Please be patient as the page loads
|
COA2_HUMAN
|
||||||
| SwissProt Accessions | O00763, Q16852 | Gene names | ACACB, ACC2, ACCB | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 2 (EC 6.4.1.2) (ACC-beta) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
COA1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.968167 (rank : 2) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13085 | Gene names | ACACA, ACAC, ACC1, ACCA | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 1 (EC 6.4.1.2) (ACC-alpha) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
COA2_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 184 | |
| SwissProt Accessions | O00763, Q16852 | Gene names | ACACB, ACC2, ACCB | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 2 (EC 6.4.1.2) (ACC-beta) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
PCCA_HUMAN
|
||||||
| θ value | 2.10721e-57 (rank : 3) | NC score | 0.790805 (rank : 3) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P05165, Q15979 | Gene names | PCCA | |||
|
Domain Architecture |

|
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
PCCA_MOUSE
|
||||||
| θ value | 2.32979e-56 (rank : 4) | NC score | 0.790193 (rank : 4) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91ZA3, Q80VU5, Q922N3 | Gene names | Pcca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
MCCA_HUMAN
|
||||||
| θ value | 1.51013e-55 (rank : 5) | NC score | 0.785866 (rank : 6) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96RQ3, Q59ES4, Q9H959, Q9NS97 | Gene names | MCCC1, MCCA | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
MCCA_MOUSE
|
||||||
| θ value | 6.34462e-54 (rank : 6) | NC score | 0.786731 (rank : 5) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99MR8, Q3TGU0, Q9D8R2 | Gene names | Mccc1, Mcca | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
PYC_MOUSE
|
||||||
| θ value | 9.48078e-50 (rank : 7) | NC score | 0.760277 (rank : 7) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05920 | Gene names | Pc, Pcx | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
|
PYC_HUMAN
|
||||||
| θ value | 2.1121e-49 (rank : 8) | NC score | 0.759553 (rank : 8) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P11498, Q16705 | Gene names | PC | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
|
PCCB_MOUSE
|
||||||
| θ value | 7.58209e-15 (rank : 9) | NC score | 0.363728 (rank : 9) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99MN9 | Gene names | Pccb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Propionyl-CoA carboxylase beta chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit beta) (Propanoyl-CoA:carbon dioxide ligase subunit beta). | |||||
|
PCCB_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 10) | NC score | 0.358422 (rank : 10) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P05166, Q16813 | Gene names | PCCB | |||
|
Domain Architecture |

|
|||||
| Description | Propionyl-CoA carboxylase beta chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit beta) (Propanoyl-CoA:carbon dioxide ligase subunit beta). | |||||
|
PYR1_HUMAN
|
||||||
| θ value | 7.34386e-10 (rank : 11) | NC score | 0.351412 (rank : 11) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
|
CPSM_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 12) | NC score | 0.344931 (rank : 13) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P31327, O43774, Q7Z5I5 | Gene names | CPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
CPSM_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 13) | NC score | 0.344981 (rank : 12) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C196, Q6NX75 | Gene names | Cps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
MCCC2_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 14) | NC score | 0.249906 (rank : 14) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCC0, Q96C27, Q9Y4L7 | Gene names | MCCC2, MCCB | |||
|
Domain Architecture |
|
|||||
| Description | Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial precursor (EC 6.4.1.4) (3-Methylcrotonyl-CoA carboxylase 2) (MCCase subunit beta) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit beta) (3- Methylcrotonyl-CoA carboxylase non-biotin-containing subunit). | |||||
|
SYN2_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 15) | NC score | 0.082046 (rank : 15) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q64332, Q6NZR0, Q9QWV7 | Gene names | Syn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
SYN2_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 16) | NC score | 0.078893 (rank : 16) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92777 | Gene names | SYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 17) | NC score | 0.035040 (rank : 73) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
PININ_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 18) | NC score | 0.057596 (rank : 33) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
SH3K1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 19) | NC score | 0.042663 (rank : 53) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96B97, Q8IWX6, Q8IX98, Q96RN4, Q9NYR0 | Gene names | SH3KBP1, CIN85 | |||
|
Domain Architecture |

|
|||||
| Description | SH3-domain kinase-binding protein 1 (Cbl-interacting protein of 85 kDa) (Human Src-family kinase-binding protein 1) (HSB-1) (CD2-binding protein 3) (CD2BP3). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 20) | NC score | 0.073050 (rank : 18) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 21) | NC score | 0.076758 (rank : 17) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
XRCC1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 22) | NC score | 0.057658 (rank : 32) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 23) | NC score | 0.064092 (rank : 26) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
PO6F2_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 24) | NC score | 0.028019 (rank : 96) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
SH3K1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 25) | NC score | 0.041095 (rank : 58) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R550, Q8CAL8, Q8CEF6, Q8R545, Q8R546, Q8R547, Q8R548, Q8R549, Q8R551, Q9CTQ9, Q9DC14, Q9JKC3 | Gene names | Sh3kbp1, Ruk, Seta | |||
|
Domain Architecture |

|
|||||
| Description | SH3-domain kinase-binding protein 1 (SH3-containing, expressed in tumorigenic astrocytes) (Regulator of ubiquitous kinase) (Ruk). | |||||
|
KCNK4_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 26) | NC score | 0.030386 (rank : 87) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 27) | NC score | 0.070718 (rank : 21) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
MYCB2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 28) | NC score | 0.039051 (rank : 60) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
PININ_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 29) | NC score | 0.048962 (rank : 41) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 30) | NC score | 0.035718 (rank : 70) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 31) | NC score | 0.072032 (rank : 19) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
ELOA3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 32) | NC score | 0.047363 (rank : 44) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NG57 | Gene names | TCEB3C, TCEB3L2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A3 (Elongin-A3) (EloA3) (Transcription elongation factor B polypeptide 3C). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 33) | NC score | 0.059069 (rank : 31) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.125558 (rank : 34) | NC score | 0.064686 (rank : 25) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.125558 (rank : 35) | NC score | 0.045396 (rank : 51) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 0.125558 (rank : 36) | NC score | 0.042326 (rank : 54) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 37) | NC score | 0.071301 (rank : 20) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 0.125558 (rank : 38) | NC score | 0.032601 (rank : 83) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
FOXGB_MOUSE
|
||||||
| θ value | 0.163984 (rank : 39) | NC score | 0.016226 (rank : 127) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60987, Q80VP3 | Gene names | Foxg1b, Fkhl1, Foxg1, Hfhbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein G1B (Forkhead-related protein FKHL1) (Transcription factor BF-1) (Brain factor 1) (BF1). | |||||
|
CJ086_HUMAN
|
||||||
| θ value | 0.21417 (rank : 40) | NC score | 0.064889 (rank : 24) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NXX6, Q5SQQ5, Q6P673, Q8WY66, Q9BS90 | Gene names | C10orf86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf86. | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 0.21417 (rank : 41) | NC score | 0.057180 (rank : 34) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
SREC2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 42) | NC score | 0.021818 (rank : 111) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
TCF19_MOUSE
|
||||||
| θ value | 0.279714 (rank : 43) | NC score | 0.046970 (rank : 46) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99KJ5, Q8BPA3, Q9CT93 | Gene names | Tcf19, Sc1 | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor 19-like protein (SC1 protein homolog). | |||||
|
WIRE_MOUSE
|
||||||
| θ value | 0.279714 (rank : 44) | NC score | 0.068883 (rank : 22) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
HCFC1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 45) | NC score | 0.037579 (rank : 65) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
ITCH_MOUSE
|
||||||
| θ value | 0.365318 (rank : 46) | NC score | 0.020048 (rank : 114) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 47) | NC score | 0.053373 (rank : 37) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
HORN_MOUSE
|
||||||
| θ value | 0.47712 (rank : 48) | NC score | 0.028074 (rank : 95) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
PDC6I_HUMAN
|
||||||
| θ value | 0.47712 (rank : 49) | NC score | 0.041779 (rank : 56) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WUM4, Q9BX86, Q9NUN0, Q9P2H2, Q9UKL5 | Gene names | PDCD6IP, AIP1, ALIX, KIAA1375 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | 0.47712 (rank : 50) | NC score | 0.066009 (rank : 23) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 0.62314 (rank : 51) | NC score | 0.055552 (rank : 36) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
VATA1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.032636 (rank : 82) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50516 | Gene names | Atp6v1a, Atp6a1, Atp6a2, Atp6v1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar ATP synthase catalytic subunit A, ubiquitous isoform (EC 3.6.3.14) (V-ATPase subunit A 1) (Vacuolar proton pump alpha subunit 1) (V-ATPase 69 kDa subunit 1). | |||||
|
ADA29_HUMAN
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.009905 (rank : 149) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKF5, Q9UHP1, Q9UKF3, Q9UKF4 | Gene names | ADAM29 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 29 precursor (A disintegrin and metalloproteinase domain 29). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 54) | NC score | 0.059666 (rank : 30) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
APC_HUMAN
|
||||||
| θ value | 0.813845 (rank : 55) | NC score | 0.045765 (rank : 49) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
ERBB2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 56) | NC score | 0.002644 (rank : 176) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70424, Q61525, Q6ZPE0 | Gene names | Erbb2, Kiaa3023, Neu | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene). | |||||
|
ITCH_HUMAN
|
||||||
| θ value | 0.813845 (rank : 57) | NC score | 0.018924 (rank : 118) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
NEB2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 58) | NC score | 0.017275 (rank : 122) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 1.06291 (rank : 59) | NC score | 0.048259 (rank : 43) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
GNAS3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 60) | NC score | 0.060379 (rank : 28) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95467, O95417 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 1.06291 (rank : 61) | NC score | 0.030589 (rank : 86) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
HCFC1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 62) | NC score | 0.034741 (rank : 75) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 63) | NC score | 0.035053 (rank : 72) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MAST1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 64) | NC score | 0.004705 (rank : 168) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 1.06291 (rank : 65) | NC score | 0.037900 (rank : 64) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
PI5PA_MOUSE
|
||||||
| θ value | 1.06291 (rank : 66) | NC score | 0.035893 (rank : 69) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 1.06291 (rank : 67) | NC score | 0.031983 (rank : 84) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RFIP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 68) | NC score | 0.031875 (rank : 85) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6WKZ4, Q6AZK4, Q6WKZ2, Q6WKZ6, Q86YV4, Q8TDL1, Q9H642 | Gene names | RAB11FIP1, RCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 69) | NC score | 0.032954 (rank : 81) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
VINEX_MOUSE
|
||||||
| θ value | 1.06291 (rank : 70) | NC score | 0.020964 (rank : 113) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9R1Z8, Q62423 | Gene names | Sorbs3, Scam1, Sh3d4 | |||
|
Domain Architecture |

|
|||||
| Description | Vinexin (Sorbin and SH3 domain-containing protein 3) (SH3-containing adapter molecule 1) (SCAM-1) (SH3 domain-containing protein SH3P3). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 71) | NC score | 0.048290 (rank : 42) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
5NT1B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 72) | NC score | 0.026817 (rank : 97) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96P26, Q8N9W3, Q8NA26, Q96DU5, Q96KE6, Q96M25, Q96SA3 | Gene names | NT5C1B, AIRP | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic 5'-nucleotidase 1B (EC 3.1.3.5) (Cytosolic 5'-nucleotidase IB) (cN1B) (cN-IB) (Autoimmune infertility-related protein). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 73) | NC score | 0.033535 (rank : 79) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
H6ST3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 74) | NC score | 0.019910 (rank : 115) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZP7, Q5W0L0, Q68CW6 | Gene names | HS6ST3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3). | |||||
|
KI21A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 75) | NC score | 0.006236 (rank : 165) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
MAGI1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 76) | NC score | 0.019649 (rank : 117) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 77) | NC score | 0.050784 (rank : 40) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
NEUM_HUMAN
|
||||||
| θ value | 1.38821 (rank : 78) | NC score | 0.041703 (rank : 57) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
NKX61_HUMAN
|
||||||
| θ value | 1.38821 (rank : 79) | NC score | 0.010922 (rank : 145) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P78426 | Gene names | NKX6-1, NKX6A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-6.1. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 80) | NC score | 0.041880 (rank : 55) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
SEBP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 81) | NC score | 0.029380 (rank : 91) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96T21, Q8IYC0 | Gene names | SECISBP2, SBP2 | |||
|
Domain Architecture |

|
|||||
| Description | SECIS-binding protein 2 (Selenocysteine insertion sequence-binding protein 2). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 82) | NC score | 0.038599 (rank : 61) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
VATA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 83) | NC score | 0.029741 (rank : 89) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P38606, Q96DY6, Q9UHY3 | Gene names | ATP6V1A, ATP6A1, ATP6V1A1, VPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar ATP synthase catalytic subunit A, ubiquitous isoform (EC 3.6.3.14) (V-ATPase subunit A 1) (Vacuolar proton pump alpha subunit 1) (V-ATPase 69 kDa subunit 1) (Isoform VA68). | |||||
|
ZN406_MOUSE
|
||||||
| θ value | 1.38821 (rank : 84) | NC score | 0.000062 (rank : 181) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TS63 | Gene names | Znf406, Zfp406 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 406. | |||||
|
APC_MOUSE
|
||||||
| θ value | 1.81305 (rank : 85) | NC score | 0.036639 (rank : 67) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
GRIN1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 86) | NC score | 0.045849 (rank : 48) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
MAML3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 87) | NC score | 0.029029 (rank : 93) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
PDC6I_MOUSE
|
||||||
| θ value | 1.81305 (rank : 88) | NC score | 0.033197 (rank : 80) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WU78, O88695, O89014, Q8BSL8, Q8R0H5, Q99LR3, Q9QZN8 | Gene names | Pdcd6ip, Aip1, Alix | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (ALG-2-interacting protein X) (ALG-2-interacting protein 1) (E2F1-inducible protein) (Eig2). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 89) | NC score | 0.038054 (rank : 63) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
RIN3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 90) | NC score | 0.023153 (rank : 105) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P59729 | Gene names | Rin3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 91) | NC score | 0.028953 (rank : 94) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SREC2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 92) | NC score | 0.016500 (rank : 125) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 93) | NC score | 0.023627 (rank : 103) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
3BP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 94) | NC score | 0.022206 (rank : 110) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
AB1IP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 95) | NC score | 0.024929 (rank : 102) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z5R6, Q8IWS8, Q8IZZ7 | Gene names | APBB1IP, PREL1, RARP1, RIAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Rap1-GTP- interacting adapter molecule) (RIAM) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 73) (Retinoic acid-responsive proline- rich protein 1) (RARP-1). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 96) | NC score | 0.046611 (rank : 47) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
CI047_HUMAN
|
||||||
| θ value | 2.36792 (rank : 97) | NC score | 0.038375 (rank : 62) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZRZ4, Q5SQD7, Q7Z568, Q8N1V4 | Gene names | C9orf47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf47 precursor. | |||||
|
EGLN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 98) | NC score | 0.016911 (rank : 123) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9GZT9, Q8N3M8, Q9BZS8, Q9BZT0 | Gene names | EGLN1, C1orf12 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (Prolyl hydroxylase domain-containing protein 2) (PHD2) (SM-20). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 99) | NC score | 0.045440 (rank : 50) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 100) | NC score | 0.035445 (rank : 71) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
RENT1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 101) | NC score | 0.013721 (rank : 134) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EPU0, Q99PR4 | Gene names | Upf1, Rent1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (mUpf1). | |||||
|
YAP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 102) | NC score | 0.026035 (rank : 99) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P46937 | Gene names | YAP1, YAP65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 103) | NC score | 0.044629 (rank : 52) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 104) | NC score | 0.022994 (rank : 106) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MYLK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 105) | NC score | 0.001465 (rank : 177) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H1R3, Q96I84 | Gene names | MYLK2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain kinase 2, skeletal/cardiac muscle (EC 2.7.11.18) (MLCK2). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 3.0926 (rank : 106) | NC score | 0.056401 (rank : 35) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NRP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 107) | NC score | 0.009344 (rank : 152) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35375, O35373, O35374, O35376, O35377, O35378 | Gene names | Nrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 108) | NC score | 0.059682 (rank : 29) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TTBK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 109) | NC score | 0.009177 (rank : 153) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
WRIP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 110) | NC score | 0.026483 (rank : 98) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
WRIP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 111) | NC score | 0.026022 (rank : 100) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91XU0, Q3TCT7, Q6PDF0, Q8BUW5, Q8BWP6, Q8BY55, Q921W3, Q9EQL3 | Gene names | Wrnip1, Whip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
ADA2A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 112) | NC score | -0.001218 (rank : 184) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 900 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01338 | Gene names | Adra2a | |||
|
Domain Architecture |
|
|||||
| Description | Alpha-2A adrenergic receptor (Alpha-2A adrenoceptor) (Alpha-2A adrenoreceptor) (Alpha-2AAR). | |||||
|
AT2B1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 113) | NC score | 0.005517 (rank : 166) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20020, Q12992, Q12993, Q13819, Q13820, Q13821, Q16504, Q93082 | Gene names | ATP2B1, PMCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 1 (EC 3.6.3.8) (PMCA1) (Plasma membrane calcium pump isoform 1) (Plasma membrane calcium ATPase isoform 1). | |||||
|
ATX2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.033634 (rank : 78) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
CRYL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 115) | NC score | 0.017827 (rank : 120) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99KP3, Q542R9, Q8R4W7 | Gene names | Cryl1, Cry | |||
|
Domain Architecture |
|
|||||
| Description | Lambda-crystallin homolog. | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | 0.025384 (rank : 101) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
F104A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 117) | NC score | 0.033665 (rank : 77) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q969W3 | Gene names | FAM104A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM104A. | |||||
|
JPH4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 118) | NC score | 0.010974 (rank : 144) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80WT0, Q8BMI1 | Gene names | Jph4, Jphl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
KLC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 119) | NC score | 0.008436 (rank : 158) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H0B6, Q9H9C8, Q9HA20 | Gene names | KLC2 | |||
|
Domain Architecture |
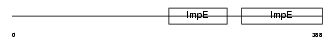
|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 4.03905 (rank : 120) | NC score | 0.034889 (rank : 74) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NRP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 121) | NC score | 0.008707 (rank : 157) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60462, O14820, O14821 | Gene names | NRP2, VEGF165R2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
PXK_MOUSE
|
||||||
| θ value | 4.03905 (rank : 122) | NC score | 0.013698 (rank : 135) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BX57, Q3TD60, Q3TEL7, Q3TZZ1, Q3U197, Q3U773, Q3UCS5, Q4FBH7, Q91WB6 | Gene names | Pxk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PX domain-containing protein kinase-like protein (Modulator of Na,K- ATPase) (MONaKA). | |||||
|
ROBO4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 123) | NC score | 0.008397 (rank : 159) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C310, Q9DBW1 | Gene names | Robo4 | |||
|
Domain Architecture |
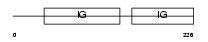
|
|||||
| Description | Roundabout homolog 4 precursor. | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 4.03905 (rank : 124) | NC score | 0.015501 (rank : 128) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 125) | NC score | 0.053067 (rank : 38) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TRRAP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 126) | NC score | 0.013752 (rank : 133) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
ZFY16_MOUSE
|
||||||
| θ value | 4.03905 (rank : 127) | NC score | 0.010713 (rank : 147) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80U44, Q8BRD2, Q8CG97 | Gene names | Zfyve16, Kiaa0305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 16 (Endofin) (Endosomal- associated FYVE domain protein). | |||||
|
ZN524_MOUSE
|
||||||
| θ value | 4.03905 (rank : 128) | NC score | -0.001598 (rank : 185) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D0B1, Q9DCQ0 | Gene names | Znf524, Zfp524 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 524. | |||||
|
ATX2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 129) | NC score | 0.029052 (rank : 92) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
CCD37_MOUSE
|
||||||
| θ value | 5.27518 (rank : 130) | NC score | 0.022296 (rank : 109) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80VN0, Q6AXD6 | Gene names | Ccdc37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 37. | |||||
|
CD99_MOUSE
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.061484 (rank : 27) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VCN6, Q4L299, Q4L2A4, Q8BLN1 | Gene names | Cd99, Pilrl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD99 antigen precursor (Paired immunoglobin-like type 2 receptor- ligand) (PILR-L). | |||||
|
CDT1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.019797 (rank : 116) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R4E9, Q8BUQ1, Q8BX29, Q8R3V0, Q8R4E8 | Gene names | Cdt1, Ris2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication factor Cdt1 (Double parked homolog) (DUP) (Retroviral insertion site 2 protein). | |||||
|
DUET_HUMAN
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.001219 (rank : 178) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y2A5, Q8TBQ5, Q9NSZ4 | Gene names | DUET, TRAD | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Duet (EC 2.7.11.1) (Serine/threonine kinase with Dbl- and pleckstrin homology domain). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.013982 (rank : 131) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
FEZ1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 135) | NC score | 0.012838 (rank : 138) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K0X8 | Gene names | Fez1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fasciculation and elongation protein zeta 1 (Zygin-1) (Zygin I). | |||||
|
FXR2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 136) | NC score | 0.012687 (rank : 139) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WVR4, Q9WVR5 | Gene names | Fxr2, Fxr2h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
JHD2C_MOUSE
|
||||||
| θ value | 5.27518 (rank : 137) | NC score | 0.016254 (rank : 126) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q69ZK6, Q6NV48, Q8BUF5, Q8C4I5, Q8C5Q9 | Gene names | Jmjd1c, Jhdm2c, Kiaa1380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C). | |||||
|
MICA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 138) | NC score | 0.009102 (rank : 155) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TDZ2, Q7Z633, Q8IVS9, Q96G47, Q9H6X6, Q9H7I0, Q9HAA1, Q9UFF7 | Gene names | MICAL1, MICAL, NICAL | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 139) | NC score | 0.013865 (rank : 132) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 140) | NC score | 0.011384 (rank : 141) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
ROBO3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 141) | NC score | 0.004472 (rank : 169) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96MS0 | Gene names | ROBO3 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Roundabout-like protein 3). | |||||
|
SIN3A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 142) | NC score | 0.013668 (rank : 136) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96ST3, Q8N8N4, Q8NC83, Q8WV18, Q96L98, Q9UFQ1 | Gene names | SIN3A | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
UBP11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 143) | NC score | 0.009507 (rank : 151) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51784, Q8IUG6, Q9BWE1 | Gene names | USP11, UHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 11 (EC 3.1.2.15) (Ubiquitin thioesterase 11) (Ubiquitin-specific-processing protease 11) (Deubiquitinating enzyme 11). | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 144) | NC score | 0.003480 (rank : 172) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.047349 (rank : 45) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
B3A3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.008060 (rank : 160) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P16283 | Gene names | Slc4a3, Ae3 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 3 (Neuronal band 3-like protein) (Solute carrier family 4 member 3). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | 0.023533 (rank : 104) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
FOXH1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.010093 (rank : 148) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75593 | Gene names | FOXH1, FAST1, FAST2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (hFAST-1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
GLP1R_MOUSE
|
||||||
| θ value | 6.88961 (rank : 149) | NC score | 0.003637 (rank : 171) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35659 | Gene names | Glp1r | |||
|
Domain Architecture |

|
|||||
| Description | Glucagon-like peptide 1 receptor precursor (GLP-1 receptor) (GLP-1-R) (GLP-1R). | |||||
|
JADE2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 150) | NC score | 0.009153 (rank : 154) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQC1, Q6IE80, Q8TEK0, Q92513, Q96GQ6 | Gene names | PHF15, JADE2, KIAA0239 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 151) | NC score | 0.029441 (rank : 90) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 6.88961 (rank : 152) | NC score | 0.009532 (rank : 150) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
NKX61_MOUSE
|
||||||
| θ value | 6.88961 (rank : 153) | NC score | 0.007471 (rank : 161) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99MA9, Q9ERQ7 | Gene names | Nkx6-1, Nkx6.1, Nkx6a | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-6.1. | |||||
|
PCDGJ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 154) | NC score | 0.000145 (rank : 180) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5F8, Q9UN63 | Gene names | PCDHGB7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B7 precursor (PCDH-gamma-B7). | |||||
|
PCQAP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 155) | NC score | 0.022580 (rank : 108) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
RANB9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 156) | NC score | 0.015349 (rank : 129) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P69566 | Gene names | Ranbp9, Ranbpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 9 (RanBP9) (Ran-binding protein M) (RanBPM) (B cell antigen receptor Ig beta-associated protein 1) (IBAP-1). | |||||
|
ROR2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 157) | NC score | -0.000791 (rank : 182) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q01974, Q59GF5, Q5SPI5, Q9HAY7, Q9HB61 | Gene names | ROR2, NTRKR2 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase transmembrane receptor ROR2 precursor (EC 2.7.10.1) (Neurotrophic tyrosine kinase, receptor-related 2). | |||||
|
TBX2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 158) | NC score | 0.006303 (rank : 164) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60707 | Gene names | Tbx2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
|
TCF20_HUMAN
|
||||||
| θ value | 6.88961 (rank : 159) | NC score | 0.021618 (rank : 112) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
TMPSD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 160) | NC score | 0.003219 (rank : 173) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
VILL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 161) | NC score | 0.005481 (rank : 167) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91YD6, P97809, Q8BIS5, Q8R211, Q8VDI8 | Gene names | Vill, Villp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Villin-like protein (EF-6). | |||||
|
ZN592_MOUSE
|
||||||
| θ value | 6.88961 (rank : 162) | NC score | 0.000665 (rank : 179) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BHZ4, Q80XM1 | Gene names | Znf592, Kiaa0211, Zfp592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 592 (Zfp-592). | |||||
|
4ET_HUMAN
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | 0.011137 (rank : 143) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NRA8, Q8NCF2, Q9H708 | Gene names | EIF4ENIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
AFF3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | 0.037012 (rank : 66) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
E2AK2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | -0.001198 (rank : 183) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q03963, Q61742, Q62026 | Gene names | Eif2ak2, Pkr, Prkr, Tik | |||
|
Domain Architecture |
|
|||||
| Description | Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase) (Serine/threonine-protein kinase TIK). | |||||
|
ENAM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | 0.018786 (rank : 119) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
HERC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | 0.010772 (rank : 146) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15034 | Gene names | HERC3, KIAA0032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain protein 3. | |||||
|
JHD2A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | 0.012322 (rank : 140) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y4C1, Q53S72, Q68D47, Q68UT9, Q6N050, Q8IY08 | Gene names | JMJD1A, JHDM2A, JMJD1, KIAA0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
KIF11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | 0.002750 (rank : 174) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P52732, Q15716, Q5VWX0 | Gene names | KIF11, EG5, KNSL1 | |||
|
Domain Architecture |
|
|||||
| Description | Kinesin-like protein KIF11 (Kinesin-related motor protein Eg5) (Kinesin-like spindle protein HKSP) (Thyroid receptor-interacting protein 5) (TRIP5) (Kinesin-like protein 1). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.017552 (rank : 121) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
PHLB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | 0.013341 (rank : 137) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
PKP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | 0.007209 (rank : 163) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q68FH0, Q640N0, Q68G56, Q8BK47, Q8BVH1, Q9CRE3 | Gene names | Pkp4, Armrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plakophilin-4 (Armadillo-related protein). | |||||
|
RANB9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | 0.016594 (rank : 124) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96S59, O94764, Q6P3T7, Q7LBR2, Q7Z7F9 | Gene names | RANBP9, RANBPM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 9 (RanBP9) (RanBP7) (Ran-binding protein M) (RanBPM) (BPM90) (BPM-L). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.034740 (rank : 76) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SGPP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 175) | NC score | 0.008884 (rank : 156) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JI99 | Gene names | Sgpp1, Spp1, Spph1 | |||
|
Domain Architecture |
|
|||||
| Description | Sphingosine-1-phosphate phosphatase 1 (EC 3.1.3.-) (Sphingosine-1- phosphatase 1) (SPPase1) (SPP) (mSPP1). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 176) | NC score | 0.039702 (rank : 59) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 177) | NC score | 0.022809 (rank : 107) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TACC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 178) | NC score | 0.014237 (rank : 130) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 179) | NC score | 0.030157 (rank : 88) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TMPSD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 180) | NC score | 0.002735 (rank : 175) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BYE2, Q86YM4, Q96JY8, Q9BYE1 | Gene names | TMPRSS13, MSP, TMPRSS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
TTBK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 181) | NC score | 0.004189 (rank : 170) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6IQ55, O94932, Q6ZN52, Q8IVV1 | Gene names | TTBK2, KIAA0847 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2 (EC 2.7.11.1). | |||||
|
VCY1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 182) | NC score | 0.036096 (rank : 68) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14598 | Gene names | VCY, BPY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific basic protein Y 1 (Variably charged protein Y). | |||||
|
X3CL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 183) | NC score | 0.011211 (rank : 142) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35188, O35933 | Gene names | Cx3cl1, Cx3c, Fkn, Scyd1 | |||
|
Domain Architecture |
|
|||||
| Description | Fractalkine precursor (CX3CL1) (Neurotactin) (CX3C membrane-anchored chemokine) (Small inducible cytokine D1). | |||||
|
ZFYV1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 184) | NC score | 0.007234 (rank : 162) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HBF4, Q8WYX7, Q96K57, Q9BXP9, Q9HCI3 | Gene names | ZFYVE1, DFCP1, KIAA1589, TAFF1, ZNFN2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 1 (Double FYVE-containing protein 1) (Tandem FYVE fingers-1) (SR3). | |||||
|
TARA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.051234 (rank : 39) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
COA2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 184 | |
| SwissProt Accessions | O00763, Q16852 | Gene names | ACACB, ACC2, ACCB | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 2 (EC 6.4.1.2) (ACC-beta) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
COA1_HUMAN
|
||||||
| NC score | 0.968167 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13085 | Gene names | ACACA, ACAC, ACC1, ACCA | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 1 (EC 6.4.1.2) (ACC-alpha) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
PCCA_HUMAN
|
||||||
| NC score | 0.790805 (rank : 3) | θ value | 2.10721e-57 (rank : 3) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P05165, Q15979 | Gene names | PCCA | |||
|
Domain Architecture |

|
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
PCCA_MOUSE
|
||||||
| NC score | 0.790193 (rank : 4) | θ value | 2.32979e-56 (rank : 4) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91ZA3, Q80VU5, Q922N3 | Gene names | Pcca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Propionyl-CoA carboxylase alpha chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit alpha) (Propanoyl-CoA:carbon dioxide ligase subunit alpha). | |||||
|
MCCA_MOUSE
|
||||||
| NC score | 0.786731 (rank : 5) | θ value | 6.34462e-54 (rank : 6) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99MR8, Q3TGU0, Q9D8R2 | Gene names | Mccc1, Mcca | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
MCCA_HUMAN
|
||||||
| NC score | 0.785866 (rank : 6) | θ value | 1.51013e-55 (rank : 5) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96RQ3, Q59ES4, Q9H959, Q9NS97 | Gene names | MCCC1, MCCA | |||
|
Domain Architecture |

|
|||||
| Description | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial precursor (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 1) (MCCase subunit alpha) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit alpha) (3- methylcrotonyl-CoA carboxylase biotin-containing subunit). | |||||
|
PYC_MOUSE
|
||||||
| NC score | 0.760277 (rank : 7) | θ value | 9.48078e-50 (rank : 7) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q05920 | Gene names | Pc, Pcx | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
|
PYC_HUMAN
|
||||||
| NC score | 0.759553 (rank : 8) | θ value | 2.1121e-49 (rank : 8) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P11498, Q16705 | Gene names | PC | |||
|
Domain Architecture |

|
|||||
| Description | Pyruvate carboxylase, mitochondrial precursor (EC 6.4.1.1) (Pyruvic carboxylase) (PCB). | |||||
|
PCCB_MOUSE
|
||||||
| NC score | 0.363728 (rank : 9) | θ value | 7.58209e-15 (rank : 9) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99MN9 | Gene names | Pccb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Propionyl-CoA carboxylase beta chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit beta) (Propanoyl-CoA:carbon dioxide ligase subunit beta). | |||||
|
PCCB_HUMAN
|
||||||
| NC score | 0.358422 (rank : 10) | θ value | 2.88119e-14 (rank : 10) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P05166, Q16813 | Gene names | PCCB | |||
|
Domain Architecture |

|
|||||
| Description | Propionyl-CoA carboxylase beta chain, mitochondrial precursor (EC 6.4.1.3) (PCCase subunit beta) (Propanoyl-CoA:carbon dioxide ligase subunit beta). | |||||
|
PYR1_HUMAN
|
||||||
| NC score | 0.351412 (rank : 11) | θ value | 7.34386e-10 (rank : 11) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P27708, Q6P0Q0 | Gene names | CAD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAD protein [Includes: Glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5); Aspartate carbamoyltransferase (EC 2.1.3.2); Dihydroorotase (EC 3.5.2.3)]. | |||||
|
CPSM_MOUSE
|
||||||
| NC score | 0.344981 (rank : 12) | θ value | 1.43324e-05 (rank : 13) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C196, Q6NX75 | Gene names | Cps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
CPSM_HUMAN
|
||||||
| NC score | 0.344931 (rank : 13) | θ value | 1.69304e-06 (rank : 12) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P31327, O43774, Q7Z5I5 | Gene names | CPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor (EC 6.3.4.16) (Carbamoyl-phosphate synthetase I) (CPSase I). | |||||
|
MCCC2_HUMAN
|
||||||
| NC score | 0.249906 (rank : 14) | θ value | 9.29e-05 (rank : 14) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCC0, Q96C27, Q9Y4L7 | Gene names | MCCC2, MCCB | |||
|
Domain Architecture |
|
|||||
| Description | Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial precursor (EC 6.4.1.4) (3-Methylcrotonyl-CoA carboxylase 2) (MCCase subunit beta) (3-methylcrotonyl-CoA:carbon dioxide ligase subunit beta) (3- Methylcrotonyl-CoA carboxylase non-biotin-containing subunit). | |||||
|
SYN2_MOUSE
|
||||||
| NC score | 0.082046 (rank : 15) | θ value | 0.000461057 (rank : 15) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q64332, Q6NZR0, Q9QWV7 | Gene names | Syn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
SYN2_HUMAN
|
||||||
| NC score | 0.078893 (rank : 16) | θ value | 0.000602161 (rank : 16) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92777 | Gene names | SYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.076758 (rank : 17) | θ value | 0.0330416 (rank : 21) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.073050 (rank : 18) | θ value | 0.0330416 (rank : 20) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.072032 (rank : 19) | θ value | 0.0736092 (rank : 31) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.071301 (rank : 20) | θ value | 0.125558 (rank : 37) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.070718 (rank : 21) | θ value | 0.0563607 (rank : 27) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
WIRE_MOUSE
|
||||||
| NC score | 0.068883 (rank : 22) | θ value | 0.279714 (rank : 44) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.066009 (rank : 23) | θ value | 0.47712 (rank : 50) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
CJ086_HUMAN
|
||||||
| NC score | 0.064889 (rank : 24) | θ value | 0.21417 (rank : 40) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NXX6, Q5SQQ5, Q6P673, Q8WY66, Q9BS90 | Gene names | C10orf86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf86. | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.064686 (rank : 25) | θ value | 0.125558 (rank : 34) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.064092 (rank : 26) | θ value | 0.0431538 (rank : 23) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
CD99_MOUSE
|
||||||
| NC score | 0.061484 (rank : 27) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VCN6, Q4L299, Q4L2A4, Q8BLN1 | Gene names | Cd99, Pilrl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CD99 antigen precursor (Paired immunoglobin-like type 2 receptor- ligand) (PILR-L). | |||||
|
GNAS3_HUMAN
|
||||||
| NC score | 0.060379 (rank : 28) | θ value | 1.06291 (rank : 60) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95467, O95417 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.059682 (rank : 29) | θ value | 3.0926 (rank : 108) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.059666 (rank : 30) | θ value | 0.813845 (rank : 54) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.059069 (rank : 31) | θ value | 0.0961366 (rank : 33) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
XRCC1_MOUSE
|
||||||
| NC score | 0.057658 (rank : 32) | θ value | 0.0330416 (rank : 22) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q60596, Q3THC5, Q5U435, Q7TNQ5 | Gene names | Xrcc1, Xrcc-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC1 (X-ray repair cross-complementing protein 1). | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.057596 (rank : 33) | θ value | 0.0113563 (rank : 18) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.057180 (rank : 34) | θ value | 0.21417 (rank : 41) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.056401 (rank : 35) | θ value | 3.0926 (rank : 106) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.055552 (rank : 36) | θ value | 0.62314 (rank : 51) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.053373 (rank : 37) | θ value | 0.365318 (rank : 47) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.053067 (rank : 38) | θ value | 4.03905 (rank : 125) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.051234 (rank : 39) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.050784 (rank : 40) | θ value | 1.38821 (rank : 77) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.048962 (rank : 41) | θ value | 0.0736092 (rank : 29) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.048290 (rank : 42) | θ value | 1.06291 (rank : 71) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.048259 (rank : 43) | θ value | 1.06291 (rank : 59) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
ELOA3_HUMAN
|
||||||
| NC score | 0.047363 (rank : 44) | θ value | 0.0961366 (rank : 32) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NG57 | Gene names | TCEB3C, TCEB3L2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A3 (Elongin-A3) (EloA3) (Transcription elongation factor B polypeptide 3C). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.047349 (rank : 45) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
TCF19_MOUSE
|
||||||
| NC score | 0.046970 (rank : 46) | θ value | 0.279714 (rank : 43) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99KJ5, Q8BPA3, Q9CT93 | Gene names | Tcf19, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 19-like protein (SC1 protein homolog). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.046611 (rank : 47) | θ value | 2.36792 (rank : 96) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
GRIN1_MOUSE
|
||||||
| NC score | 0.045849 (rank : 48) | θ value | 1.81305 (rank : 86) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
APC_HUMAN
|
||||||
| NC score | 0.045765 (rank : 49) | θ value | 0.813845 (rank : 55) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.045440 (rank : 50) | θ value | 2.36792 (rank : 99) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.045396 (rank : 51) | θ value | 0.125558 (rank : 35) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.044629 (rank : 52) | θ value | 3.0926 (rank : 103) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
SH3K1_HUMAN
|
||||||
| NC score | 0.042663 (rank : 53) | θ value | 0.0252991 (rank : 19) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96B97, Q8IWX6, Q8IX98, Q96RN4, Q9NYR0 | Gene names | SH3KBP1, CIN85 | |||
|
Domain Architecture |

|
|||||
| Description | SH3-domain kinase-binding protein 1 (Cbl-interacting protein of 85 kDa) (Human Src-family kinase-binding protein 1) (HSB-1) (CD2-binding protein 3) (CD2BP3). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.042326 (rank : 54) | θ value | 0.125558 (rank : 36) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.041880 (rank : 55) | θ value | 1.38821 (rank : 80) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PDC6I_HUMAN
|
||||||
| NC score | 0.041779 (rank : 56) | θ value | 0.47712 (rank : 49) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WUM4, Q9BX86, Q9NUN0, Q9P2H2, Q9UKL5 | Gene names | PDCD6IP, AIP1, ALIX, KIAA1375 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95). | |||||
|
NEUM_HUMAN
|
||||||
| NC score | 0.041703 (rank : 57) | θ value | 1.38821 (rank : 78) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
SH3K1_MOUSE
|
||||||
| NC score | 0.041095 (rank : 58) | θ value | 0.0431538 (rank : 25) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R550, Q8CAL8, Q8CEF6, Q8R545, Q8R546, Q8R547, Q8R548, Q8R549, Q8R551, Q9CTQ9, Q9DC14, Q9JKC3 | Gene names | Sh3kbp1, Ruk, Seta | |||
|
Domain Architecture |

|
|||||
| Description | SH3-domain kinase-binding protein 1 (SH3-containing, expressed in tumorigenic astrocytes) (Regulator of ubiquitous kinase) (Ruk). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.039702 (rank : 59) | θ value | 8.99809 (rank : 176) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
MYCB2_MOUSE
|
||||||
| NC score | 0.039051 (rank : 60) | θ value | 0.0736092 (rank : 28) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.038599 (rank : 61) | θ value | 1.38821 (rank : 82) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CI047_HUMAN
|
||||||
| NC score | 0.038375 (rank : 62) | θ value | 2.36792 (rank : 97) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZRZ4, Q5SQD7, Q7Z568, Q8N1V4 | Gene names | C9orf47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf47 precursor. | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.038054 (rank : 63) | θ value | 1.81305 (rank : 89) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.037900 (rank : 64) | θ value | 1.06291 (rank : 65) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
HCFC1_MOUSE
|
||||||
| NC score | 0.037579 (rank : 65) | θ value | 0.365318 (rank : 45) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61191, Q684R1, Q7TSB0, Q8C2D0, Q9QWH2 | Gene names | Hcfc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N- terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C-terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
AFF3_HUMAN
|
||||||
| NC score | 0.037012 (rank : 66) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
APC_MOUSE
|
||||||
| NC score | 0.036639 (rank : 67) | θ value | 1.81305 (rank : 85) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
VCY1_HUMAN
|
||||||
| NC score | 0.036096 (rank : 68) | θ value | 8.99809 (rank : 182) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14598 | Gene names | VCY, BPY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific basic protein Y 1 (Variably charged protein Y). | |||||
|
PI5PA_MOUSE
|
||||||
| NC score | 0.035893 (rank : 69) | θ value | 1.06291 (rank : 66) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.035718 (rank : 70) | θ value | 0.0736092 (rank : 30) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.035445 (rank : 71) | θ value | 2.36792 (rank : 100) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.035053 (rank : 72) | θ value | 1.06291 (rank : 63) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.035040 (rank : 73) | θ value | 0.0113563 (rank : 17) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.034889 (rank : 74) | θ value | 4.03905 (rank : 120) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
HCFC1_HUMAN
|
||||||
| NC score | 0.034741 (rank : 75) | θ value | 1.06291 (rank : 62) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51610, Q6P4G5 | Gene names | HCFC1, HCF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Host cell factor (HCF) (HCF-1) (C1 factor) (VP16 accessory protein) (VCAF) (CFF) [Contains: HCF N-terminal chain 1; HCF N-terminal chain 2; HCF N-terminal chain 3; HCF N-terminal chain 4; HCF N-terminal chain 5; HCF N-terminal chain 6; HCF C-terminal chain 1; HCF C- terminal chain 2; HCF C-terminal chain 3; HCF C-terminal chain 4; HCF C-terminal chain 5; HCF C-terminal chain 6]. | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.034740 (rank : 76) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
F104A_HUMAN
|
||||||
| NC score | 0.033665 (rank : 77) | θ value | 4.03905 (rank : 117) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q969W3 | Gene names | FAM104A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM104A. | |||||
|
ATX2_MOUSE
|
||||||
| NC score | 0.033634 (rank : 78) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.033535 (rank : 79) | θ value | 1.38821 (rank : 73) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
PDC6I_MOUSE
|
||||||
| NC score | 0.033197 (rank : 80) | θ value | 1.81305 (rank : 88) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WU78, O88695, O89014, Q8BSL8, Q8R0H5, Q99LR3, Q9QZN8 | Gene names | Pdcd6ip, Aip1, Alix | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (ALG-2-interacting protein X) (ALG-2-interacting protein 1) (E2F1-inducible protein) (Eig2). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.032954 (rank : 81) | θ value | 1.06291 (rank : 69) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
VATA1_MOUSE
|
||||||
| NC score | 0.032636 (rank : 82) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50516 | Gene names | Atp6v1a, Atp6a1, Atp6a2, Atp6v1a1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar ATP synthase catalytic subunit A, ubiquitous isoform (EC 3.6.3.14) (V-ATPase subunit A 1) (Vacuolar proton pump alpha subunit 1) (V-ATPase 69 kDa subunit 1). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.032601 (rank : 83) | θ value | 0.125558 (rank : 38) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.031983 (rank : 84) | θ value | 1.06291 (rank : 67) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RFIP1_HUMAN
|
||||||
| NC score | 0.031875 (rank : 85) | θ value | 1.06291 (rank : 68) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6WKZ4, Q6AZK4, Q6WKZ2, Q6WKZ6, Q86YV4, Q8TDL1, Q9H642 | Gene names | RAB11FIP1, RCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.030589 (rank : 86) | θ value | 1.06291 (rank : 61) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
KCNK4_HUMAN
|
||||||
| NC score | 0.030386 (rank : 87) | θ value | 0.0563607 (rank : 26) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NYG8, Q96T94 | Gene names | KCNK4, TRAAK | |||
|
Domain Architecture |

|
|||||
| Description | Potassium channel subfamily K member 4 (TWIK-related arachidonic acid- stimulated potassium channel protein) (TRAAK) (Two pore K(+) channel KT4.1). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.030157 (rank : 88) | θ value | 8.99809 (rank : 179) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
VATA1_HUMAN
|
||||||
| NC score | 0.029741 (rank : 89) | θ value | 1.38821 (rank : 83) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P38606, Q96DY6, Q9UHY3 | Gene names | ATP6V1A, ATP6A1, ATP6V1A1, VPP2 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar ATP synthase catalytic subunit A, ubiquitous isoform (EC 3.6.3.14) (V-ATPase subunit A 1) (Vacuolar proton pump alpha subunit 1) (V-ATPase 69 kDa subunit 1) (Isoform VA68). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.029441 (rank : 90) | θ value | 6.88961 (rank : 151) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
SEBP2_HUMAN
|
||||||
| NC score | 0.029380 (rank : 91) | θ value | 1.38821 (rank : 81) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96T21, Q8IYC0 | Gene names | SECISBP2, SBP2 | |||
|
Domain Architecture |

|
|||||
| Description | SECIS-binding protein 2 (Selenocysteine insertion sequence-binding protein 2). | |||||
|
ATX2_HUMAN
|
||||||
| NC score | 0.029052 (rank : 92) | θ value | 5.27518 (rank : 129) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
MAML3_HUMAN
|
||||||
| NC score | 0.029029 (rank : 93) | θ value | 1.81305 (rank : 87) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.028953 (rank : 94) | θ value | 1.81305 (rank : 91) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
HORN_MOUSE
|
||||||
| NC score | 0.028074 (rank : 95) | θ value | 0.47712 (rank : 48) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
PO6F2_MOUSE
|
||||||
| NC score | 0.028019 (rank : 96) | θ value | 0.0431538 (rank : 24) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BJI4 | Gene names | Pou6f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | POU domain, class 6, transcription factor 2. | |||||
|
5NT1B_HUMAN
|
||||||
| NC score | 0.026817 (rank : 97) | θ value | 1.38821 (rank : 72) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96P26, Q8N9W3, Q8NA26, Q96DU5, Q96KE6, Q96M25, Q96SA3 | Gene names | NT5C1B, AIRP | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic 5'-nucleotidase 1B (EC 3.1.3.5) (Cytosolic 5'-nucleotidase IB) (cN1B) (cN-IB) (Autoimmune infertility-related protein). | |||||
|
WRIP1_HUMAN
|
||||||
| NC score | 0.026483 (rank : 98) | θ value | 3.0926 (rank : 110) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96S55, Q53EP6, Q59ET8, Q5W0E2, Q5W0E4, Q8WV26, Q9H681, Q9NRJ6 | Gene names | WRNIP1, WHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
YAP1_HUMAN
|
||||||
| NC score | 0.026035 (rank : 99) | θ value | 2.36792 (rank : 102) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P46937 | Gene names | YAP1, YAP65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
WRIP1_MOUSE
|
||||||
| NC score | 0.026022 (rank : 100) | θ value | 3.0926 (rank : 111) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91XU0, Q3TCT7, Q6PDF0, Q8BUW5, Q8BWP6, Q8BY55, Q921W3, Q9EQL3 | Gene names | Wrnip1, Whip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase WRNIP1 (Werner helicase-interacting protein 1). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.025384 (rank : 101) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
AB1IP_HUMAN
|
||||||
| NC score | 0.024929 (rank : 102) | θ value | 2.36792 (rank : 95) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z5R6, Q8IWS8, Q8IZZ7 | Gene names | APBB1IP, PREL1, RARP1, RIAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Rap1-GTP- interacting adapter molecule) (RIAM) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 73) (Retinoic acid-responsive proline- rich protein 1) (RARP-1). | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.023627 (rank : 103) | θ value | 1.81305 (rank : 93) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.023533 (rank : 104) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
RIN3_MOUSE
|
||||||
| NC score | 0.023153 (rank : 105) | θ value | 1.81305 (rank : 90) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P59729 | Gene names | Rin3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.022994 (rank : 106) | θ value | 3.0926 (rank : 104) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.022809 (rank : 107) | θ value | 8.99809 (rank : 177) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
PCQAP_MOUSE
|
||||||
| NC score | 0.022580 (rank : 108) | θ value | 6.88961 (rank : 155) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
CCD37_MOUSE
|
||||||
| NC score | 0.022296 (rank : 109) | θ value | 5.27518 (rank : 130) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80VN0, Q6AXD6 | Gene names | Ccdc37 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 37. | |||||
|
3BP1_HUMAN
|
||||||
| NC score | 0.022206 (rank : 110) | θ value | 2.36792 (rank : 94) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
SREC2_HUMAN
|
||||||
| NC score | 0.021818 (rank : 111) | θ value | 0.21417 (rank : 42) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
TCF20_HUMAN
|
||||||
| NC score | 0.021618 (rank : 112) | θ value | 6.88961 (rank : 159) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
VINEX_MOUSE
|
||||||
| NC score | 0.020964 (rank : 113) | θ value | 1.06291 (rank : 70) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9R1Z8, Q62423 | Gene names | Sorbs3, Scam1, Sh3d4 | |||
|
Domain Architecture |

|
|||||
| Description | Vinexin (Sorbin and SH3 domain-containing protein 3) (SH3-containing adapter molecule 1) (SCAM-1) (SH3 domain-containing protein SH3P3). | |||||
|
ITCH_MOUSE
|
||||||
| NC score | 0.020048 (rank : 114) | θ value | 0.365318 (rank : 46) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8C863, O54971 | Gene names | Itch | |||
|
Domain Architecture |

|
|||||
| Description | Itchy E3 ubiquitin protein ligase (EC 6.3.2.-). | |||||
|
H6ST3_HUMAN
|
||||||
| NC score | 0.019910 (rank : 115) | θ value | 1.38821 (rank : 74) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZP7, Q5W0L0, Q68CW6 | Gene names | HS6ST3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heparan-sulfate 6-O-sulfotransferase 3 (EC 2.8.2.-) (HS6ST-3). | |||||
|
CDT1_MOUSE
|
||||||
| NC score | 0.019797 (rank : 116) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R4E9, Q8BUQ1, Q8BX29, Q8R3V0, Q8R4E8 | Gene names | Cdt1, Ris2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication factor Cdt1 (Double parked homolog) (DUP) (Retroviral insertion site 2 protein). | |||||
|
MAGI1_MOUSE
|
||||||
| NC score | 0.019649 (rank : 117) | θ value | 1.38821 (rank : 76) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
ITCH_HUMAN
|
||||||
| NC score | 0.018924 (rank : 118) | θ value | 0.813845 (rank : 57) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
ENAM_HUMAN
|
||||||
| NC score | 0.018786 (rank : 119) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NRM1, Q9H3D1 | Gene names | ENAM | |||
|
Domain Architecture |

|
|||||
| Description | Enamelin precursor. | |||||
|
CRYL1_MOUSE
|
||||||
| NC score | 0.017827 (rank : 120) | θ value | 4.03905 (rank : 115) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99KP3, Q542R9, Q8R4W7 | Gene names | Cryl1, Cry | |||
|
Domain Architecture |

|
|||||
| Description | Lambda-crystallin homolog. | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.017552 (rank : 121) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
NEB2_MOUSE
|
||||||
| NC score | 0.017275 (rank : 122) | θ value | 0.813845 (rank : 58) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
EGLN1_HUMAN
|
||||||
| NC score | 0.016911 (rank : 123) | θ value | 2.36792 (rank : 98) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9GZT9, Q8N3M8, Q9BZS8, Q9BZT0 | Gene names | EGLN1, C1orf12 | |||
|
Domain Architecture |

|
|||||
| Description | Egl nine homolog 1 (EC 1.14.11.-) (Hypoxia-inducible factor prolyl hydroxylase 2) (HIF-prolyl hydroxylase 2) (HIF-PH2) (HPH-2) (Prolyl hydroxylase domain-containing protein 2) (PHD2) (SM-20). | |||||
|
RANB9_HUMAN
|
||||||
| NC score | 0.016594 (rank : 124) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96S59, O94764, Q6P3T7, Q7LBR2, Q7Z7F9 | Gene names | RANBP9, RANBPM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 9 (RanBP9) (RanBP7) (Ran-binding protein M) (RanBPM) (BPM90) (BPM-L). | |||||
|
SREC2_MOUSE
|
||||||
| NC score | 0.016500 (rank : 125) | θ value | 1.81305 (rank : 92) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
JHD2C_MOUSE
|
||||||
| NC score | 0.016254 (rank : 126) | θ value | 5.27518 (rank : 137) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q69ZK6, Q6NV48, Q8BUF5, Q8C4I5, Q8C5Q9 | Gene names | Jmjd1c, Jhdm2c, Kiaa1380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C). | |||||
|
FOXGB_MOUSE
|
||||||
| NC score | 0.016226 (rank : 127) | θ value | 0.163984 (rank : 39) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60987, Q80VP3 | Gene names | Foxg1b, Fkhl1, Foxg1, Hfhbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein G1B (Forkhead-related protein FKHL1) (Transcription factor BF-1) (Brain factor 1) (BF1). | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.015501 (rank : 128) | θ value | 4.03905 (rank : 124) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
RANB9_MOUSE
|
||||||
| NC score | 0.015349 (rank : 129) | θ value | 6.88961 (rank : 156) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P69566 | Gene names | Ranbp9, Ranbpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 9 (RanBP9) (Ran-binding protein M) (RanBPM) (B cell antigen receptor Ig beta-associated protein 1) (IBAP-1). | |||||
|
TACC2_HUMAN
|
||||||
| NC score | 0.014237 (rank : 130) | θ value | 8.99809 (rank : 178) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O95359, Q9NZ41, Q9NZR5 | Gene names | TACC2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2 (Anti Zuai-1) (AZU-1). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.013982 (rank : 131) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.013865 (rank : 132) | θ value | 5.27518 (rank : 139) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
TRRAP_HUMAN
|
||||||
| NC score | 0.013752 (rank : 133) | θ value | 4.03905 (rank : 126) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
RENT1_MOUSE
|
||||||
| NC score | 0.013721 (rank : 134) | θ value | 2.36792 (rank : 101) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EPU0, Q99PR4 | Gene names | Upf1, Rent1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of nonsense transcripts 1 (EC 3.6.1.-) (ATP-dependent helicase RENT1) (Nonsense mRNA reducing factor 1) (NORF1) (Up- frameshift suppressor 1 homolog) (mUpf1). | |||||
|
PXK_MOUSE
|
||||||
| NC score | 0.013698 (rank : 135) | θ value | 4.03905 (rank : 122) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BX57, Q3TD60, Q3TEL7, Q3TZZ1, Q3U197, Q3U773, Q3UCS5, Q4FBH7, Q91WB6 | Gene names | Pxk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PX domain-containing protein kinase-like protein (Modulator of Na,K- ATPase) (MONaKA). | |||||
|
SIN3A_HUMAN
|
||||||
| NC score | 0.013668 (rank : 136) | θ value | 5.27518 (rank : 142) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96ST3, Q8N8N4, Q8NC83, Q8WV18, Q96L98, Q9UFQ1 | Gene names | SIN3A | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
PHLB1_MOUSE
|
||||||
| NC score | 0.013341 (rank : 137) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 678 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6PDH0, Q80TV2 | Gene names | Phldb1, Kiaa0638, Ll5a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 1 (Protein LL5-alpha). | |||||
|
FEZ1_MOUSE
|
||||||
| NC score | 0.012838 (rank : 138) | θ value | 5.27518 (rank : 135) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K0X8 | Gene names | Fez1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fasciculation and elongation protein zeta 1 (Zygin-1) (Zygin I). | |||||
|
FXR2_MOUSE
|
||||||
| NC score | 0.012687 (rank : 139) | θ value | 5.27518 (rank : 136) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WVR4, Q9WVR5 | Gene names | Fxr2, Fxr2h | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
JHD2A_HUMAN
|
||||||
| NC score | 0.012322 (rank : 140) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y4C1, Q53S72, Q68D47, Q68UT9, Q6N050, Q8IY08 | Gene names | JMJD1A, JHDM2A, JMJD1, KIAA0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.011384 (rank : 141) | θ value | 5.27518 (rank : 140) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
X3CL1_MOUSE
|
||||||
| NC score | 0.011211 (rank : 142) | θ value | 8.99809 (rank : 183) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35188, O35933 | Gene names | Cx3cl1, Cx3c, Fkn, Scyd1 | |||
|
Domain Architecture |

|
|||||
| Description | Fractalkine precursor (CX3CL1) (Neurotactin) (CX3C membrane-anchored chemokine) (Small inducible cytokine D1). | |||||
|
4ET_HUMAN
|
||||||
| NC score | 0.011137 (rank : 143) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NRA8, Q8NCF2, Q9H708 | Gene names | EIF4ENIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4E transporter (eIF4E transporter) (4E-T) (Eukaryotic translation initiation factor 4E nuclear import factor 1). | |||||
|
JPH4_MOUSE
|
||||||
| NC score | 0.010974 (rank : 144) | θ value | 4.03905 (rank : 118) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80WT0, Q8BMI1 | Gene names | Jph4, Jphl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junctophilin-4 (Junctophilin-like 1 protein). | |||||
|
NKX61_HUMAN
|
||||||
| NC score | 0.010922 (rank : 145) | θ value | 1.38821 (rank : 79) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P78426 | Gene names | NKX6-1, NKX6A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-6.1. | |||||
|
HERC3_HUMAN
|
||||||
| NC score | 0.010772 (rank : 146) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15034 | Gene names | HERC3, KIAA0032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT domain and RCC1-like domain protein 3. | |||||
|
ZFY16_MOUSE
|
||||||
| NC score | 0.010713 (rank : 147) | θ value | 4.03905 (rank : 127) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80U44, Q8BRD2, Q8CG97 | Gene names | Zfyve16, Kiaa0305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 16 (Endofin) (Endosomal- associated FYVE domain protein). | |||||
|
FOXH1_HUMAN
|
||||||
| NC score | 0.010093 (rank : 148) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75593 | Gene names | FOXH1, FAST1, FAST2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (hFAST-1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
ADA29_HUMAN
|
||||||
| NC score | 0.009905 (rank : 149) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKF5, Q9UHP1, Q9UKF3, Q9UKF4 | Gene names | ADAM29 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 29 precursor (A disintegrin and metalloproteinase domain 29). | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.009532 (rank : 150) | θ value | 6.88961 (rank : 152) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
UBP11_HUMAN
|
||||||
| NC score | 0.009507 (rank : 151) | θ value | 5.27518 (rank : 143) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51784, Q8IUG6, Q9BWE1 | Gene names | USP11, UHX1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 11 (EC 3.1.2.15) (Ubiquitin thioesterase 11) (Ubiquitin-specific-processing protease 11) (Deubiquitinating enzyme 11). | |||||
|
NRP2_MOUSE
|
||||||
| NC score | 0.009344 (rank : 152) | θ value | 3.0926 (rank : 107) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35375, O35373, O35374, O35376, O35377, O35378 | Gene names | Nrp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
TTBK1_HUMAN
|
||||||
| NC score | 0.009177 (rank : 153) | θ value | 3.0926 (rank : 109) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1058 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5TCY1, Q2L6C6, Q8N444, Q96JH2 | Gene names | TTBK1, BDTK, KIAA1855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 1 (EC 2.7.11.1) (Brain-derived tau kinase). | |||||
|
JADE2_HUMAN
|
||||||
| NC score | 0.009153 (rank : 154) | θ value | 6.88961 (rank : 150) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQC1, Q6IE80, Q8TEK0, Q92513, Q96GQ6 | Gene names | PHF15, JADE2, KIAA0239 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
MICA1_HUMAN
|
||||||
| NC score | 0.009102 (rank : 155) | θ value | 5.27518 (rank : 138) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TDZ2, Q7Z633, Q8IVS9, Q96G47, Q9H6X6, Q9H7I0, Q9HAA1, Q9UFF7 | Gene names | MICAL1, MICAL, NICAL | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
SGPP1_MOUSE
|
||||||
| NC score | 0.008884 (rank : 156) | θ value | 8.99809 (rank : 175) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JI99 | Gene names | Sgpp1, Spp1, Spph1 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosine-1-phosphate phosphatase 1 (EC 3.1.3.-) (Sphingosine-1- phosphatase 1) (SPPase1) (SPP) (mSPP1). | |||||
|
NRP2_HUMAN
|
||||||
| NC score | 0.008707 (rank : 157) | θ value | 4.03905 (rank : 121) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60462, O14820, O14821 | Gene names | NRP2, VEGF165R2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropilin-2 precursor (Vascular endothelial cell growth factor 165 receptor 2). | |||||
|
KLC2_HUMAN
|
||||||
| NC score | 0.008436 (rank : 158) | θ value | 4.03905 (rank : 119) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H0B6, Q9H9C8, Q9HA20 | Gene names | KLC2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 2 (KLC 2). | |||||
|
ROBO4_MOUSE
|
||||||
| NC score | 0.008397 (rank : 159) | θ value | 4.03905 (rank : 123) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C310, Q9DBW1 | Gene names | Robo4 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 4 precursor. | |||||
|
B3A3_MOUSE
|
||||||
| NC score | 0.008060 (rank : 160) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P16283 | Gene names | Slc4a3, Ae3 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 3 (Neuronal band 3-like protein) (Solute carrier family 4 member 3). | |||||
|
NKX61_MOUSE
|
||||||
| NC score | 0.007471 (rank : 161) | θ value | 6.88961 (rank : 153) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99MA9, Q9ERQ7 | Gene names | Nkx6-1, Nkx6.1, Nkx6a | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-6.1. | |||||
|
ZFYV1_HUMAN
|
||||||
| NC score | 0.007234 (rank : 162) | θ value | 8.99809 (rank : 184) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HBF4, Q8WYX7, Q96K57, Q9BXP9, Q9HCI3 | Gene names | ZFYVE1, DFCP1, KIAA1589, TAFF1, ZNFN2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 1 (Double FYVE-containing protein 1) (Tandem FYVE fingers-1) (SR3). | |||||
|
PKP4_MOUSE
|
||||||
| NC score | 0.007209 (rank : 163) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q68FH0, Q640N0, Q68G56, Q8BK47, Q8BVH1, Q9CRE3 | Gene names | Pkp4, Armrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plakophilin-4 (Armadillo-related protein). | |||||
|
TBX2_MOUSE
|
||||||
| NC score | 0.006303 (rank : 164) | θ value | 6.88961 (rank : 158) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60707 | Gene names | Tbx2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
|
KI21A_MOUSE
|
||||||
| NC score | 0.006236 (rank : 165) | θ value | 1.38821 (rank : 75) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
AT2B1_HUMAN
|
||||||
| NC score | 0.005517 (rank : 166) | θ value | 4.03905 (rank : 113) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20020, Q12992, Q12993, Q13819, Q13820, Q13821, Q16504, Q93082 | Gene names | ATP2B1, PMCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma membrane calcium-transporting ATPase 1 (EC 3.6.3.8) (PMCA1) (Plasma membrane calcium pump isoform 1) (Plasma membrane calcium ATPase isoform 1). | |||||
|
VILL_MOUSE
|
||||||
| NC score | 0.005481 (rank : 167) | θ value | 6.88961 (rank : 161) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91YD6, P97809, Q8BIS5, Q8R211, Q8VDI8 | Gene names | Vill, Villp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Villin-like protein (EF-6). | |||||
|
MAST1_HUMAN
|
||||||
| NC score | 0.004705 (rank : 168) | θ value | 1.06291 (rank : 64) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
ROBO3_HUMAN
|
||||||
| NC score | 0.004472 (rank : 169) | θ value | 5.27518 (rank : 141) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96MS0 | Gene names | ROBO3 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 3 precursor (Roundabout-like protein 3). | |||||
|
TTBK2_HUMAN
|
||||||
| NC score | 0.004189 (rank : 170) | θ value | 8.99809 (rank : 181) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6IQ55, O94932, Q6ZN52, Q8IVV1 | Gene names | TTBK2, KIAA0847 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2 (EC 2.7.11.1). | |||||
|
GLP1R_MOUSE
|
||||||
| NC score | 0.003637 (rank : 171) | θ value | 6.88961 (rank : 149) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35659 | Gene names | Glp1r | |||
|
Domain Architecture |

|
|||||
| Description | Glucagon-like peptide 1 receptor precursor (GLP-1 receptor) (GLP-1-R) (GLP-1R). | |||||
|
WNK1_MOUSE
|
||||||
| NC score | 0.003480 (rank : 172) | θ value | 5.27518 (rank : 144) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
TMPSD_MOUSE
|
||||||
| NC score | 0.003219 (rank : 173) | θ value | 6.88961 (rank : 160) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
KIF11_HUMAN
|
||||||
| NC score | 0.002750 (rank : 174) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P52732, Q15716, Q5VWX0 | Gene names | KIF11, EG5, KNSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF11 (Kinesin-related motor protein Eg5) (Kinesin-like spindle protein HKSP) (Thyroid receptor-interacting protein 5) (TRIP5) (Kinesin-like protein 1). | |||||
|
TMPSD_HUMAN
|
||||||
| NC score | 0.002735 (rank : 175) | θ value | 8.99809 (rank : 180) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BYE2, Q86YM4, Q96JY8, Q9BYE1 | Gene names | TMPRSS13, MSP, TMPRSS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
ERBB2_MOUSE
|
||||||
| NC score | 0.002644 (rank : 176) | θ value | 0.813845 (rank : 56) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70424, Q61525, Q6ZPE0 | Gene names | Erbb2, Kiaa3023, Neu | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene). | |||||
|
MYLK2_HUMAN
|
||||||
| NC score | 0.001465 (rank : 177) | θ value | 3.0926 (rank : 105) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H1R3, Q96I84 | Gene names | MYLK2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin light chain kinase 2, skeletal/cardiac muscle (EC 2.7.11.18) (MLCK2). | |||||
|
DUET_HUMAN
|
||||||
| NC score | 0.001219 (rank : 178) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y2A5, Q8TBQ5, Q9NSZ4 | Gene names | DUET, TRAD | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Duet (EC 2.7.11.1) (Serine/threonine kinase with Dbl- and pleckstrin homology domain). | |||||
|
ZN592_MOUSE
|
||||||
| NC score | 0.000665 (rank : 179) | θ value | 6.88961 (rank : 162) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BHZ4, Q80XM1 | Gene names | Znf592, Kiaa0211, Zfp592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 592 (Zfp-592). | |||||
|
PCDGJ_HUMAN
|
||||||
| NC score | 0.000145 (rank : 180) | θ value | 6.88961 (rank : 154) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5F8, Q9UN63 | Gene names | PCDHGB7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B7 precursor (PCDH-gamma-B7). | |||||
|
ZN406_MOUSE
|
||||||
| NC score | 0.000062 (rank : 181) | θ value | 1.38821 (rank : 84) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7TS63 | Gene names | Znf406, Zfp406 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 406. | |||||
|
ROR2_HUMAN
|
||||||
| NC score | -0.000791 (rank : 182) | θ value | 6.88961 (rank : 157) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q01974, Q59GF5, Q5SPI5, Q9HAY7, Q9HB61 | Gene names | ROR2, NTRKR2 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase transmembrane receptor ROR2 precursor (EC 2.7.10.1) (Neurotrophic tyrosine kinase, receptor-related 2). | |||||
|
E2AK2_MOUSE
|
||||||
| NC score | -0.001198 (rank : 183) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q03963, Q61742, Q62026 | Gene names | Eif2ak2, Pkr, Prkr, Tik | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced, double-stranded RNA-activated protein kinase (EC 2.7.11.1) (Interferon-inducible RNA-dependent protein kinase) (Protein kinase RNA-activated) (PKR) (p68 kinase) (P1/eIF-2A protein kinase) (Serine/threonine-protein kinase TIK). | |||||
|
ADA2A_MOUSE
|
||||||
| NC score | -0.001218 (rank : 184) | θ value | 4.03905 (rank : 112) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 900 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01338 | Gene names | Adra2a | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2A adrenergic receptor (Alpha-2A adrenoceptor) (Alpha-2A adrenoreceptor) (Alpha-2AAR). | |||||
|
ZN524_MOUSE
|
||||||
| NC score | -0.001598 (rank : 185) | θ value | 4.03905 (rank : 128) | |||
| Query Neighborhood Hits | 184 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D0B1, Q9DCQ0 | Gene names | Znf524, Zfp524 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 524. | |||||