Please be patient as the page loads
|
ITA3_MOUSE
|
||||||
| SwissProt Accessions | Q62470, Q08441, Q08442 | Gene names | Itga3 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-3 precursor (Galactoprotein B3) (GAPB3) (VLA-3 alpha chain) (CD49c antigen) [Contains: Integrin alpha-3 heavy chain; Integrin alpha-3 light chain]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ITA3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998578 (rank : 2) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P26006 | Gene names | ITGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-3 precursor (Galactoprotein B3) (GAPB3) (VLA-3 alpha chain) (FRP-2) (CD49c antigen) [Contains: Integrin alpha-3 heavy chain; Integrin alpha-3 light chain]. | |||||
|
ITA3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q62470, Q08441, Q08442 | Gene names | Itga3 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-3 precursor (Galactoprotein B3) (GAPB3) (VLA-3 alpha chain) (CD49c antigen) [Contains: Integrin alpha-3 heavy chain; Integrin alpha-3 light chain]. | |||||
|
ITA6_MOUSE
|
||||||
| θ value | 5.01279e-176 (rank : 3) | NC score | 0.977628 (rank : 3) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61739 | Gene names | Itga6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-6 precursor (VLA-6) (CD49f antigen) [Contains: Integrin alpha-6 heavy chain; Integrin alpha-6 light chain]. | |||||
|
ITA6_HUMAN
|
||||||
| θ value | 1.56188e-169 (rank : 4) | NC score | 0.976427 (rank : 4) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P23229, Q08443, Q14646, Q16508, Q9UN03 | Gene names | ITGA6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-6 precursor (VLA-6) (CD49f antigen) [Contains: Integrin alpha-6 heavy chain; Integrin alpha-6 light chain]. | |||||
|
ITA7_HUMAN
|
||||||
| θ value | 3.26429e-159 (rank : 5) | NC score | 0.973821 (rank : 5) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13683, O43197, Q9NY89, Q9UET0, Q9UEV2 | Gene names | ITGA7 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-7 precursor [Contains: Integrin alpha-7 heavy chain; Integrin alpha-7 light chain]. | |||||
|
ITA7_MOUSE
|
||||||
| θ value | 1.37464e-149 (rank : 6) | NC score | 0.973192 (rank : 6) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61738, O88731, O88732, P70350, Q61737, Q61741 | Gene names | Itga7 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-7 precursor [Contains: Integrin alpha-7 heavy chain; Integrin alpha-7 light chain]. | |||||
|
ITAV_MOUSE
|
||||||
| θ value | 1.5555e-68 (rank : 7) | NC score | 0.927066 (rank : 10) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P43406 | Gene names | Itgav | |||
|
Domain Architecture |
|
|||||
| Description | Integrin alpha-V precursor (Vitronectin receptor subunit alpha) (CD51 antigen) [Contains: Integrin alpha-V heavy chain; Integrin alpha-V light chain]. | |||||
|
ITA9_HUMAN
|
||||||
| θ value | 7.71989e-68 (rank : 8) | NC score | 0.945531 (rank : 7) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13797, Q14638 | Gene names | ITGA9 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-9 precursor (Integrin alpha-RLC). | |||||
|
ITA4_MOUSE
|
||||||
| θ value | 8.53533e-67 (rank : 9) | NC score | 0.935759 (rank : 8) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q00651 | Gene names | Itga4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-4 precursor (Integrin alpha-IV) (VLA-4) (Lymphocyte Peyer patch adhesion molecules subunit alpha) (LPAM subunit alpha) (CD49d antigen). | |||||
|
ITA8_HUMAN
|
||||||
| θ value | 9.43692e-66 (rank : 10) | NC score | 0.926924 (rank : 11) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P53708, Q5VX94 | Gene names | ITGA8 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-8 precursor [Contains: Integrin alpha-8 heavy chain; Integrin alpha-8 light chain]. | |||||
|
ITA4_HUMAN
|
||||||
| θ value | 2.74572e-65 (rank : 11) | NC score | 0.935596 (rank : 9) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P13612 | Gene names | ITGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-4 precursor (Integrin alpha-IV) (VLA-4) (CD49d antigen). | |||||
|
ITAV_HUMAN
|
||||||
| θ value | 2.32439e-64 (rank : 12) | NC score | 0.923796 (rank : 12) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P06756 | Gene names | ITGAV, VNRA | |||
|
Domain Architecture |
|
|||||
| Description | Integrin alpha-V precursor (Vitronectin receptor subunit alpha) (CD51 antigen) [Contains: Integrin alpha-V heavy chain; Integrin alpha-V light chain]. | |||||
|
ITA2B_HUMAN
|
||||||
| θ value | 1.7238e-59 (rank : 13) | NC score | 0.916576 (rank : 15) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P08514, O95366, Q14443 | Gene names | ITGA2B, GP2B, ITGAB | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-IIb precursor (Platelet membrane glycoprotein IIb) (GPalpha IIb) (GPIIb) (CD41 antigen) [Contains: Integrin alpha-IIb heavy chain; Integrin alpha-IIb light chain]. | |||||
|
ITA2B_MOUSE
|
||||||
| θ value | 8.55514e-59 (rank : 14) | NC score | 0.915062 (rank : 16) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QUM0, Q64229, Q9Z2M0 | Gene names | Itga2b | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-IIb precursor (Platelet membrane glycoprotein IIb) (GPalpha IIb) (GPIIb) (CD41 antigen) [Contains: Integrin alpha-IIb heavy chain; Integrin alpha-IIb light chain]. | |||||
|
ITA5_MOUSE
|
||||||
| θ value | 8.28633e-54 (rank : 15) | NC score | 0.917666 (rank : 13) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P11688 | Gene names | Itga5 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-5 precursor (Fibronectin receptor subunit alpha) (Integrin alpha-F) (VLA-5) (CD49e antigen) [Contains: Integrin alpha-5 heavy chain; Integrin alpha-5 light chain]. | |||||
|
ITA5_HUMAN
|
||||||
| θ value | 1.56274e-52 (rank : 16) | NC score | 0.917110 (rank : 14) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P08648, Q96HA5 | Gene names | ITGA5, FNRA | |||
|
Domain Architecture |
|
|||||
| Description | Integrin alpha-5 precursor (Fibronectin receptor subunit alpha) (Integrin alpha-F) (VLA-5) (CD49e antigen) [Contains: Integrin alpha-5 heavy chain; Integrin alpha-5 light chain]. | |||||
|
ITA1_HUMAN
|
||||||
| θ value | 1.04822e-48 (rank : 17) | NC score | 0.844331 (rank : 17) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P56199 | Gene names | ITGA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrin alpha-1 (Laminin and collagen receptor) (VLA-1) (CD49a antigen). | |||||
|
ITAE_MOUSE
|
||||||
| θ value | 1.41672e-45 (rank : 18) | NC score | 0.832659 (rank : 20) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q60677 | Gene names | Itgae | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-E precursor (Integrin alpha M290) (CD103 antigen) [Contains: Integrin alpha-E light chain; Integrin alpha-E heavy chain]. | |||||
|
ITA11_MOUSE
|
||||||
| θ value | 4.122e-45 (rank : 19) | NC score | 0.831932 (rank : 21) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P61622 | Gene names | Itga11 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-11 precursor. | |||||
|
ITAE_HUMAN
|
||||||
| θ value | 2.95404e-43 (rank : 20) | NC score | 0.831334 (rank : 22) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P38570, Q9NZU9 | Gene names | ITGAE | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-E precursor (Mucosal lymphocyte 1 antigen) (HML-1 antigen) (Integrin alpha-IEL) (CD103 antigen) [Contains: Integrin alpha-E light chain; Integrin alpha-E heavy chain]. | |||||
|
ITA11_HUMAN
|
||||||
| θ value | 3.85808e-43 (rank : 21) | NC score | 0.828290 (rank : 23) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UKX5, Q8WYI8, Q9UKQ1 | Gene names | ITGA11 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-11 precursor. | |||||
|
ITAM_HUMAN
|
||||||
| θ value | 1.12253e-42 (rank : 22) | NC score | 0.805944 (rank : 27) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P11215 | Gene names | ITGAM, CD11B, CR3A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (Neutrophil adherence receptor) (CD11b antigen). | |||||
|
ITAL_MOUSE
|
||||||
| θ value | 3.26607e-42 (rank : 23) | NC score | 0.827236 (rank : 24) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P24063 | Gene names | Itgal, Lfa-1, Ly-15 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-L precursor (Leukocyte adhesion glycoprotein LFA-1 alpha chain) (LFA-1A) (Leukocyte function-associated molecule 1 alpha chain) (Lymphocyte antigen Ly-15) (CD11a antigen). | |||||
|
ITAL_HUMAN
|
||||||
| θ value | 1.62093e-41 (rank : 24) | NC score | 0.822756 (rank : 26) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P20701, O43746, Q45H73, Q9UBC8 | Gene names | ITGAL, CD11A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-L precursor (Leukocyte adhesion glycoprotein LFA-1 alpha chain) (LFA-1A) (Leukocyte function-associated molecule 1 alpha chain) (CD11a antigen). | |||||
|
ITAD_HUMAN
|
||||||
| θ value | 3.61106e-41 (rank : 25) | NC score | 0.797006 (rank : 30) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13349, Q15575, Q15576 | Gene names | ITGAD | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-D precursor (Leukointegrin alpha D) (ADB2) (CD11d antigen). | |||||
|
ITA10_HUMAN
|
||||||
| θ value | 3.05695e-40 (rank : 26) | NC score | 0.836414 (rank : 18) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75578, Q6UXJ6, Q9UHZ8 | Gene names | ITGA10 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-10 precursor. | |||||
|
ITAX_MOUSE
|
||||||
| θ value | 5.21438e-40 (rank : 27) | NC score | 0.798352 (rank : 28) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QXH4 | Gene names | Itgax | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-X precursor (Leukocyte adhesion glycoprotein p150,95 alpha chain) (Leukocyte adhesion receptor p150,95) (CD11c antigen). | |||||
|
ITAM_MOUSE
|
||||||
| θ value | 3.37985e-39 (rank : 28) | NC score | 0.790736 (rank : 31) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P05555, Q8CA73 | Gene names | Itgam | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (CD11b antigen). | |||||
|
ITAD_MOUSE
|
||||||
| θ value | 1.28434e-38 (rank : 29) | NC score | 0.797332 (rank : 29) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q3V0T4 | Gene names | Itgad | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrin alpha-D precursor (CD11d antigen). | |||||
|
ITA2_HUMAN
|
||||||
| θ value | 1.6774e-38 (rank : 30) | NC score | 0.833673 (rank : 19) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P17301, Q14595 | Gene names | ITGA2 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-2 precursor (Platelet membrane glycoprotein Ia) (GPIa) (Collagen receptor) (VLA-2 alpha chain) (CD49b antigen). | |||||
|
ITA2_MOUSE
|
||||||
| θ value | 5.39604e-37 (rank : 31) | NC score | 0.826794 (rank : 25) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q62469, Q62163 | Gene names | Itga2 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-2 precursor (Platelet membrane glycoprotein Ia) (GPIa) (Collagen receptor) (VLA-2 alpha chain) (CD49b antigen). | |||||
|
ITAX_HUMAN
|
||||||
| θ value | 3.27365e-34 (rank : 32) | NC score | 0.788438 (rank : 32) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P20702, Q8IVA6 | Gene names | ITGAX, CD11C | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-X precursor (Leukocyte adhesion glycoprotein p150,95 alpha chain) (Leukocyte adhesion receptor p150,95) (Leu M5) (CD11c antigen). | |||||
|
PHLD1_HUMAN
|
||||||
| θ value | 8.67504e-11 (rank : 33) | NC score | 0.578101 (rank : 34) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P80108, Q15128 | Gene names | GPLD1, PIGPLD1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-glycan-specific phospholipase D 1 precursor (EC 3.1.4.50) (PI-G PLD) (Glycoprotein phospholipase D) (Glycosyl- phosphatidylinositol-specific phospholipase D). | |||||
|
PHLD2_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 34) | NC score | 0.572229 (rank : 35) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15127 | Gene names | GPLD2, PIGPLD2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-glycan-specific phospholipase D 2 precursor (EC 3.1.4.50) (PI-G PLD) (Glycoprotein phospholipase D) (Glycosyl- phosphatidylinositol-specific phospholipase D). | |||||
|
PHLD_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 35) | NC score | 0.585491 (rank : 33) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O70362 | Gene names | Gpld1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-glycan-specific phospholipase D 1 precursor (EC 3.1.4.50) (PI-G PLD) (Glycoprotein phospholipase D) (Glycosyl- phosphatidylinositol-specific phospholipase D). | |||||
|
LHX3_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 36) | NC score | 0.019335 (rank : 63) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBR4, Q9NZB5, Q9P0I8, Q9P0I9 | Gene names | LHX3 | |||
|
Domain Architecture |
|
|||||
| Description | LIM/homeobox protein Lhx3. | |||||
|
PLXB3_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 37) | NC score | 0.017717 (rank : 64) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QY40, Q80TH8 | Gene names | Plxnb3, Kiaa1206, Plxn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor (Plexin-6). | |||||
|
ITB6_HUMAN
|
||||||
| θ value | 0.163984 (rank : 38) | NC score | 0.012463 (rank : 66) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P18564, Q16500 | Gene names | ITGB6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
MYOM2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 39) | NC score | 0.009184 (rank : 70) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54296 | Gene names | MYOM2 | |||
|
Domain Architecture |

|
|||||
| Description | Myomesin-2 (M-protein) (165 kDa titin-associated protein) (165 kDa connectin-associated protein). | |||||
|
PO121_MOUSE
|
||||||
| θ value | 0.21417 (rank : 40) | NC score | 0.017662 (rank : 65) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
PSB5_HUMAN
|
||||||
| θ value | 0.279714 (rank : 41) | NC score | 0.033339 (rank : 59) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28074, Q16242, Q6PEW2 | Gene names | PSMB5, LMPX | |||
|
Domain Architecture |

|
|||||
| Description | Proteasome subunit beta type 5 precursor (EC 3.4.25.1) (Proteasome epsilon chain) (Macropain epsilon chain) (Multicatalytic endopeptidase complex epsilon chain) (Proteasome subunit X) (Proteasome chain 6) (Proteasome subunit MB1). | |||||
|
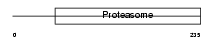
PSB5_MOUSE
|
||||||
| θ value | 0.365318 (rank : 42) | NC score | 0.029991 (rank : 60) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O55234, Q9R1P2 | Gene names | Psmb5 | |||
|
Domain Architecture |
|
|||||
| Description | Proteasome subunit beta type 5 precursor (EC 3.4.25.1) (Proteasome epsilon chain) (Macropain epsilon chain) (Multicatalytic endopeptidase complex epsilon chain) (Proteasome subunit X) (Proteasome chain 6). | |||||
|
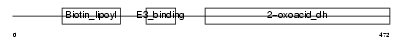
ODB2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 43) | NC score | 0.023599 (rank : 61) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53395 | Gene names | Dbt | |||
|
Domain Architecture |
|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ITB6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.009845 (rank : 68) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0T9 | Gene names | Itgb6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
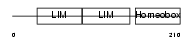
LHX3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.009299 (rank : 69) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50481, Q61800, Q61801 | Gene names | Lhx3, Lim-3, Lim3, Plim | |||
|
Domain Architecture |
|
|||||
| Description | LIM/homeobox protein Lhx3 (Homeobox protein LIM-3) (Homeobox protein P-LIM) (LIM homeobox protein 3). | |||||
|
FANCM_HUMAN
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.010572 (rank : 67) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
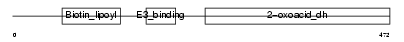
ODB2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.019387 (rank : 62) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11182 | Gene names | DBT, BCATE2 | |||
|
Domain Architecture |
|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ZN445_MOUSE
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | -0.003875 (rank : 77) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R2V3, Q8K216 | Gene names | Znf445, Zfp445 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 445. | |||||
|
CRP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.008336 (rank : 71) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P02741, Q8WW75 | Gene names | CRP, PTX1 | |||
|
Domain Architecture |
|
|||||
| Description | C-reactive protein precursor [Contains: C-reactive protein(1-205)]. | |||||
|
DNJA3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.006383 (rank : 72) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96EY1, O75472, Q8WUJ6, Q8WXJ3, Q96D76, Q96IV1, Q9NYH8 | Gene names | DNAJA3, TID1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (hTid-1). | |||||
|
CSMD2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.001481 (rank : 75) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
DNJA3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.005798 (rank : 73) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99M87, Q8BSM0, Q99L09, Q99P71, Q99P76, Q9CT11, Q9DBJ7, Q9DC44 | Gene names | Dnaja3, Tid1 | |||
|
Domain Architecture |
|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (mTid-1). | |||||
|
DPOLZ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.005252 (rank : 74) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60673, O43214 | Gene names | REV3L, POLZ, REV3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (hREV3). | |||||
|
5HT5A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | -0.002756 (rank : 76) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 615 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30966 | Gene names | Htr5a, 5ht5a | |||
|
Domain Architecture |
|
|||||
| Description | 5-hydroxytryptamine 5A receptor (5-HT-5A) (Serotonin receptor 5A) (5- HT-5). | |||||
|
CO6A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.059233 (rank : 58) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P12110, Q13909, Q13910, Q13911, Q14049, Q16259, Q16597, Q9UML3 | Gene names | COL6A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(VI) chain precursor. | |||||
|
CO6A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.061477 (rank : 57) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02788, Q05505 | Gene names | Col6a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(VI) chain precursor. | |||||
|
CO6A3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.138741 (rank : 51) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12111, Q16501 | Gene names | COL6A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(VI) chain precursor. | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.085851 (rank : 56) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
COCA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.141561 (rank : 48) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99715, Q99716 | Gene names | COL12A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
COCA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.141413 (rank : 49) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60847, P70322 | Gene names | Col12a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
COCH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.190481 (rank : 40) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43405 | Gene names | COCH, COCH5B2 | |||
|
Domain Architecture |

|
|||||
| Description | Cochlin precursor (COCH-5B2). | |||||
|
COCH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.190132 (rank : 41) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62507, Q3TAF5, Q9QWK6 | Gene names | Coch | |||
|
Domain Architecture |

|
|||||
| Description | Cochlin precursor (COCH-5B2). | |||||
|
COEA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.146085 (rank : 43) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q05707, O00260, O00261, O00262, Q05708, Q5XJ18, Q96C67 | Gene names | COL14A1, UND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor (Undulin). | |||||
|
COEA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.148569 (rank : 42) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80X19, Q8C6X3, Q9WV05 | Gene names | Col14a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor. | |||||
|
COKA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.141604 (rank : 47) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
MATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.207305 (rank : 38) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P21941 | Gene names | MATN1, CMP, CRTM | |||
|
Domain Architecture |
|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
MATN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.207807 (rank : 37) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P51942 | Gene names | Matn1, Cmp, Crtm | |||
|
Domain Architecture |
|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
MATN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.120652 (rank : 53) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00339, Q6UWA5, Q7Z5X1, Q8NDE6, Q96FT5, Q9NSZ1 | Gene names | MATN2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
MATN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.124086 (rank : 52) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O08746 | Gene names | Matn2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
MATN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.145654 (rank : 44) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15232 | Gene names | MATN3 | |||
|
Domain Architecture |
|
|||||
| Description | Matrilin-3 precursor. | |||||
|
MATN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.143556 (rank : 46) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O35701, Q9JHM0 | Gene names | Matn3 | |||
|
Domain Architecture |
|
|||||
| Description | Matrilin-3 precursor. | |||||
|
MATN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.139253 (rank : 50) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95460, Q5QPU2, Q5QPU3, Q5QPU4, Q8N2M5, Q8N2M7, Q9H1F8, Q9H1F9 | Gene names | MATN4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor. | |||||
|
MATN4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.144434 (rank : 45) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O89029, O89030, Q9QWS3 | Gene names | Matn4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor (MAT-4). | |||||
|
VITRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.204930 (rank : 39) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6UXI7, Q6P7T3, Q96DT1 | Gene names | VIT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vitrin precursor. | |||||
|
VITRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.209882 (rank : 36) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VHI5, Q3TZ47, Q8BQ41, Q8K047, Q9CYZ1 | Gene names | Vit | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vitrin precursor. | |||||
|
VWF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.099095 (rank : 55) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
VWF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.108678 (rank : 54) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
ITA3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q62470, Q08441, Q08442 | Gene names | Itga3 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-3 precursor (Galactoprotein B3) (GAPB3) (VLA-3 alpha chain) (CD49c antigen) [Contains: Integrin alpha-3 heavy chain; Integrin alpha-3 light chain]. | |||||
|
ITA3_HUMAN
|
||||||
| NC score | 0.998578 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P26006 | Gene names | ITGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-3 precursor (Galactoprotein B3) (GAPB3) (VLA-3 alpha chain) (FRP-2) (CD49c antigen) [Contains: Integrin alpha-3 heavy chain; Integrin alpha-3 light chain]. | |||||
|
ITA6_MOUSE
|
||||||
| NC score | 0.977628 (rank : 3) | θ value | 5.01279e-176 (rank : 3) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61739 | Gene names | Itga6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-6 precursor (VLA-6) (CD49f antigen) [Contains: Integrin alpha-6 heavy chain; Integrin alpha-6 light chain]. | |||||
|
ITA6_HUMAN
|
||||||
| NC score | 0.976427 (rank : 4) | θ value | 1.56188e-169 (rank : 4) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P23229, Q08443, Q14646, Q16508, Q9UN03 | Gene names | ITGA6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-6 precursor (VLA-6) (CD49f antigen) [Contains: Integrin alpha-6 heavy chain; Integrin alpha-6 light chain]. | |||||
|
ITA7_HUMAN
|
||||||
| NC score | 0.973821 (rank : 5) | θ value | 3.26429e-159 (rank : 5) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13683, O43197, Q9NY89, Q9UET0, Q9UEV2 | Gene names | ITGA7 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-7 precursor [Contains: Integrin alpha-7 heavy chain; Integrin alpha-7 light chain]. | |||||
|
ITA7_MOUSE
|
||||||
| NC score | 0.973192 (rank : 6) | θ value | 1.37464e-149 (rank : 6) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61738, O88731, O88732, P70350, Q61737, Q61741 | Gene names | Itga7 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-7 precursor [Contains: Integrin alpha-7 heavy chain; Integrin alpha-7 light chain]. | |||||
|
ITA9_HUMAN
|
||||||
| NC score | 0.945531 (rank : 7) | θ value | 7.71989e-68 (rank : 8) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13797, Q14638 | Gene names | ITGA9 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-9 precursor (Integrin alpha-RLC). | |||||
|
ITA4_MOUSE
|
||||||
| NC score | 0.935759 (rank : 8) | θ value | 8.53533e-67 (rank : 9) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q00651 | Gene names | Itga4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-4 precursor (Integrin alpha-IV) (VLA-4) (Lymphocyte Peyer patch adhesion molecules subunit alpha) (LPAM subunit alpha) (CD49d antigen). | |||||
|
ITA4_HUMAN
|
||||||
| NC score | 0.935596 (rank : 9) | θ value | 2.74572e-65 (rank : 11) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P13612 | Gene names | ITGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-4 precursor (Integrin alpha-IV) (VLA-4) (CD49d antigen). | |||||
|
ITAV_MOUSE
|
||||||
| NC score | 0.927066 (rank : 10) | θ value | 1.5555e-68 (rank : 7) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P43406 | Gene names | Itgav | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-V precursor (Vitronectin receptor subunit alpha) (CD51 antigen) [Contains: Integrin alpha-V heavy chain; Integrin alpha-V light chain]. | |||||
|
ITA8_HUMAN
|
||||||
| NC score | 0.926924 (rank : 11) | θ value | 9.43692e-66 (rank : 10) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P53708, Q5VX94 | Gene names | ITGA8 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-8 precursor [Contains: Integrin alpha-8 heavy chain; Integrin alpha-8 light chain]. | |||||
|
ITAV_HUMAN
|
||||||
| NC score | 0.923796 (rank : 12) | θ value | 2.32439e-64 (rank : 12) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P06756 | Gene names | ITGAV, VNRA | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-V precursor (Vitronectin receptor subunit alpha) (CD51 antigen) [Contains: Integrin alpha-V heavy chain; Integrin alpha-V light chain]. | |||||
|
ITA5_MOUSE
|
||||||
| NC score | 0.917666 (rank : 13) | θ value | 8.28633e-54 (rank : 15) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P11688 | Gene names | Itga5 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-5 precursor (Fibronectin receptor subunit alpha) (Integrin alpha-F) (VLA-5) (CD49e antigen) [Contains: Integrin alpha-5 heavy chain; Integrin alpha-5 light chain]. | |||||
|
ITA5_HUMAN
|
||||||
| NC score | 0.917110 (rank : 14) | θ value | 1.56274e-52 (rank : 16) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P08648, Q96HA5 | Gene names | ITGA5, FNRA | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-5 precursor (Fibronectin receptor subunit alpha) (Integrin alpha-F) (VLA-5) (CD49e antigen) [Contains: Integrin alpha-5 heavy chain; Integrin alpha-5 light chain]. | |||||
|
ITA2B_HUMAN
|
||||||
| NC score | 0.916576 (rank : 15) | θ value | 1.7238e-59 (rank : 13) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P08514, O95366, Q14443 | Gene names | ITGA2B, GP2B, ITGAB | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-IIb precursor (Platelet membrane glycoprotein IIb) (GPalpha IIb) (GPIIb) (CD41 antigen) [Contains: Integrin alpha-IIb heavy chain; Integrin alpha-IIb light chain]. | |||||
|
ITA2B_MOUSE
|
||||||
| NC score | 0.915062 (rank : 16) | θ value | 8.55514e-59 (rank : 14) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QUM0, Q64229, Q9Z2M0 | Gene names | Itga2b | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-IIb precursor (Platelet membrane glycoprotein IIb) (GPalpha IIb) (GPIIb) (CD41 antigen) [Contains: Integrin alpha-IIb heavy chain; Integrin alpha-IIb light chain]. | |||||
|
ITA1_HUMAN
|
||||||
| NC score | 0.844331 (rank : 17) | θ value | 1.04822e-48 (rank : 17) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P56199 | Gene names | ITGA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrin alpha-1 (Laminin and collagen receptor) (VLA-1) (CD49a antigen). | |||||
|
ITA10_HUMAN
|
||||||
| NC score | 0.836414 (rank : 18) | θ value | 3.05695e-40 (rank : 26) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75578, Q6UXJ6, Q9UHZ8 | Gene names | ITGA10 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-10 precursor. | |||||
|
ITA2_HUMAN
|
||||||
| NC score | 0.833673 (rank : 19) | θ value | 1.6774e-38 (rank : 30) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P17301, Q14595 | Gene names | ITGA2 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-2 precursor (Platelet membrane glycoprotein Ia) (GPIa) (Collagen receptor) (VLA-2 alpha chain) (CD49b antigen). | |||||
|
ITAE_MOUSE
|
||||||
| NC score | 0.832659 (rank : 20) | θ value | 1.41672e-45 (rank : 18) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q60677 | Gene names | Itgae | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-E precursor (Integrin alpha M290) (CD103 antigen) [Contains: Integrin alpha-E light chain; Integrin alpha-E heavy chain]. | |||||
|
ITA11_MOUSE
|
||||||
| NC score | 0.831932 (rank : 21) | θ value | 4.122e-45 (rank : 19) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P61622 | Gene names | Itga11 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-11 precursor. | |||||
|
ITAE_HUMAN
|
||||||
| NC score | 0.831334 (rank : 22) | θ value | 2.95404e-43 (rank : 20) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P38570, Q9NZU9 | Gene names | ITGAE | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-E precursor (Mucosal lymphocyte 1 antigen) (HML-1 antigen) (Integrin alpha-IEL) (CD103 antigen) [Contains: Integrin alpha-E light chain; Integrin alpha-E heavy chain]. | |||||
|
ITA11_HUMAN
|
||||||
| NC score | 0.828290 (rank : 23) | θ value | 3.85808e-43 (rank : 21) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UKX5, Q8WYI8, Q9UKQ1 | Gene names | ITGA11 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-11 precursor. | |||||
|
ITAL_MOUSE
|
||||||
| NC score | 0.827236 (rank : 24) | θ value | 3.26607e-42 (rank : 23) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P24063 | Gene names | Itgal, Lfa-1, Ly-15 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-L precursor (Leukocyte adhesion glycoprotein LFA-1 alpha chain) (LFA-1A) (Leukocyte function-associated molecule 1 alpha chain) (Lymphocyte antigen Ly-15) (CD11a antigen). | |||||
|
ITA2_MOUSE
|
||||||
| NC score | 0.826794 (rank : 25) | θ value | 5.39604e-37 (rank : 31) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q62469, Q62163 | Gene names | Itga2 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-2 precursor (Platelet membrane glycoprotein Ia) (GPIa) (Collagen receptor) (VLA-2 alpha chain) (CD49b antigen). | |||||
|
ITAL_HUMAN
|
||||||
| NC score | 0.822756 (rank : 26) | θ value | 1.62093e-41 (rank : 24) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P20701, O43746, Q45H73, Q9UBC8 | Gene names | ITGAL, CD11A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-L precursor (Leukocyte adhesion glycoprotein LFA-1 alpha chain) (LFA-1A) (Leukocyte function-associated molecule 1 alpha chain) (CD11a antigen). | |||||
|
ITAM_HUMAN
|
||||||
| NC score | 0.805944 (rank : 27) | θ value | 1.12253e-42 (rank : 22) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P11215 | Gene names | ITGAM, CD11B, CR3A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (Neutrophil adherence receptor) (CD11b antigen). | |||||
|
ITAX_MOUSE
|
||||||
| NC score | 0.798352 (rank : 28) | θ value | 5.21438e-40 (rank : 27) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QXH4 | Gene names | Itgax | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-X precursor (Leukocyte adhesion glycoprotein p150,95 alpha chain) (Leukocyte adhesion receptor p150,95) (CD11c antigen). | |||||
|
ITAD_MOUSE
|
||||||
| NC score | 0.797332 (rank : 29) | θ value | 1.28434e-38 (rank : 29) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q3V0T4 | Gene names | Itgad | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrin alpha-D precursor (CD11d antigen). | |||||
|
ITAD_HUMAN
|
||||||
| NC score | 0.797006 (rank : 30) | θ value | 3.61106e-41 (rank : 25) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q13349, Q15575, Q15576 | Gene names | ITGAD | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-D precursor (Leukointegrin alpha D) (ADB2) (CD11d antigen). | |||||
|
ITAM_MOUSE
|
||||||
| NC score | 0.790736 (rank : 31) | θ value | 3.37985e-39 (rank : 28) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P05555, Q8CA73 | Gene names | Itgam | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-M precursor (Cell surface glycoprotein MAC-1 alpha subunit) (CR-3 alpha chain) (Leukocyte adhesion receptor MO1) (CD11b antigen). | |||||
|
ITAX_HUMAN
|
||||||
| NC score | 0.788438 (rank : 32) | θ value | 3.27365e-34 (rank : 32) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P20702, Q8IVA6 | Gene names | ITGAX, CD11C | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-X precursor (Leukocyte adhesion glycoprotein p150,95 alpha chain) (Leukocyte adhesion receptor p150,95) (Leu M5) (CD11c antigen). | |||||
|
PHLD_MOUSE
|
||||||
| NC score | 0.585491 (rank : 33) | θ value | 4.30538e-10 (rank : 35) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O70362 | Gene names | Gpld1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-glycan-specific phospholipase D 1 precursor (EC 3.1.4.50) (PI-G PLD) (Glycoprotein phospholipase D) (Glycosyl- phosphatidylinositol-specific phospholipase D). | |||||
|
PHLD1_HUMAN
|
||||||
| NC score | 0.578101 (rank : 34) | θ value | 8.67504e-11 (rank : 33) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P80108, Q15128 | Gene names | GPLD1, PIGPLD1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-glycan-specific phospholipase D 1 precursor (EC 3.1.4.50) (PI-G PLD) (Glycoprotein phospholipase D) (Glycosyl- phosphatidylinositol-specific phospholipase D). | |||||
|
PHLD2_HUMAN
|
||||||
| NC score | 0.572229 (rank : 35) | θ value | 1.9326e-10 (rank : 34) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15127 | Gene names | GPLD2, PIGPLD2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-glycan-specific phospholipase D 2 precursor (EC 3.1.4.50) (PI-G PLD) (Glycoprotein phospholipase D) (Glycosyl- phosphatidylinositol-specific phospholipase D). | |||||
|
VITRN_MOUSE
|
||||||
| NC score | 0.209882 (rank : 36) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VHI5, Q3TZ47, Q8BQ41, Q8K047, Q9CYZ1 | Gene names | Vit | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vitrin precursor. | |||||
|
MATN1_MOUSE
|
||||||
| NC score | 0.207807 (rank : 37) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P51942 | Gene names | Matn1, Cmp, Crtm | |||
|
Domain Architecture |

|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
MATN1_HUMAN
|
||||||
| NC score | 0.207305 (rank : 38) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P21941 | Gene names | MATN1, CMP, CRTM | |||
|
Domain Architecture |

|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
VITRN_HUMAN
|
||||||
| NC score | 0.204930 (rank : 39) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6UXI7, Q6P7T3, Q96DT1 | Gene names | VIT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vitrin precursor. | |||||
|
COCH_HUMAN
|
||||||
| NC score | 0.190481 (rank : 40) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43405 | Gene names | COCH, COCH5B2 | |||
|
Domain Architecture |

|
|||||
| Description | Cochlin precursor (COCH-5B2). | |||||
|
COCH_MOUSE
|
||||||
| NC score | 0.190132 (rank : 41) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62507, Q3TAF5, Q9QWK6 | Gene names | Coch | |||
|
Domain Architecture |

|
|||||
| Description | Cochlin precursor (COCH-5B2). | |||||
|
COEA1_MOUSE
|
||||||
| NC score | 0.148569 (rank : 42) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80X19, Q8C6X3, Q9WV05 | Gene names | Col14a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor. | |||||
|
COEA1_HUMAN
|
||||||
| NC score | 0.146085 (rank : 43) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q05707, O00260, O00261, O00262, Q05708, Q5XJ18, Q96C67 | Gene names | COL14A1, UND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor (Undulin). | |||||
|
MATN3_HUMAN
|
||||||
| NC score | 0.145654 (rank : 44) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15232 | Gene names | MATN3 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-3 precursor. | |||||
|
MATN4_MOUSE
|
||||||
| NC score | 0.144434 (rank : 45) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O89029, O89030, Q9QWS3 | Gene names | Matn4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor (MAT-4). | |||||
|
MATN3_MOUSE
|
||||||
| NC score | 0.143556 (rank : 46) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O35701, Q9JHM0 | Gene names | Matn3 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-3 precursor. | |||||
|
COKA1_HUMAN
|
||||||
| NC score | 0.141604 (rank : 47) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
COCA1_HUMAN
|
||||||
| NC score | 0.141561 (rank : 48) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99715, Q99716 | Gene names | COL12A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
COCA1_MOUSE
|
||||||
| NC score | 0.141413 (rank : 49) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q60847, P70322 | Gene names | Col12a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
MATN4_HUMAN
|
||||||
| NC score | 0.139253 (rank : 50) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95460, Q5QPU2, Q5QPU3, Q5QPU4, Q8N2M5, Q8N2M7, Q9H1F8, Q9H1F9 | Gene names | MATN4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor. | |||||
|
CO6A3_HUMAN
|
||||||
| NC score | 0.138741 (rank : 51) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P12111, Q16501 | Gene names | COL6A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(VI) chain precursor. | |||||
|
MATN2_MOUSE
|
||||||
| NC score | 0.124086 (rank : 52) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O08746 | Gene names | Matn2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
MATN2_HUMAN
|
||||||
| NC score | 0.120652 (rank : 53) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00339, Q6UWA5, Q7Z5X1, Q8NDE6, Q96FT5, Q9NSZ1 | Gene names | MATN2 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-2 precursor. | |||||
|
VWF_MOUSE
|
||||||
| NC score | 0.108678 (rank : 54) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CIZ8, Q60863, Q6XUV6, Q8BIU9, Q8CGN0, Q9JK16 | Gene names | Vwf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
VWF_HUMAN
|
||||||
| NC score | 0.099095 (rank : 55) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P04275 | Gene names | VWF, F8VWF | |||
|
Domain Architecture |

|
|||||
| Description | von Willebrand factor precursor (vWF) [Contains: von Willebrand antigen 2 (von Willebrand antigen II)]. | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.085851 (rank : 56) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
CO6A2_MOUSE
|
||||||
| NC score | 0.061477 (rank : 57) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02788, Q05505 | Gene names | Col6a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(VI) chain precursor. | |||||
|
CO6A2_HUMAN
|
||||||
| NC score | 0.059233 (rank : 58) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P12110, Q13909, Q13910, Q13911, Q14049, Q16259, Q16597, Q9UML3 | Gene names | COL6A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(VI) chain precursor. | |||||
|
PSB5_HUMAN
|
||||||
| NC score | 0.033339 (rank : 59) | θ value | 0.279714 (rank : 41) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28074, Q16242, Q6PEW2 | Gene names | PSMB5, LMPX | |||
|
Domain Architecture |

|
|||||
| Description | Proteasome subunit beta type 5 precursor (EC 3.4.25.1) (Proteasome epsilon chain) (Macropain epsilon chain) (Multicatalytic endopeptidase complex epsilon chain) (Proteasome subunit X) (Proteasome chain 6) (Proteasome subunit MB1). | |||||
|
PSB5_MOUSE
|
||||||
| NC score | 0.029991 (rank : 60) | θ value | 0.365318 (rank : 42) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O55234, Q9R1P2 | Gene names | Psmb5 | |||
|
Domain Architecture |

|
|||||
| Description | Proteasome subunit beta type 5 precursor (EC 3.4.25.1) (Proteasome epsilon chain) (Macropain epsilon chain) (Multicatalytic endopeptidase complex epsilon chain) (Proteasome subunit X) (Proteasome chain 6). | |||||
|
ODB2_MOUSE
|
||||||
| NC score | 0.023599 (rank : 61) | θ value | 0.47712 (rank : 43) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53395 | Gene names | Dbt | |||
|
Domain Architecture |

|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
ODB2_HUMAN
|
||||||
| NC score | 0.019387 (rank : 62) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11182 | Gene names | DBT, BCATE2 | |||
|
Domain Architecture |

|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
LHX3_HUMAN
|
||||||
| NC score | 0.019335 (rank : 63) | θ value | 0.00298849 (rank : 36) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBR4, Q9NZB5, Q9P0I8, Q9P0I9 | Gene names | LHX3 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx3. | |||||
|
PLXB3_MOUSE
|
||||||
| NC score | 0.017717 (rank : 64) | θ value | 0.0431538 (rank : 37) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QY40, Q80TH8 | Gene names | Plxnb3, Kiaa1206, Plxn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B3 precursor (Plexin-6). | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.017662 (rank : 65) | θ value | 0.21417 (rank : 40) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
ITB6_HUMAN
|
||||||
| NC score | 0.012463 (rank : 66) | θ value | 0.163984 (rank : 38) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P18564, Q16500 | Gene names | ITGB6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
FANCM_HUMAN
|
||||||
| NC score | 0.010572 (rank : 67) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IYD8, Q3YFH9, Q8N9X6, Q9HCH6 | Gene names | FANCM, KIAA1596 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group M protein (EC 3.6.1.-) (ATP-dependent RNA helicase FANCM) (Protein FACM) (Fanconi anemia-associated polypeptide of 250 kDa) (FAAP250) (Protein Hef ortholog). | |||||
|
ITB6_MOUSE
|
||||||
| NC score | 0.009845 (rank : 68) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0T9 | Gene names | Itgb6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
LHX3_MOUSE
|
||||||
| NC score | 0.009299 (rank : 69) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50481, Q61800, Q61801 | Gene names | Lhx3, Lim-3, Lim3, Plim | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx3 (Homeobox protein LIM-3) (Homeobox protein P-LIM) (LIM homeobox protein 3). | |||||
|
MYOM2_HUMAN
|
||||||
| NC score | 0.009184 (rank : 70) | θ value | 0.21417 (rank : 39) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54296 | Gene names | MYOM2 | |||
|
Domain Architecture |

|
|||||
| Description | Myomesin-2 (M-protein) (165 kDa titin-associated protein) (165 kDa connectin-associated protein). | |||||
|
CRP_HUMAN
|
||||||
| NC score | 0.008336 (rank : 71) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P02741, Q8WW75 | Gene names | CRP, PTX1 | |||
|
Domain Architecture |

|
|||||
| Description | C-reactive protein precursor [Contains: C-reactive protein(1-205)]. | |||||
|
DNJA3_HUMAN
|
||||||
| NC score | 0.006383 (rank : 72) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96EY1, O75472, Q8WUJ6, Q8WXJ3, Q96D76, Q96IV1, Q9NYH8 | Gene names | DNAJA3, TID1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (hTid-1). | |||||
|
DNJA3_MOUSE
|
||||||
| NC score | 0.005798 (rank : 73) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99M87, Q8BSM0, Q99L09, Q99P71, Q99P76, Q9CT11, Q9DBJ7, Q9DC44 | Gene names | Dnaja3, Tid1 | |||
|
Domain Architecture |

|
|||||
| Description | DnaJ homolog subfamily A member 3, mitochondrial precursor (Tumorous imaginal discs protein Tid56 homolog) (DnaJ protein Tid-1) (mTid-1). | |||||
|
DPOLZ_HUMAN
|
||||||
| NC score | 0.005252 (rank : 74) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60673, O43214 | Gene names | REV3L, POLZ, REV3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (hREV3). | |||||
|
CSMD2_HUMAN
|
||||||
| NC score | 0.001481 (rank : 75) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z408, Q5VT59, Q8N963, Q96Q03, Q9H4V7, Q9H4V8, Q9H4V9, Q9H4W0, Q9H4W1, Q9H4W2, Q9H4W3, Q9H4W4, Q9HCY5, Q9HCY6, Q9HCY7 | Gene names | CSMD2, KIAA1884 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 2 (CUB and sushi multiple domains protein 2). | |||||
|
5HT5A_MOUSE
|
||||||
| NC score | -0.002756 (rank : 76) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 615 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P30966 | Gene names | Htr5a, 5ht5a | |||
|
Domain Architecture |

|
|||||
| Description | 5-hydroxytryptamine 5A receptor (5-HT-5A) (Serotonin receptor 5A) (5- HT-5). | |||||
|
ZN445_MOUSE
|
||||||
| NC score | -0.003875 (rank : 77) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R2V3, Q8K216 | Gene names | Znf445, Zfp445 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 445. | |||||