : 23) | θ value | 0.279714 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.034249 (rank : 24) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.032922 (rank : 25) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.032068 (rank : 26) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.029906 (rank : 27) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
PO6F1_MOUSE
|
||||||
| NC score | 0.029159 (rank : 28) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q07916, Q91XK3 | Gene names | Pou6f1, Emb | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 1 (Octamer-binding transcription factor EMB) (Transcription regulatory protein MCP-1). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.028948 (rank : 29) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.025961 (rank : 30) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
GORS2_MOUSE
|
||||||
| NC score | 0.025849 (rank : 31) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99JX3, Q3U9D2, Q8CCD0, Q91W68 | Gene names | Gorasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55). | |||||
|
CHMP6_HUMAN
|
||||||
| NC score | 0.025202 (rank : 32) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96FZ7, Q53FU4, Q9HAE8 | Gene names | CHMP6, VPS20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Charged multivesicular body protein 6 (Chromatin-modifying protein 6) (Vacuolar protein sorting-associated protein 20) (hVps20). | |||||
|
CS021_HUMAN
|
||||||
| NC score | 0.023999 (rank : 33) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IVT2 | Gene names | C19orf21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf21. | |||||
|
ALMS1_HUMAN
|
||||||
| NC score | 0.022614 (rank : 34) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.022524 (rank : 35) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
DEAF1_MOUSE
|
||||||
| NC score | 0.022464 (rank : 36) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z1T5 | Gene names | Deaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.022127 (rank : 37) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CC28B_HUMAN
|
||||||
| NC score | 0.021588 (rank : 38) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BUN5, Q8TBV8 | Gene names | CCDC28B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 28B. | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.021506 (rank : 39) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.021434 (rank : 40) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
TACC2_HUMAN
|
||||||
| NC score | 0.020124 (rank : 41) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.018528 (rank : 42) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
COIA1_MOUSE
|
||||||
| NC score | 0.016947 (rank : 43) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.016687 (rank : 44) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
HGS_HUMAN
|
||||||
| NC score | 0.016673 (rank : 45) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
PHLB1_HUMAN
|
||||||
| NC score | 0.014488 (rank : 46) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1038 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q86UU1, O75133, Q4KMF8, Q8TEQ2 | Gene names | PHLDB1, DLNB07, KIAA0638, LL5A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
G3BP_MOUSE
|
||||||
| NC score | 0.012636 (rank : 47) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97855 | Gene names | G3bp, G3bp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 1 (EC 3.6.1.-) (ATP- dependent DNA helicase VIII) (GAP SH3-domain-binding protein 1) (G3BP- 1) (HDH-VIII). | |||||
|
REL_MOUSE
|
||||||
| NC score | 0.011423 (rank : 48) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P15307 | Gene names | Rel | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
LAMC1_HUMAN
|
||||||
| NC score | 0.010704 (rank : 49) | θ value | 1.06291 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P11047 | Gene names | LAMC1, LAMB2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.010453 (rank : 50) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
WNK1_HUMAN
|
||||||
| NC score | 0.010139 (rank : 51) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
UBP10_HUMAN
|
||||||
| NC score | 0.009814 (rank : 52) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14694, Q9BWG7, Q9NSL7 | Gene names | USP10, KIAA0190 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
NOTC2_MOUSE
|
||||||
| NC score | 0.009438 (rank : 53) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.007407 (rank : 54) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
FREA_HUMAN
|
||||||
| NC score | 0.006280 (rank : 55) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43638, Q96D28 | Gene names | FKHL18, FREAC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead-related transcription factor 10 (FREAC-10) (Forkhead-like 18 protein). | |||||
|
DYNA_HUMAN
|
||||||
| NC score | 0.006238 (rank : 56) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
MYLK_MOUSE
|
||||||
| NC score | 0.005697 (rank : 57) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
ZBT16_HUMAN
|
||||||
| NC score | 0.000301 (rank : 58) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||