Please be patient as the page loads
|
TRRAP_HUMAN
|
||||||
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TRRAP_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
TRRAP_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.990457 (rank : 2) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80YV3, Q8C0Z5, Q8K104 | Gene names | Trrap | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (Tra1 homolog). | |||||
|
ATR_HUMAN
|
||||||
| θ value | 1.13037e-18 (rank : 3) | NC score | 0.468437 (rank : 3) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13535, Q7KYL3, Q93051, Q9BXK4 | Gene names | ATR, FRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein) (FRAP-related protein 1). | |||||
|
PRKDC_MOUSE
|
||||||
| θ value | 1.47631e-18 (rank : 4) | NC score | 0.443930 (rank : 5) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97313, O88187, P97928, Q307W9, Q3V2W8, Q8C2A7, Q9Z341 | Gene names | Prkdc, Xrcc7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (P460). | |||||
|
ATR_MOUSE
|
||||||
| θ value | 2.35696e-16 (rank : 5) | NC score | 0.455913 (rank : 4) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JKK8 | Gene names | Atr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein). | |||||
|
FRAP_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 6) | NC score | 0.417920 (rank : 7) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JLN9, Q811J5, Q9CST1 | Gene names | Frap1, Frap | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
PRKDC_HUMAN
|
||||||
| θ value | 5.43371e-13 (rank : 7) | NC score | 0.418417 (rank : 6) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
FRAP_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 8) | NC score | 0.415045 (rank : 8) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P42345, Q6LE87, Q96QG3, Q9Y4I3 | Gene names | FRAP1, FRAP, FRAP2 | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
ATM_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 9) | NC score | 0.337551 (rank : 9) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13315, O15429, Q12758, Q16551, Q93007, Q9NP02, Q9UCX7 | Gene names | ATM | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated) (A-T, mutated). | |||||
|
SMG1_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 10) | NC score | 0.333823 (rank : 11) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
SMG1_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 11) | NC score | 0.335841 (rank : 10) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
ATM_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 12) | NC score | 0.325332 (rank : 12) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62388 | Gene names | Atm | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated homolog) (A-T, mutated homolog). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 13) | NC score | 0.027073 (rank : 52) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
ZN365_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 14) | NC score | 0.107737 (rank : 15) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BG89, Q80TQ4, Q8BK39, Q8BXT2 | Gene names | Znf365, Kiaa0844, Zfp365 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF365 (Su48). | |||||
|
REV1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 15) | NC score | 0.047772 (rank : 33) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q920Q2, Q9QXV2 | Gene names | Rev1l, Rev1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein REV1 (EC 2.7.7.-) (Rev1-like terminal deoxycytidyl transferase). | |||||
|
PANK1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 16) | NC score | 0.075315 (rank : 17) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TE04, Q7RTX6, Q7Z495, Q8TBQ8 | Gene names | PANK1, PANK | |||
|
Domain Architecture |
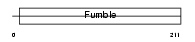
|
|||||
| Description | Pantothenate kinase 1 (EC 2.7.1.33) (Pantothenic acid kinase 1) (hPanK1) (hPanK). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 17) | NC score | 0.037401 (rank : 39) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.030819 (rank : 45) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
IPO4_MOUSE
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.059061 (rank : 29) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VI75, Q3TBW0, Q99J52 | Gene names | Ipo4, Imp4a, Ranbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Importin-4 (Importin 4a) (Imp4a) (Ran-binding protein 4) (RanBP4). | |||||
|
UTY_HUMAN
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.073028 (rank : 18) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14607, O14608 | Gene names | UTY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed Y chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the Y chromosome). | |||||
|
PANK1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 21) | NC score | 0.068586 (rank : 21) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K4K6, Q9D3K1, Q9QXM8 | Gene names | Pank1, Pank | |||
|
Domain Architecture |
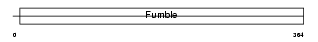
|
|||||
| Description | Pantothenate kinase 1 (EC 2.7.1.33) (Pantothenic acid kinase 1) (mPank1) (mPank). | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.040857 (rank : 36) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.014896 (rank : 83) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
ZN365_HUMAN
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.088033 (rank : 16) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q70YC5, O94930, Q5HYE6, Q68SG8, Q6NSK2, Q6P9D4, Q70YC6, Q70YC7 | Gene names | ZNF365, KIAA0844 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF365. | |||||
|
ARMC4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.037523 (rank : 38) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T2S8, Q9H0C0 | Gene names | ARMC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 4. | |||||
|
KCNQ1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.017054 (rank : 77) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.023796 (rank : 56) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
UTX_HUMAN
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.061918 (rank : 27) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15550 | Gene names | UTX | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed X chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the X chromosome). | |||||
|
CLC4M_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.021050 (rank : 63) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H2X3, Q69F40, Q969M4, Q96QP3, Q96QP4, Q96QP5, Q96QP6, Q9BXS3, Q9H2Q9, Q9H8F0, Q9Y2A8 | Gene names | CLEC4M, CD209L, CD209L1 | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 4 member M (CD209 antigen-like protein 1) (Dendritic cell-specific ICAM-3-grabbing nonintegrin 2) (DC-SIGN2) (DC-SIGN-related protein) (DC-SIGNR) (Liver/lymph node-specific ICAM- 3-grabbing nonintegrin) (L-SIGN) (CD299 antigen). | |||||
|
ITPR2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.032163 (rank : 42) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14571, O94773 | Gene names | ITPR2 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2). | |||||
|
ITPR2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.031841 (rank : 44) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z329, P70226, Q5DWM3, Q5DWM5, Q61744, Q8R3B0 | Gene names | Itpr2, Itpr5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2) (Inositol 1,4,5-trisphosphate type V receptor). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.025640 (rank : 53) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.007811 (rank : 111) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
CEP55_MOUSE
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.014095 (rank : 89) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BT07, Q8C2J0, Q8K2I8, Q8R2Y4, Q9DBZ8 | Gene names | Cep55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.023318 (rank : 58) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
PANK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.053203 (rank : 30) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BZ23, Q5T7I2, Q8N7Q4, Q8TCR5, Q9BYW5, Q9HAF2 | Gene names | PANK2, C20orf48 | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 2, mitochondrial precursor (EC 2.7.1.33) (Pantothenic acid kinase 2) (hPANK2). | |||||
|
SPR1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.042693 (rank : 35) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
SPR1A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.036471 (rank : 41) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.025472 (rank : 54) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.014275 (rank : 86) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.027372 (rank : 50) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
CD209_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.014646 (rank : 84) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NNX6, Q96QP7, Q96QP8, Q96QP9, Q96QQ0, Q96QQ1, Q96QQ2, Q96QQ3, Q96QQ4, Q96QQ5, Q96QQ6, Q96QQ7, Q96QQ8 | Gene names | CD209, CLEC4L | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen (Dendritic cell-specific ICAM-3-grabbing nonintegrin 1) (DC-SIGN1) (DC-SIGN) (C-type lectin domain family 4 member L). | |||||
|
EPHB6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.002273 (rank : 123) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15197 | Gene names | EPHB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 6 precursor (Tyrosine-protein kinase-defective receptor EPH-6) (HEP). | |||||
|
HEAT2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.037232 (rank : 40) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86Y56, Q69YL1, Q96FI9, Q9NX75 | Gene names | HEATR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HEAT repeat-containing protein 2. | |||||
|
IPO4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.048537 (rank : 32) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TEX9, Q86TZ9, Q8NCG8, Q96SJ3, Q9BTI4, Q9H5L0 | Gene names | IPO4, IMP4B, RANBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Importin-4 (Importin 4b) (Imp4b) (Ran-binding protein 4) (RanBP4). | |||||
|
NAGPA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.020954 (rank : 64) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BJ48, Q3UUT5, Q8CHQ8, Q9QZE6 | Gene names | Nagpa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase precursor (EC 3.1.4.45) (Phosphodiester alpha-GlcNAcase) (Mannose 6- phosphate-uncovering enzyme). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.009278 (rank : 107) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PK3C3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.112414 (rank : 14) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NEB9, Q15134 | Gene names | PIK3C3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3) (Phosphatidylinositol 3-kinase p100 subunit). | |||||
|
PK3C3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.113124 (rank : 13) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PF93, Q8R3S8 | Gene names | Pik3c3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.014413 (rank : 85) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
TM108_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.027812 (rank : 49) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
3BP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.020943 (rank : 65) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
COA2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.013752 (rank : 91) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00763, Q16852 | Gene names | ACACB, ACC2, ACCB | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 2 (EC 6.4.1.2) (ACC-beta) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
IL3B2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.023597 (rank : 57) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
KI21A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.007528 (rank : 112) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
KI21A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.007940 (rank : 110) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
MBB1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.019243 (rank : 68) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
NUMBL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.017395 (rank : 76) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.012421 (rank : 96) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.011892 (rank : 98) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
SPR1B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.039346 (rank : 37) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.021397 (rank : 62) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
UBP53_MOUSE
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.022526 (rank : 59) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P15975, Q8CB37, Q8R251 | Gene names | Usp53, Phxr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53) (Per-hexamer repeat protein 3). | |||||
|
ACRBP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.028413 (rank : 47) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3V140, Q62253, Q62254, Q8C621, Q91VQ1 | Gene names | Acrbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acrosin-binding protein precursor (Proacrosin-binding protein sp32). | |||||
|
ANFY1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.008797 (rank : 108) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q810B6, O54807, Q80TG6, Q80UH8 | Gene names | Ankfy1, Ankhzn, Kiaa1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
BMX_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.001517 (rank : 125) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51813, O60564, Q12871 | Gene names | BMX | |||
|
Domain Architecture |

|
|||||
| Description | Cytoplasmic tyrosine-protein kinase BMX (EC 2.7.10.2) (Bone marrow tyrosine kinase gene in chromosome X protein) (Epithelial and endothelial tyrosine kinase) (ETK) (NTK38). | |||||
|
BSND_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.021501 (rank : 61) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WZ55, Q6NT28 | Gene names | BSND, BART | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Barttin. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.015981 (rank : 81) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
GABR1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.011501 (rank : 99) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
GALC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.017455 (rank : 75) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54803, Q8J030 | Gene names | GALC | |||
|
Domain Architecture |

|
|||||
| Description | Galactocerebrosidase precursor (EC 3.2.1.46) (GALCERase) (Galactosylceramidase) (Galactosylceramide beta-galactosidase) (Galactocerebroside beta-galactosidase). | |||||
|
IL3RB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.020696 (rank : 66) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
IRF3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.011420 (rank : 100) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
ITPR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.028731 (rank : 46) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14643, Q14660, Q99897 | Gene names | ITPR1, INSP3R1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (IP3R). | |||||
|
ITPR1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.027198 (rank : 51) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11881, P20943, Q99LG5 | Gene names | Itpr1, Insp3r, Pcd6, Pcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (Inositol 1,4,5-trisphosphate-binding protein P400) (Purkinje cell protein 1) (Protein PCD-6). | |||||
|
KLK13_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.005049 (rank : 120) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UKR3, Q9Y433 | Gene names | KLK13, KLKL4 | |||
|
Domain Architecture |
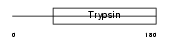
|
|||||
| Description | Kallikrein-13 precursor (EC 3.4.21.-) (Kallikrein-like protein 4) (KLK-L4). | |||||
|
LTBP3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.005742 (rank : 118) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NS15, O15107, Q96HB9, Q9H7K2, Q9UFN4 | Gene names | LTBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
LTBP3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.006309 (rank : 115) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61810, Q8BNQ6 | Gene names | Ltbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
MICA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.013461 (rank : 92) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BML1, Q3UPU6 | Gene names | Mical2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.018640 (rank : 69) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.009338 (rank : 106) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
UTP11_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.019592 (rank : 67) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CZJ1, Q3UI21, Q9CVB5 | Gene names | Utp11l | |||
|
Domain Architecture |
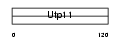
|
|||||
| Description | Probable U3 small nucleolar RNA-associated protein 11 (U3 snoRNA- associated protein 11) (UTP11-like protein). | |||||
|
ZN395_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.025244 (rank : 55) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H8N7, Q6F6H2, Q9BY72, Q9NPB2, Q9NS57, Q9NS58, Q9NS59 | Gene names | ZNF395, HDBP2, PBF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 395 (Papillomavirus-binding factor) (Papillomavirus regulatory factor 1) (PRF-1) (Huntington disease gene regulatory region-binding protein 2) (HDBP-2) (HD gene regulatory region-binding protein 2) (HD-regulating factor 2) (HDRF-2). | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.010050 (rank : 104) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
EPN1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.017978 (rank : 72) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.009785 (rank : 105) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
KI13A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.007183 (rank : 113) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9EQW7, O35062 | Gene names | Kif13a | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13A. | |||||
|
NCKX4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.014246 (rank : 87) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CGQ8, Q8BLL5, Q8BLL7 | Gene names | Slc24a4, Nckx4 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 4 precursor (Na(+)/K(+)/Ca(2+)- exchange protein 4). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.017708 (rank : 74) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RPA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.010239 (rank : 102) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95602, Q9UEH0, Q9UFT9 | Gene names | POLR1A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
SC6A5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.003990 (rank : 122) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q761V0, Q8CFM5, Q91ZQ2 | Gene names | Slc6a5, Glyt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium- and chloride-dependent glycine transporter 2 (GlyT2) (GlyT-2) (Solute carrier family 6 member 5). | |||||
|
UTY_MOUSE
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.045651 (rank : 34) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P79457, O97979 | Gene names | Uty | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed Y chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein ON the Y chromosome) (Male- specific histocompatibility antigen H-YDB). | |||||
|
ZN385_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.015252 (rank : 82) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
ANFY1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.006871 (rank : 114) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P2R3, Q9ULG5 | Gene names | ANKFY1, ANKHZN, KIAA1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
ATX7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.016371 (rank : 79) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
AXN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.016902 (rank : 78) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y2T1, Q9H3M6, Q9UH84 | Gene names | AXIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
AXN2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.016281 (rank : 80) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88566, Q9QXJ6 | Gene names | Axin2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.018386 (rank : 71) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.018388 (rank : 70) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.017875 (rank : 73) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.010190 (rank : 103) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.031960 (rank : 43) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
KINH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.004741 (rank : 121) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.008261 (rank : 109) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
LYST_MOUSE
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.006254 (rank : 116) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97412, Q62403, Q8VBS6 | Gene names | Lyst, Bg, Chs1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige protein) (CHS1 homolog). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.014210 (rank : 88) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MNT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.021938 (rank : 60) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
NUFP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.013787 (rank : 90) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UHK0, Q8WVM5, Q96SG1 | Gene names | NUFIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 1 (Nuclear FMRP-interacting protein 1). | |||||
|
PRD13_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | -0.000274 (rank : 126) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H4Q3, Q5TGC1 | Gene names | PRDM13, PFM10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 13 (PR domain-containing protein 13). | |||||
|
PSMD5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.013101 (rank : 93) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BJY1, Q3UFV2, Q8BMA9, Q8VEE9 | Gene names | Psmd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 5 (26S proteasome subunit S5B) (26S protease subunit S5 basic). | |||||
|
SON_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.028080 (rank : 48) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
SPRL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.013005 (rank : 94) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
STA13_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.005512 (rank : 119) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
|
TMPS2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.001961 (rank : 124) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JIQ8, Q9JKC4, Q9QY82 | Gene names | Tmprss2 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane protease, serine 2 (EC 3.4.21.-) (Epitheliasin) (Plasmic transmembrane protein X) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
TREF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.012459 (rank : 95) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
TSH3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.006065 (rank : 117) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CGV9, Q5DTX4 | Gene names | Tshz3, Kiaa1474, Tsh3, Zfp537, Znf537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 3 (Zinc finger protein 537). | |||||
|
TSKS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.011041 (rank : 101) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54887 | Gene names | Stk22s1, Tsks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific serine kinase substrate (Testis-specific kinase substrate) (STK22 substrate 1). | |||||
|
VEGFB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.012104 (rank : 97) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49766, Q3UG04, Q5D0B1, Q64290 | Gene names | Vegfb, Vrf | |||
|
Domain Architecture |
|
|||||
| Description | Vascular endothelial growth factor B precursor (VEGF-B) (VEGF-related factor) (VRF). | |||||
|
PI4KA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.050510 (rank : 31) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P42356, Q7Z625, Q9UPG2 | Gene names | PIK4CA, PIK4 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase alpha (EC 2.7.1.67) (PI4-kinase alpha) (PtdIns-4-kinase alpha) (PI4K-alpha). | |||||
|
PI4KB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.071677 (rank : 19) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UBF8, O15096, P78405, Q9BWR6 | Gene names | PIK4CB, PI4KB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta) (NPIK) (PI4K92). | |||||
|
PI4KB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.069994 (rank : 20) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BKC8, Q8C146 | Gene names | Pik4cb | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta). | |||||
|
PK3CB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.064566 (rank : 23) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P42338 | Gene names | PIK3CB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3CB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.064888 (rank : 22) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BTI9 | Gene names | Pik3cb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3CD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.063532 (rank : 25) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00329, O15445 | Gene names | PIK3CD | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PK3CD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.064202 (rank : 24) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35904 | Gene names | Pik3cd | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PK3CG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.062453 (rank : 26) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P48736, Q8IV23, Q9BZC8 | Gene names | PIK3CG | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
PK3CG_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.061282 (rank : 28) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JHG7 | Gene names | Pik3cg, Pi3kg1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
TRRAP_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
TRRAP_MOUSE
|
||||||
| NC score | 0.990457 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80YV3, Q8C0Z5, Q8K104 | Gene names | Trrap | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (Tra1 homolog). | |||||
|
ATR_HUMAN
|
||||||
| NC score | 0.468437 (rank : 3) | θ value | 1.13037e-18 (rank : 3) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13535, Q7KYL3, Q93051, Q9BXK4 | Gene names | ATR, FRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein) (FRAP-related protein 1). | |||||
|
ATR_MOUSE
|
||||||
| NC score | 0.455913 (rank : 4) | θ value | 2.35696e-16 (rank : 5) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JKK8 | Gene names | Atr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein). | |||||
|
PRKDC_MOUSE
|
||||||
| NC score | 0.443930 (rank : 5) | θ value | 1.47631e-18 (rank : 4) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97313, O88187, P97928, Q307W9, Q3V2W8, Q8C2A7, Q9Z341 | Gene names | Prkdc, Xrcc7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (P460). | |||||
|
PRKDC_HUMAN
|
||||||
| NC score | 0.418417 (rank : 6) | θ value | 5.43371e-13 (rank : 7) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
FRAP_MOUSE
|
||||||
| NC score | 0.417920 (rank : 7) | θ value | 2.43908e-13 (rank : 6) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JLN9, Q811J5, Q9CST1 | Gene names | Frap1, Frap | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
FRAP_HUMAN
|
||||||
| NC score | 0.415045 (rank : 8) | θ value | 7.09661e-13 (rank : 8) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P42345, Q6LE87, Q96QG3, Q9Y4I3 | Gene names | FRAP1, FRAP, FRAP2 | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
ATM_HUMAN
|
||||||
| NC score | 0.337551 (rank : 9) | θ value | 3.77169e-06 (rank : 9) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13315, O15429, Q12758, Q16551, Q93007, Q9NP02, Q9UCX7 | Gene names | ATM | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated) (A-T, mutated). | |||||
|
SMG1_MOUSE
|
||||||
| NC score | 0.335841 (rank : 10) | θ value | 3.77169e-06 (rank : 11) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
SMG1_HUMAN
|
||||||
| NC score | 0.333823 (rank : 11) | θ value | 3.77169e-06 (rank : 10) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
ATM_MOUSE
|
||||||
| NC score | 0.325332 (rank : 12) | θ value | 0.000121331 (rank : 12) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62388 | Gene names | Atm | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated homolog) (A-T, mutated homolog). | |||||
|
PK3C3_MOUSE
|
||||||
| NC score | 0.113124 (rank : 13) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6PF93, Q8R3S8 | Gene names | Pik3c3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3). | |||||
|
PK3C3_HUMAN
|
||||||
| NC score | 0.112414 (rank : 14) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NEB9, Q15134 | Gene names | PIK3C3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3) (Phosphatidylinositol 3-kinase p100 subunit). | |||||
|
ZN365_MOUSE
|
||||||
| NC score | 0.107737 (rank : 15) | θ value | 0.0330416 (rank : 14) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BG89, Q80TQ4, Q8BK39, Q8BXT2 | Gene names | Znf365, Kiaa0844, Zfp365 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF365 (Su48). | |||||
|
ZN365_HUMAN
|
||||||
| NC score | 0.088033 (rank : 16) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q70YC5, O94930, Q5HYE6, Q68SG8, Q6NSK2, Q6P9D4, Q70YC6, Q70YC7 | Gene names | ZNF365, KIAA0844 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF365. | |||||
|
PANK1_HUMAN
|
||||||
| NC score | 0.075315 (rank : 17) | θ value | 0.0961366 (rank : 16) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TE04, Q7RTX6, Q7Z495, Q8TBQ8 | Gene names | PANK1, PANK | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 1 (EC 2.7.1.33) (Pantothenic acid kinase 1) (hPanK1) (hPanK). | |||||
|
UTY_HUMAN
|
||||||
| NC score | 0.073028 (rank : 18) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14607, O14608 | Gene names | UTY | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed Y chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the Y chromosome). | |||||
|
PI4KB_HUMAN
|
||||||
| NC score | 0.071677 (rank : 19) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UBF8, O15096, P78405, Q9BWR6 | Gene names | PIK4CB, PI4KB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta) (NPIK) (PI4K92). | |||||
|
PI4KB_MOUSE
|
||||||
| NC score | 0.069994 (rank : 20) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BKC8, Q8C146 | Gene names | Pik4cb | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta). | |||||
|
PANK1_MOUSE
|
||||||
| NC score | 0.068586 (rank : 21) | θ value | 0.279714 (rank : 21) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K4K6, Q9D3K1, Q9QXM8 | Gene names | Pank1, Pank | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 1 (EC 2.7.1.33) (Pantothenic acid kinase 1) (mPank1) (mPank). | |||||
|
PK3CB_MOUSE
|
||||||
| NC score | 0.064888 (rank : 22) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BTI9 | Gene names | Pik3cb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3CB_HUMAN
|
||||||
| NC score | 0.064566 (rank : 23) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P42338 | Gene names | PIK3CB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3CD_MOUSE
|
||||||
| NC score | 0.064202 (rank : 24) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35904 | Gene names | Pik3cd | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PK3CD_HUMAN
|
||||||
| NC score | 0.063532 (rank : 25) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00329, O15445 | Gene names | PIK3CD | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PK3CG_HUMAN
|
||||||
| NC score | 0.062453 (rank : 26) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P48736, Q8IV23, Q9BZC8 | Gene names | PIK3CG | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
UTX_HUMAN
|
||||||
| NC score | 0.061918 (rank : 27) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15550 | Gene names | UTX | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed X chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein on the X chromosome). | |||||
|
PK3CG_MOUSE
|
||||||
| NC score | 0.061282 (rank : 28) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JHG7 | Gene names | Pik3cg, Pi3kg1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
IPO4_MOUSE
|
||||||
| NC score | 0.059061 (rank : 29) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VI75, Q3TBW0, Q99J52 | Gene names | Ipo4, Imp4a, Ranbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Importin-4 (Importin 4a) (Imp4a) (Ran-binding protein 4) (RanBP4). | |||||
|
PANK2_HUMAN
|
||||||
| NC score | 0.053203 (rank : 30) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BZ23, Q5T7I2, Q8N7Q4, Q8TCR5, Q9BYW5, Q9HAF2 | Gene names | PANK2, C20orf48 | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 2, mitochondrial precursor (EC 2.7.1.33) (Pantothenic acid kinase 2) (hPANK2). | |||||
|
PI4KA_HUMAN
|
||||||
| NC score | 0.050510 (rank : 31) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P42356, Q7Z625, Q9UPG2 | Gene names | PIK4CA, PIK4 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase alpha (EC 2.7.1.67) (PI4-kinase alpha) (PtdIns-4-kinase alpha) (PI4K-alpha). | |||||
|
IPO4_HUMAN
|
||||||
| NC score | 0.048537 (rank : 32) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TEX9, Q86TZ9, Q8NCG8, Q96SJ3, Q9BTI4, Q9H5L0 | Gene names | IPO4, IMP4B, RANBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Importin-4 (Importin 4b) (Imp4b) (Ran-binding protein 4) (RanBP4). | |||||
|
REV1_MOUSE
|
||||||
| NC score | 0.047772 (rank : 33) | θ value | 0.0736092 (rank : 15) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q920Q2, Q9QXV2 | Gene names | Rev1l, Rev1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein REV1 (EC 2.7.7.-) (Rev1-like terminal deoxycytidyl transferase). | |||||
|
UTY_MOUSE
|
||||||
| NC score | 0.045651 (rank : 34) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P79457, O97979 | Gene names | Uty | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitously transcribed Y chromosome tetratricopeptide repeat protein (Ubiquitously transcribed TPR protein ON the Y chromosome) (Male- specific histocompatibility antigen H-YDB). | |||||
|
SPR1A_HUMAN
|
||||||
| NC score | 0.042693 (rank : 35) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35321, Q9UDG4 | Gene names | SPRR1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein IA) (SPR-IA) (SPRK) (19 kDa pancornulin). | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.040857 (rank : 36) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
SPR1B_HUMAN
|
||||||
| NC score | 0.039346 (rank : 37) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22528, P22529, P22530, Q5T524 | Gene names | SPRR1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin B (Small proline-rich protein IB) (SPR-IB) (14.9 kDa pancornulin). | |||||
|
ARMC4_HUMAN
|
||||||
| NC score | 0.037523 (rank : 38) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T2S8, Q9H0C0 | Gene names | ARMC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 4. | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.037401 (rank : 39) | θ value | 0.125558 (rank : 17) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
HEAT2_HUMAN
|
||||||
| NC score | 0.037232 (rank : 40) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86Y56, Q69YL1, Q96FI9, Q9NX75 | Gene names | HEATR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HEAT repeat-containing protein 2. | |||||
|
SPR1A_MOUSE
|
||||||
| NC score | 0.036471 (rank : 41) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
ITPR2_HUMAN
|
||||||
| NC score | 0.032163 (rank : 42) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14571, O94773 | Gene names | ITPR2 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.031960 (rank : 43) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
ITPR2_MOUSE
|
||||||
| NC score | 0.031841 (rank : 44) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z329, P70226, Q5DWM3, Q5DWM5, Q61744, Q8R3B0 | Gene names | Itpr2, Itpr5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 2 (Type 2 inositol 1,4,5- trisphosphate receptor) (Type 2 InsP3 receptor) (IP3 receptor isoform 2) (InsP3R2) (Inositol 1,4,5-trisphosphate type V receptor). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.030819 (rank : 45) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
ITPR1_HUMAN
|
||||||
| NC score | 0.028731 (rank : 46) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14643, Q14660, Q99897 | Gene names | ITPR1, INSP3R1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (IP3R). | |||||
|
ACRBP_MOUSE
|
||||||
| NC score | 0.028413 (rank : 47) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3V140, Q62253, Q62254, Q8C621, Q91VQ1 | Gene names | Acrbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acrosin-binding protein precursor (Proacrosin-binding protein sp32). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.028080 (rank : 48) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
TM108_MOUSE
|
||||||
| NC score | 0.027812 (rank : 49) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.027372 (rank : 50) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
ITPR1_MOUSE
|
||||||
| NC score | 0.027198 (rank : 51) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11881, P20943, Q99LG5 | Gene names | Itpr1, Insp3r, Pcd6, Pcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (Inositol 1,4,5-trisphosphate-binding protein P400) (Purkinje cell protein 1) (Protein PCD-6). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.027073 (rank : 52) | θ value | 0.0252991 (rank : 13) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.025640 (rank : 53) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.025472 (rank : 54) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
ZN395_HUMAN
|
||||||
| NC score | 0.025244 (rank : 55) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H8N7, Q6F6H2, Q9BY72, Q9NPB2, Q9NS57, Q9NS58, Q9NS59 | Gene names | ZNF395, HDBP2, PBF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 395 (Papillomavirus-binding factor) (Papillomavirus regulatory factor 1) (PRF-1) (Huntington disease gene regulatory region-binding protein 2) (HDBP-2) (HD gene regulatory region-binding protein 2) (HD-regulating factor 2) (HDRF-2). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.023796 (rank : 56) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
IL3B2_MOUSE
|
||||||
| NC score | 0.023597 (rank : 57) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.023318 (rank : 58) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
UBP53_MOUSE
|
||||||
| NC score | 0.022526 (rank : 59) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P15975, Q8CB37, Q8R251 | Gene names | Usp53, Phxr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive ubiquitin carboxyl-terminal hydrolase 53 (Inactive ubiquitin- specific peptidase 53) (Per-hexamer repeat protein 3). | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.021938 (rank : 60) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
BSND_HUMAN
|
||||||
| NC score | 0.021501 (rank : 61) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WZ55, Q6NT28 | Gene names | BSND, BART | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Barttin. | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.021397 (rank : 62) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
CLC4M_HUMAN
|
||||||
| NC score | 0.021050 (rank : 63) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H2X3, Q69F40, Q969M4, Q96QP3, Q96QP4, Q96QP5, Q96QP6, Q9BXS3, Q9H2Q9, Q9H8F0, Q9Y2A8 | Gene names | CLEC4M, CD209L, CD209L1 | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 4 member M (CD209 antigen-like protein 1) (Dendritic cell-specific ICAM-3-grabbing nonintegrin 2) (DC-SIGN2) (DC-SIGN-related protein) (DC-SIGNR) (Liver/lymph node-specific ICAM- 3-grabbing nonintegrin) (L-SIGN) (CD299 antigen). | |||||
|
NAGPA_MOUSE
|
||||||
| NC score | 0.020954 (rank : 64) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BJ48, Q3UUT5, Q8CHQ8, Q9QZE6 | Gene names | Nagpa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase precursor (EC 3.1.4.45) (Phosphodiester alpha-GlcNAcase) (Mannose 6- phosphate-uncovering enzyme). | |||||
|
3BP2_HUMAN
|
||||||
| NC score | 0.020943 (rank : 65) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
IL3RB_MOUSE
|
||||||
| NC score | 0.020696 (rank : 66) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
UTP11_MOUSE
|
||||||
| NC score | 0.019592 (rank : 67) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CZJ1, Q3UI21, Q9CVB5 | Gene names | Utp11l | |||
|
Domain Architecture |

|
|||||
| Description | Probable U3 small nucleolar RNA-associated protein 11 (U3 snoRNA- associated protein 11) (UTP11-like protein). | |||||
|
MBB1A_MOUSE
|
||||||
| NC score | 0.019243 (rank : 68) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TPV4, O35851, Q80Y66, Q8R4X2, Q99KP0 | Gene names | Mybbp1a, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A (Myb-binding protein of 160 kDa). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.018640 (rank : 69) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.018388 (rank : 70) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.018386 (rank : 71) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
EPN1_MOUSE
|
||||||
| NC score | 0.017978 (rank : 72) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.017875 (rank : 73) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.017708 (rank : 74) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
GALC_HUMAN
|
||||||
| NC score | 0.017455 (rank : 75) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54803, Q8J030 | Gene names | GALC | |||
|
Domain Architecture |

|
|||||
| Description | Galactocerebrosidase precursor (EC 3.2.1.46) (GALCERase) (Galactosylceramidase) (Galactosylceramide beta-galactosidase) (Galactocerebroside beta-galactosidase). | |||||
|
NUMBL_HUMAN
|
||||||
| NC score | 0.017395 (rank : 76) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
KCNQ1_MOUSE
|
||||||
| NC score | 0.017054 (rank : 77) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
AXN2_HUMAN
|
||||||
| NC score | 0.016902 (rank : 78) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y2T1, Q9H3M6, Q9UH84 | Gene names | AXIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
ATX7_MOUSE
|
||||||
| NC score | 0.016371 (rank : 79) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8R4I1, Q8BL17 | Gene names | Atxn7, Sca7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein homolog). | |||||
|
AXN2_MOUSE
|
||||||
| NC score | 0.016281 (rank : 80) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88566, Q9QXJ6 | Gene names | Axin2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.015981 (rank : 81) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
ZN385_HUMAN
|
||||||
| NC score | 0.015252 (rank : 82) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.014896 (rank : 83) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
CD209_HUMAN
|
||||||
| NC score | 0.014646 (rank : 84) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NNX6, Q96QP7, Q96QP8, Q96QP9, Q96QQ0, Q96QQ1, Q96QQ2, Q96QQ3, Q96QQ4, Q96QQ5, Q96QQ6, Q96QQ7, Q96QQ8 | Gene names | CD209, CLEC4L | |||
|
Domain Architecture |

|
|||||
| Description | CD209 antigen (Dendritic cell-specific ICAM-3-grabbing nonintegrin 1) (DC-SIGN1) (DC-SIGN) (C-type lectin domain family 4 member L). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.014413 (rank : 85) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.014275 (rank : 86) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
NCKX4_MOUSE
|
||||||
| NC score | 0.014246 (rank : 87) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CGQ8, Q8BLL5, Q8BLL7 | Gene names | Slc24a4, Nckx4 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 4 precursor (Na(+)/K(+)/Ca(2+)- exchange protein 4). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.014210 (rank : 88) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
CEP55_MOUSE
|
||||||
| NC score | 0.014095 (rank : 89) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BT07, Q8C2J0, Q8K2I8, Q8R2Y4, Q9DBZ8 | Gene names | Cep55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55. | |||||
|
NUFP1_HUMAN
|
||||||
| NC score | 0.013787 (rank : 90) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UHK0, Q8WVM5, Q96SG1 | Gene names | NUFIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 1 (Nuclear FMRP-interacting protein 1). | |||||
|
COA2_HUMAN
|
||||||
| NC score | 0.013752 (rank : 91) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00763, Q16852 | Gene names | ACACB, ACC2, ACCB | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-CoA carboxylase 2 (EC 6.4.1.2) (ACC-beta) [Includes: Biotin carboxylase (EC 6.3.4.14)]. | |||||
|
MICA2_MOUSE
|
||||||
| NC score | 0.013461 (rank : 92) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BML1, Q3UPU6 | Gene names | Mical2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein MICAL-2. | |||||
|
PSMD5_MOUSE
|
||||||
| NC score | 0.013101 (rank : 93) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BJY1, Q3UFV2, Q8BMA9, Q8VEE9 | Gene names | Psmd5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 5 (26S proteasome subunit S5B) (26S protease subunit S5 basic). | |||||
|
SPRL1_HUMAN
|
||||||
| NC score | 0.013005 (rank : 94) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
TREF1_MOUSE
|
||||||
| NC score | 0.012459 (rank : 95) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BXJ2, Q6PCQ4, Q80X25, Q810H8, Q8BY31 | Gene names | Trerf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcriptional-regulating factor 1 (Transcriptional-regulating protein 132) (Zinc finger transcription factor TReP-132). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.012421 (rank : 96) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
VEGFB_MOUSE
|
||||||
| NC score | 0.012104 (rank : 97) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49766, Q3UG04, Q5D0B1, Q64290 | Gene names | Vegfb, Vrf | |||
|
Domain Architecture |

|
|||||
| Description | Vascular endothelial growth factor B precursor (VEGF-B) (VEGF-related factor) (VRF). | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.011892 (rank : 98) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
GABR1_MOUSE
|
||||||
| NC score | 0.011501 (rank : 99) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WV18, Q9WU48, Q9WV15, Q9WV16, Q9WV17 | Gene names | Gabbr1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-aminobutyric acid type B receptor, subunit 1 precursor (GABA-B receptor 1) (GABA-B-R1) (Gb1). | |||||
|
IRF3_HUMAN
|
||||||
| NC score | 0.011420 (rank : 100) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
TSKS_MOUSE
|
||||||
| NC score | 0.011041 (rank : 101) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54887 | Gene names | Stk22s1, Tsks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-specific serine kinase substrate (Testis-specific kinase substrate) (STK22 substrate 1). | |||||
|
RPA1_HUMAN
|
||||||
| NC score | 0.010239 (rank : 102) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95602, Q9UEH0, Q9UFT9 | Gene names | POLR1A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.010190 (rank : 103) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.010050 (rank : 104) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.009785 (rank : 105) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.009338 (rank : 106) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.009278 (rank : 107) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ANFY1_MOUSE
|
||||||
| NC score | 0.008797 (rank : 108) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q810B6, O54807, Q80TG6, Q80UH8 | Gene names | Ankfy1, Ankhzn, Kiaa1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.008261 (rank : 109) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
KI21A_MOUSE
|
||||||
| NC score | 0.007940 (rank : 110) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.007811 (rank : 111) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
KI21A_HUMAN
|
||||||
| NC score | 0.007528 (rank : 112) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
KI13A_MOUSE
|
||||||
| NC score | 0.007183 (rank : 113) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9EQW7, O35062 | Gene names | Kif13a | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13A. | |||||
|
ANFY1_HUMAN
|
||||||
| NC score | 0.006871 (rank : 114) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P2R3, Q9ULG5 | Gene names | ANKFY1, ANKHZN, KIAA1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
LTBP3_MOUSE
|
||||||
| NC score | 0.006309 (rank : 115) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61810, Q8BNQ6 | Gene names | Ltbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
LYST_MOUSE
|
||||||
| NC score | 0.006254 (rank : 116) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97412, Q62403, Q8VBS6 | Gene names | Lyst, Bg, Chs1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige protein) (CHS1 homolog). | |||||
|
TSH3_MOUSE
|
||||||
| NC score | 0.006065 (rank : 117) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CGV9, Q5DTX4 | Gene names | Tshz3, Kiaa1474, Tsh3, Zfp537, Znf537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 3 (Zinc finger protein 537). | |||||
|
LTBP3_HUMAN
|
||||||
| NC score | 0.005742 (rank : 118) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NS15, O15107, Q96HB9, Q9H7K2, Q9UFN4 | Gene names | LTBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
STA13_MOUSE
|
||||||
| NC score | 0.005512 (rank : 119) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q923Q2, Q8K369 | Gene names | Stard13 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13). | |||||
|
KLK13_HUMAN
|
||||||
| NC score | 0.005049 (rank : 120) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UKR3, Q9Y433 | Gene names | KLK13, KLKL4 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein-13 precursor (EC 3.4.21.-) (Kallikrein-like protein 4) (KLK-L4). | |||||
|
KINH_HUMAN
|
||||||
| NC score | 0.004741 (rank : 121) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
SC6A5_MOUSE
|
||||||
| NC score | 0.003990 (rank : 122) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q761V0, Q8CFM5, Q91ZQ2 | Gene names | Slc6a5, Glyt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium- and chloride-dependent glycine transporter 2 (GlyT2) (GlyT-2) (Solute carrier family 6 member 5). | |||||
|
EPHB6_HUMAN
|
||||||
| NC score | 0.002273 (rank : 123) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15197 | Gene names | EPHB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 6 precursor (Tyrosine-protein kinase-defective receptor EPH-6) (HEP). | |||||
|
TMPS2_MOUSE
|
||||||
| NC score | 0.001961 (rank : 124) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JIQ8, Q9JKC4, Q9QY82 | Gene names | Tmprss2 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 2 (EC 3.4.21.-) (Epitheliasin) (Plasmic transmembrane protein X) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
BMX_HUMAN
|
||||||
| NC score | 0.001517 (rank : 125) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51813, O60564, Q12871 | Gene names | BMX | |||
|
Domain Architecture |

|
|||||
| Description | Cytoplasmic tyrosine-protein kinase BMX (EC 2.7.10.2) (Bone marrow tyrosine kinase gene in chromosome X protein) (Epithelial and endothelial tyrosine kinase) (ETK) (NTK38). | |||||
|
PRD13_HUMAN
|
||||||
| NC score | -0.000274 (rank : 126) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 117 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H4Q3, Q5TGC1 | Gene names | PRDM13, PFM10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PR domain zinc finger protein 13 (PR domain-containing protein 13). | |||||