Please be patient as the page loads
|
PCCA_MOUSE
|
||||||
| SwissProt Accessions | Q91ZA3, Q80VU5, Q922N3 | Gene names | Pcca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PCCA_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999289 (rank : 2) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P05165, Q15979 | Gene names | PCCA | |||
|
Domain Architecture |

|
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
PCCA_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91ZA3, Q80VU5, Q922N3 | Gene names | Pcca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
MCCA_MOUSE
|
||||||
| θ value | 3.07662e-133 (rank : 3) | NC score | 0.966377 (rank : 3) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99MR8, Q3TGU0, Q9D8R2 | Gene names | Mccc1, Mcca | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
MCCA_HUMAN
|
||||||
| θ value | 1.33764e-128 (rank : 4) | NC score | 0.965229 (rank : 4) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96RQ3, Q59ES4, Q9H959, Q9NS97 | Gene names | MCCC1, MCCA | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
PYC_MOUSE
|
||||||
| θ value | 4.6295e-105 (rank : 5) | NC score | 0.930869 (rank : 6) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05920 | Gene names | Pc, Pcx | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
|
PYC_HUMAN
|
||||||
| θ value | 6.68496e-104 (rank : 6) | NC score | 0.931006 (rank : 5) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P11498, Q16705 | Gene names | PC | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
|
COA1_HUMAN
|
||||||
| θ value | 1.66577e-62 (rank : 7) | NC score | 0.824099 (rank : 7) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13085 | Gene names | ACACA, ACAC, ACC1, ACCA | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 1 (EC 6.4.1.2) (ACC-alpha) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
COA2_HUMAN
|
||||||
| θ value | 2.32979e-56 (rank : 8) | NC score | 0.790193 (rank : 8) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00763, Q16852 | Gene names | ACACB, ACC2, ACCB | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 2 (EC 6.4.1.2) (ACC-beta) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
PYR1_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 9) | NC score | 0.393927 (rank : 11) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
|
CPSM_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 10) | NC score | 0.396408 (rank : 10) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P31327, O43774, Q7Z5I5 | Gene names | CPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
CPSM_MOUSE
|
||||||
| θ value | 8.97725e-08 (rank : 11) | NC score | 0.400762 (rank : 9) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C196, Q6NX75 | Gene names | Cps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
APC7_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 12) | NC score | 0.077834 (rank : 14) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WVM3, Q3UBM7, Q8BSR2, Q91W13 | Gene names | Anapc7, Apc7 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7) (Prediabetic NOD sera-reactive autoantigen). | |||||
|
APC7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.054740 (rank : 18) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJX3, Q96AC4, Q96GF4, Q9BU24, Q9NT16 | Gene names | ANAPC7, APC7 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7). | |||||
|
CDYL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.010456 (rank : 21) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WTK2 | Gene names | Cdyl | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
PUR2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.048117 (rank : 20) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22102 | Gene names | GART, PRGS | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional purine biosynthetic protein adenosine-3 [Includes: Phosphoribosylamine--glycine ligase (EC 6.3.4.13) (GARS) (Glycinamide ribonucleotide synthetase) (Phosphoribosylglycinamide synthetase); Phosphoribosylformylglycinamidine cyclo-ligase (EC 6.3.3.1) (AIRS) (Phosphoribosyl-aminoimidazole synthetase) (AIR synthase); Phosphoribosylglycinamide formyltransferase (EC 2.1.2.2) (GART) (GAR transformylase) (5'-phosphoribosylglycinamide transformylase)]. | |||||
|
ODB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 16) | NC score | 0.065231 (rank : 15) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11182 | Gene names | DBT, BCATE2 | |||
|
Domain Architecture |
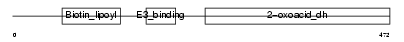
|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
MICA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 17) | NC score | 0.004958 (rank : 22) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VDP3 | Gene names | Mical1, Mical, Nical | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
ODB2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | 0.059910 (rank : 17) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P53395 | Gene names | Dbt | |||
|
Domain Architecture |
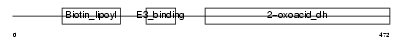
|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
HMGCL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 19) | NC score | 0.054388 (rank : 19) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35914, Q96FP8 | Gene names | HMGCL | |||
|
Domain Architecture |
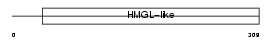
|
|||||
| Description | Hydroxymethylglutaryl-CoA lyase, mitochondrial precursor (EC 4.1.3.4) (HMG-CoA lyase) (HL) (3-hydroxy-3-methylglutarate-CoA lyase). | |||||
|
HMGCL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 20) | NC score | 0.062226 (rank : 16) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P38060 | Gene names | Hmgcl | |||
|
Domain Architecture |
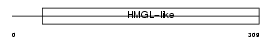
|
|||||
| Description | Hydroxymethylglutaryl-CoA lyase, mitochondrial precursor (EC 4.1.3.4) (HMG-CoA lyase) (HL) (3-hydroxy-3-methylglutarate-CoA lyase). | |||||
|
PCCB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 21) | NC score | 0.087818 (rank : 13) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P05166, Q16813 | Gene names | PCCB | |||
|
Domain Architecture |

|
|||||
| Description | Propionyl-CoA carboxylase beta chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit beta) (Propanoyl-CoA:carbon dioxide ligase subunit beta). | |||||
|
PCCB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 22) | NC score | 0.090431 (rank : 12) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99MN9 | Gene names | Pccb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Propionyl-CoA carboxylase beta chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit beta) (Propanoyl-CoA:carbon dioxide ligase subunit beta). | |||||
|
PCCA_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91ZA3, Q80VU5, Q922N3 | Gene names | Pcca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
PCCA_HUMAN
|
||||||
| NC score | 0.999289 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P05165, Q15979 | Gene names | PCCA | |||
|
Domain Architecture |

|
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
MCCA_MOUSE
|
||||||
| NC score | 0.966377 (rank : 3) | θ value | 3.07662e-133 (rank : 3) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99MR8, Q3TGU0, Q9D8R2 | Gene names | Mccc1, Mcca | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
MCCA_HUMAN
|
||||||
| NC score | 0.965229 (rank : 4) | θ value | 1.33764e-128 (rank : 4) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96RQ3, Q59ES4, Q9H959, Q9NS97 | Gene names | MCCC1, MCCA | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
PYC_HUMAN
|
||||||
| NC score | 0.931006 (rank : 5) | θ value | 6.68496e-104 (rank : 6) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P11498, Q16705 | Gene names | PC | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
|
PYC_MOUSE
|
||||||
| NC score | 0.930869 (rank : 6) | θ value | 4.6295e-105 (rank : 5) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05920 | Gene names | Pc, Pcx | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
|
COA1_HUMAN
|
||||||
| NC score | 0.824099 (rank : 7) | θ value | 1.66577e-62 (rank : 7) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q13085 | Gene names | ACACA, ACAC, ACC1, ACCA | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 1 (EC 6.4.1.2) (ACC-alpha) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
COA2_HUMAN
|
||||||
| NC score | 0.790193 (rank : 8) | θ value | 2.32979e-56 (rank : 8) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00763, Q16852 | Gene names | ACACB, ACC2, ACCB | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 2 (EC 6.4.1.2) (ACC-beta) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
CPSM_MOUSE
|
||||||
| NC score | 0.400762 (rank : 9) | θ value | 8.97725e-08 (rank : 11) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8C196, Q6NX75 | Gene names | Cps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
CPSM_HUMAN
|
||||||
| NC score | 0.396408 (rank : 10) | θ value | 4.0297e-08 (rank : 10) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P31327, O43774, Q7Z5I5 | Gene names | CPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
PYR1_HUMAN
|
||||||
| NC score | 0.393927 (rank : 11) | θ value | 1.63604e-09 (rank : 9) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
|
PCCB_MOUSE
|
||||||
| NC score | 0.090431 (rank : 12) | θ value | θ > 10 (rank : 22) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99MN9 | Gene names | Pccb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Propionyl-CoA carboxylase beta chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit beta) (Propanoyl-CoA:carbon dioxide ligase subunit beta). | |||||
|
PCCB_HUMAN
|
||||||
| NC score | 0.087818 (rank : 13) | θ value | θ > 10 (rank : 21) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P05166, Q16813 | Gene names | PCCB | |||
|
Domain Architecture |

|
|||||
| Description | Propionyl-CoA carboxylase beta chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit beta) (Propanoyl-CoA:carbon dioxide ligase subunit beta). | |||||
|
APC7_MOUSE
|
||||||
| NC score | 0.077834 (rank : 14) | θ value | 0.0736092 (rank : 12) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WVM3, Q3UBM7, Q8BSR2, Q91W13 | Gene names | Anapc7, Apc7 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7) (Prediabetic NOD sera-reactive autoantigen). | |||||
|
ODB2_HUMAN
|
||||||
| NC score | 0.065231 (rank : 15) | θ value | 5.27518 (rank : 16) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11182 | Gene names | DBT, BCATE2 | |||
|
Domain Architecture |

|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
HMGCL_MOUSE
|
||||||
| NC score | 0.062226 (rank : 16) | θ value | θ > 10 (rank : 20) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P38060 | Gene names | Hmgcl | |||
|
Domain Architecture |

|
|||||
| Description | Hydroxymethylglutaryl-CoA lyase, mitochondrial precursor (EC 4.1.3.4) (HMG-CoA lyase) (HL) (3-hydroxy-3-methylglutarate-CoA lyase). | |||||
|
ODB2_MOUSE
|
||||||
| NC score | 0.059910 (rank : 17) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P53395 | Gene names | Dbt | |||
|
Domain Architecture |

|
|||||
| Description | Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial precursor (EC 2.3.1.168) (Dihydrolipoyllysine-residue (2-methylpropanoyl)transferase) (E2) (Dihydrolipoamide branched chain transacylase) (BCKAD E2 subunit). | |||||
|
APC7_HUMAN
|
||||||
| NC score | 0.054740 (rank : 18) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJX3, Q96AC4, Q96GF4, Q9BU24, Q9NT16 | Gene names | ANAPC7, APC7 | |||
|
Domain Architecture |

|
|||||
| Description | Anaphase-promoting complex subunit 7 (APC7) (Cyclosome subunit 7). | |||||
|
HMGCL_HUMAN
|
||||||
| NC score | 0.054388 (rank : 19) | θ value | θ > 10 (rank : 19) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35914, Q96FP8 | Gene names | HMGCL | |||
|
Domain Architecture |

|
|||||
| Description | Hydroxymethylglutaryl-CoA lyase, mitochondrial precursor (EC 4.1.3.4) (HMG-CoA lyase) (HL) (3-hydroxy-3-methylglutarate-CoA lyase). | |||||
|
PUR2_HUMAN
|
||||||
| NC score | 0.048117 (rank : 20) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22102 | Gene names | GART, PRGS | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional purine biosynthetic protein adenosine-3 [Includes: Phosphoribosylamine--glycine ligase (EC 6.3.4.13) (GARS) (Glycinamide ribonucleotide synthetase) (Phosphoribosylglycinamide synthetase); Phosphoribosylformylglycinamidine cyclo-ligase (EC 6.3.3.1) (AIRS) (Phosphoribosyl-aminoimidazole synthetase) (AIR synthase); Phosphoribosylglycinamide formyltransferase (EC 2.1.2.2) (GART) (GAR transformylase) (5'-phosphoribosylglycinamide transformylase)]. | |||||
|
CDYL_MOUSE
|
||||||
| NC score | 0.010456 (rank : 21) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WTK2 | Gene names | Cdyl | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
MICA1_MOUSE
|
||||||
| NC score | 0.004958 (rank : 22) | θ value | 6.88961 (rank : 17) | |||
| Query Neighborhood Hits | 18 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VDP3 | Gene names | Mical1, Mical, Nical | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||