Please be patient as the page loads
|
ODB2_MOUSE
|
||||||
| SwissProt Accessions | P53395 | Gene names | Dbt | |||
|
Domain Architecture |
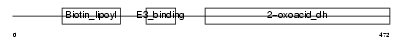
|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ODB2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999147 (rank : 2) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P11182 | Gene names | DBT, BCATE2 | |||
|
Domain Architecture |
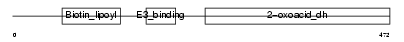
|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODB2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P53395 | Gene names | Dbt | |||
|
Domain Architecture |

|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODPX_MOUSE
|
||||||
| θ value | 5.22648e-32 (rank : 3) | NC score | 0.802154 (rank : 4) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BKZ9 | Gene names | Pdhx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X). | |||||
|
ODP2_HUMAN
|
||||||
| θ value | 4.42448e-31 (rank : 4) | NC score | 0.810752 (rank : 3) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10515, Q16783 | Gene names | DLAT, DLTA | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.12) (Pyruvate dehydrogenase complex E2 subunit) (PDCE2) (E2) (Dihydrolipoamide S- acetyltransferase component of pyruvate dehydrogenase complex) (PDC- E2) (70 kDa mitochondrial autoantigen of primary biliary cirrhosis) (PBC) (M2 antigen complex 70 kDa subunit). | |||||
|
ODPX_HUMAN
|
||||||
| θ value | 4.89182e-30 (rank : 5) | NC score | 0.800959 (rank : 5) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00330, O60221, Q96FV8, Q99783 | Gene names | PDHX, PDX1 | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X) (E3-binding protein) (E3BP) (proX). | |||||
|
ODO2_MOUSE
|
||||||
| θ value | 3.28125e-26 (rank : 6) | NC score | 0.756408 (rank : 7) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D2G2 | Gene names | Dlst | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
ODO2_HUMAN
|
||||||
| θ value | 2.12685e-25 (rank : 7) | NC score | 0.762066 (rank : 6) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P36957, Q9BQ32 | Gene names | DLST, DLTS | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
MCCA_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 8) | NC score | 0.118943 (rank : 8) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96RQ3, Q59ES4, Q9H959, Q9NS97 | Gene names | MCCC1, MCCA | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
MCCA_MOUSE
|
||||||
| θ value | 0.125558 (rank : 9) | NC score | 0.109682 (rank : 9) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99MR8, Q3TGU0, Q9D8R2 | Gene names | Mccc1, Mcca | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
ITA3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 10) | NC score | 0.023599 (rank : 14) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62470, Q08441, Q08442 | Gene names | Itga3 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-3 precursor (Galactoprotein B3) (GAPB3) (VLA-3 alpha chain) (CD49c antigen) [Contains: Integrin alpha-3 heavy chain; Integrin alpha-3 light chain]. | |||||
|
CAD10_HUMAN
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.010810 (rank : 15) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6N8, Q9ULB3 | Gene names | CDH10 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-10 precursor (T2-cadherin). | |||||
|
MCM5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 12) | NC score | 0.023968 (rank : 13) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49718 | Gene names | Mcm5, Cdc46, Mcmd5 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM5 (CDC46 homolog) (P1-CDC46). | |||||
|
PCCA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.071206 (rank : 10) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P05165, Q15979 | Gene names | PCCA | |||
|
Domain Architecture |

|
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
AOF1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.029700 (rank : 12) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CIG3, Q8C5C4, Q8CEC1 | Gene names | Aof1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
TIE2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.003392 (rank : 17) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 1192 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02763 | Gene names | TEK, TIE2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-1 receptor precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor TIE-2) (hTIE2) (Tyrosine-protein kinase receptor TEK) (p140 TEK) (Tunica interna endothelial cell kinase) (CD202b antigen). | |||||
|
PCCA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 16) | NC score | 0.059910 (rank : 11) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91ZA3, Q80VU5, Q922N3 | Gene names | Pcca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 17) | NC score | 0.010012 (rank : 16) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
TIE2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 18) | NC score | 0.002263 (rank : 18) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 1204 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02858 | Gene names | Tek, Hyk, Tie-2, Tie2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-1 receptor precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor TIE-2) (mTIE2) (Tyrosine-protein kinase receptor TEK) (p140 TEK) (Tunica interna endothelial cell kinase) (HYK) (STK1) (CD202b antigen). | |||||
|
ODB2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P53395 | Gene names | Dbt | |||
|
Domain Architecture |

|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODB2_HUMAN
|
||||||
| NC score | 0.999147 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P11182 | Gene names | DBT, BCATE2 | |||
|
Domain Architecture |

|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODP2_HUMAN
|
||||||
| NC score | 0.810752 (rank : 3) | θ value | 4.42448e-31 (rank : 4) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10515, Q16783 | Gene names | DLAT, DLTA | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.12) (Pyruvate dehydrogenase complex E2 subunit) (PDCE2) (E2) (Dihydrolipoamide S- acetyltransferase component of pyruvate dehydrogenase complex) (PDC- E2) (70 kDa mitochondrial autoantigen of primary biliary cirrhosis) (PBC) (M2 antigen complex 70 kDa subunit). | |||||
|
ODPX_MOUSE
|
||||||
| NC score | 0.802154 (rank : 4) | θ value | 5.22648e-32 (rank : 3) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BKZ9 | Gene names | Pdhx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X). | |||||
|
ODPX_HUMAN
|
||||||
| NC score | 0.800959 (rank : 5) | θ value | 4.89182e-30 (rank : 5) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00330, O60221, Q96FV8, Q99783 | Gene names | PDHX, PDX1 | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate dehydrogenase protein X component, mitochondrial precursor (Dihydrolipoamide dehydrogenase-binding protein of pyruvate dehydrogenase complex) (Lipoyl-containing pyruvate dehydrogenase complex component X) (E3-binding protein) (E3BP) (proX). | |||||
|
ODO2_HUMAN
|
||||||
| NC score | 0.762066 (rank : 6) | θ value | 2.12685e-25 (rank : 7) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P36957, Q9BQ32 | Gene names | DLST, DLTS | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
ODO2_MOUSE
|
||||||
| NC score | 0.756408 (rank : 7) | θ value | 3.28125e-26 (rank : 6) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D2G2 | Gene names | Dlst | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
MCCA_HUMAN
|
||||||
| NC score | 0.118943 (rank : 8) | θ value | 0.0113563 (rank : 8) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96RQ3, Q59ES4, Q9H959, Q9NS97 | Gene names | MCCC1, MCCA | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
MCCA_MOUSE
|
||||||
| NC score | 0.109682 (rank : 9) | θ value | 0.125558 (rank : 9) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99MR8, Q3TGU0, Q9D8R2 | Gene names | Mccc1, Mcca | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
PCCA_HUMAN
|
||||||
| NC score | 0.071206 (rank : 10) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P05165, Q15979 | Gene names | PCCA | |||
|
Domain Architecture |

|
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
PCCA_MOUSE
|
||||||
| NC score | 0.059910 (rank : 11) | θ value | 6.88961 (rank : 16) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91ZA3, Q80VU5, Q922N3 | Gene names | Pcca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
AOF1_MOUSE
|
||||||
| NC score | 0.029700 (rank : 12) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CIG3, Q8C5C4, Q8CEC1 | Gene names | Aof1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
MCM5_MOUSE
|
||||||
| NC score | 0.023968 (rank : 13) | θ value | 1.06291 (rank : 12) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49718 | Gene names | Mcm5, Cdc46, Mcmd5 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM5 (CDC46 homolog) (P1-CDC46). | |||||
|
ITA3_MOUSE
|
||||||
| NC score | 0.023599 (rank : 14) | θ value | 0.47712 (rank : 10) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62470, Q08441, Q08442 | Gene names | Itga3 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-3 precursor (Galactoprotein B3) (GAPB3) (VLA-3 alpha chain) (CD49c antigen) [Contains: Integrin alpha-3 heavy chain; Integrin alpha-3 light chain]. | |||||
|
CAD10_HUMAN
|
||||||
| NC score | 0.010810 (rank : 15) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6N8, Q9ULB3 | Gene names | CDH10 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-10 precursor (T2-cadherin). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.010012 (rank : 16) | θ value | 8.99809 (rank : 17) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
TIE2_HUMAN
|
||||||
| NC score | 0.003392 (rank : 17) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 1192 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02763 | Gene names | TEK, TIE2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-1 receptor precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor TIE-2) (hTIE2) (Tyrosine-protein kinase receptor TEK) (p140 TEK) (Tunica interna endothelial cell kinase) (CD202b antigen). | |||||
|
TIE2_MOUSE
|
||||||
| NC score | 0.002263 (rank : 18) | θ value | 8.99809 (rank : 18) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 1204 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02858 | Gene names | Tek, Hyk, Tie-2, Tie2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-1 receptor precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor TIE-2) (mTIE2) (Tyrosine-protein kinase receptor TEK) (p140 TEK) (Tunica interna endothelial cell kinase) (HYK) (STK1) (CD202b antigen). | |||||