Please be patient as the page loads
|
PANK1_HUMAN
|
||||||
| SwissProt Accessions | Q8TE04, Q7RTX6, Q7Z495, Q8TBQ8 | Gene names | PANK1, PANK | |||
|
Domain Architecture |
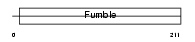
|
|||||
| Description | Pantothenate kinase 1 (EC 2.7.1.33) (Pantothenic acid kinase 1) (hPanK1) (hPanK). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PANK1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8TE04, Q7RTX6, Q7Z495, Q8TBQ8 | Gene names | PANK1, PANK | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 1 (EC 2.7.1.33) (Pantothenic acid kinase 1) (hPanK1) (hPanK). | |||||
|
PANK1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.990989 (rank : 2) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K4K6, Q9D3K1, Q9QXM8 | Gene names | Pank1, Pank | |||
|
Domain Architecture |
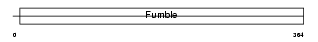
|
|||||
| Description | Pantothenate kinase 1 (EC 2.7.1.33) (Pantothenic acid kinase 1) (mPank1) (mPank). | |||||
|
PANK2_HUMAN
|
||||||
| θ value | 1.28067e-163 (rank : 3) | NC score | 0.984801 (rank : 5) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BZ23, Q5T7I2, Q8N7Q4, Q8TCR5, Q9BYW5, Q9HAF2 | Gene names | PANK2, C20orf48 | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 2, mitochondrial precursor (EC 2.7.1.33) (Pantothenic acid kinase 2) (hPANK2). | |||||
|
PANK3_HUMAN
|
||||||
| θ value | 3.16172e-154 (rank : 4) | NC score | 0.985887 (rank : 3) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H999 | Gene names | PANK3 | |||
|
Domain Architecture |
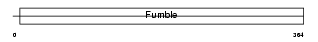
|
|||||
| Description | Pantothenate kinase 3 (EC 2.7.1.33) (Pantothenic acid kinase 3) (hPanK3). | |||||
|
PANK3_MOUSE
|
||||||
| θ value | 2.26584e-152 (rank : 5) | NC score | 0.985624 (rank : 4) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R2W9 | Gene names | Pank3 | |||
|
Domain Architecture |
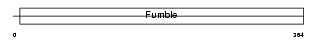
|
|||||
| Description | Pantothenate kinase 3 (EC 2.7.1.33) (Pantothenic acid kinase 3) (mPanK3). | |||||
|
PANK4_MOUSE
|
||||||
| θ value | 5.55816e-50 (rank : 6) | NC score | 0.844610 (rank : 6) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80YV4, Q7M751, Q8BQE9 | Gene names | Pank4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pantothenate kinase 4 (EC 2.7.1.33) (Pantothenic acid kinase 4) (mPanK4). | |||||
|
PANK4_HUMAN
|
||||||
| θ value | 2.58187e-47 (rank : 7) | NC score | 0.840245 (rank : 7) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NVE7, Q53EU3, Q5TA84, Q7RTX3, Q9H3X5 | Gene names | PANK4 | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 4 (EC 2.7.1.33) (Pantothenic acid kinase 4) (hPanK4). | |||||
|
TRRAP_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 8) | NC score | 0.075315 (rank : 9) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
TRRAP_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 9) | NC score | 0.077006 (rank : 8) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80YV3, Q8C0Z5, Q8K104 | Gene names | Trrap | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (Tra1 homolog). | |||||
|
VASP_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 10) | NC score | 0.048395 (rank : 11) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P70460, Q3TAP0, Q3TCD2, Q3U0C2, Q3UDF1, Q91VD2, Q9R214 | Gene names | Vasp | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
CO4A3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 11) | NC score | 0.030382 (rank : 16) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 12) | NC score | 0.006542 (rank : 67) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
VASP_HUMAN
|
||||||
| θ value | 0.125558 (rank : 13) | NC score | 0.048646 (rank : 10) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P50552, Q6PIZ1, Q93035 | Gene names | VASP | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.045229 (rank : 12) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
DBPA_MOUSE
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.025531 (rank : 31) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JKB3, Q80WG4, Q9EQF7, Q9EQF8 | Gene names | Csda, Msy4, Ybx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-binding protein A (Cold shock domain-containing protein A) (Y-box protein 3). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.028318 (rank : 19) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
CITE4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.030730 (rank : 15) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WUL8, Q66JX0, Q8R176, Q9CZV4 | Gene names | Cited4, Mrg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cbp/p300-interacting transactivator 4 (MSG1-related protein 2) (MRG- 2). | |||||
|
HNRPK_HUMAN
|
||||||
| θ value | 1.38821 (rank : 18) | NC score | 0.027899 (rank : 21) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61978, Q07244, Q15671, Q60577, Q922Y7, Q96J62 | Gene names | HNRPK, HNRNPK | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K (hnRNP K) (Transformation up-regulated nuclear protein) (TUNP). | |||||
|
HNRPK_MOUSE
|
||||||
| θ value | 1.38821 (rank : 19) | NC score | 0.027899 (rank : 22) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61979, Q07244, Q15671, Q60577, Q8BGQ8, Q922Y7, Q96J62 | Gene names | Hnrpk, Hnrnpk | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K. | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.009359 (rank : 56) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.042006 (rank : 13) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
KLF16_MOUSE
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.002146 (rank : 72) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P58334, Q3U3Y4, Q8C8S2 | Gene names | Klf16, Bteb4, Drrf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 16 (Transcription factor BTEB4) (Basic transcription element-binding protein 4) (BTE-binding protein 4) (Dopamine receptor-regulating factor). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.028483 (rank : 18) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
CO4A2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.026476 (rank : 27) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
CO9A1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.028091 (rank : 20) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
SYN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.026141 (rank : 28) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
ZNHI4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.025038 (rank : 34) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9C086 | Gene names | ZNHIT4, HMGA1L4, PAPA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger HIT domain-containing protein 4 (PAP-1-associated protein 1) (PAPA-1) (High mobility group AT-hook 1-like 4). | |||||
|
CO1A1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.026989 (rank : 25) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
COCA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.018251 (rank : 41) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60847, P70322 | Gene names | Col12a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
CYH1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.011501 (rank : 53) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15438, Q9P123, Q9P124 | Gene names | PSCD1, D17S811E | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (SEC7 homolog B2-1). | |||||
|
DIAP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.009821 (rank : 54) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60879, O60878, Q8WX06, Q8WX48, Q9UJL2 | Gene names | DIAPH2, DIA | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2). | |||||
|
EVX1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.005094 (rank : 69) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49640 | Gene names | EVX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox even-skipped homolog protein 1 (EVX-1). | |||||
|
K0460_MOUSE
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.015822 (rank : 45) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
KLF16_HUMAN
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.001764 (rank : 73) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BXK1 | Gene names | KLF16, BTEB4, NSLP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 16 (Transcription factor BTEB4) (Basic transcription element-binding protein 4) (BTE-binding protein 4) (Novel Sp1-like zinc finger transcription factor 2) (Transcription factor NSLP2). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.015616 (rank : 47) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
PP1RA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.030143 (rank : 17) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80W00, Q811B6, Q8C6T7, Q8K2U8 | Gene names | Ppp1r10, Cat53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (MHC class I region proline-rich protein CAT53). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.008316 (rank : 59) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
SYN2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.025459 (rank : 32) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92777 | Gene names | SYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
CN004_MOUSE
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.017956 (rank : 43) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K3X4, Q3USE5, Q8BUS1, Q8C011 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4 homolog. | |||||
|
COBA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.027041 (rank : 24) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P12107, Q14034, Q9UIT4, Q9UIT5, Q9UIT6 | Gene names | COL11A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
COBA1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.027289 (rank : 23) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61245, Q64047 | Gene names | Col11a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
DAXX_MOUSE
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.016895 (rank : 44) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35613, Q9QWT8, Q9QWV3 | Gene names | Daxx | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx). | |||||
|
EPS8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.009795 (rank : 55) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12929, Q8N6J0 | Gene names | EPS8 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor kinase substrate 8. | |||||
|
IRS2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.014721 (rank : 49) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
IRX5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.008741 (rank : 57) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P78411, P78416, Q7Z2E1 | Gene names | IRX5, IRX2A, IRXB2 | |||
|
Domain Architecture |
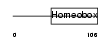
|
|||||
| Description | Iroquois-class homeodomain protein IRX-5 (Iroquois homeobox protein 5) (Homeodomain protein IRXB2) (IRX-2A). | |||||
|
LRRF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.013585 (rank : 50) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
PRR12_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.019490 (rank : 40) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
3BP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.008292 (rank : 60) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
ADA19_MOUSE
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.006936 (rank : 65) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35674 | Gene names | Adam19, Mltnb | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 19) (Meltrin beta). | |||||
|
CO9A1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.026749 (rank : 26) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
COCA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.017981 (rank : 42) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99715, Q99716 | Gene names | COL12A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
FGRL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.003965 (rank : 70) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N441, Q6PJN1, Q9BXN7, Q9H4D7 | Gene names | FGFRL1, FGFR5, FHFR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor-like 1 precursor (FGF receptor-like protein 1) (Fibroblast growth factor receptor 5) (FGFR-like protein) (FGF homologous factor receptor). | |||||
|
FOXD1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.003458 (rank : 71) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61345 | Gene names | Foxd1, Fkhl8, Freac4, Hfhbf2 | |||
|
Domain Architecture |
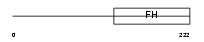
|
|||||
| Description | Forkhead box protein D1 (Forkhead-related protein FKHL8) (Forkhead- related transcription factor 4) (FREAC-4) (Brain factor 2) (HFH-BF-2). | |||||
|
KCNH6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.007349 (rank : 63) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
PPAC3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.013563 (rank : 51) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NBV4, Q5T6P0, Q96SS7, Q9BRC3 | Gene names | PPAPDC3, C9orf67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable lipid phosphate phosphatase PPAPDC3 (EC 3.1.3.-) (Phosphatidic acid phosphatase type 2 domain-containing protein 3). | |||||
|
PRPC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.024907 (rank : 35) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
PTPRV_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.007482 (rank : 61) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70289 | Gene names | Ptprv, Esp | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase V precursor (EC 3.1.3.48) (Embryonic stem cell protein-tyrosine phosphatase) (ES cell phosphatase). | |||||
|
WDR33_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.013101 (rank : 52) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 638 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9C0J8 | Gene names | WDR33, WDC146 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 33 (WD repeat protein WDC146). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.015730 (rank : 46) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
CO1A1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.025984 (rank : 29) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P11087, Q53WT0, Q60635, Q61367, Q61427, Q63919, Q6PCL3, Q810J9 | Gene names | Col1a1, Cola1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
CO3A1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.025676 (rank : 30) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
COFA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.022467 (rank : 38) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35206, Q9EQD9 | Gene names | Col15a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV)]. | |||||
|
DYR1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.000278 (rank : 74) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y463, O75258, O75788, O75789 | Gene names | DYRK1B, MIRK | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity tyrosine-phosphorylation-regulated kinase 1B (EC 2.7.12.1) (Mirk protein kinase) (Minibrain-related kinase). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.008354 (rank : 58) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
RAPSN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.015006 (rank : 48) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P12672 | Gene names | Rapsn | |||
|
Domain Architecture |
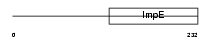
|
|||||
| Description | 43 kDa receptor-associated protein of the synapse (RAPsyn) (Acetylcholine receptor-associated 43 kDa protein) (43 kDa postsynaptic protein). | |||||
|
SYN2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.023092 (rank : 37) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q64332, Q6NZR0, Q9QWV7 | Gene names | Syn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
ZBT16_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | -0.000816 (rank : 75) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||
|
CO3A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.025398 (rank : 33) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
COHA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.022358 (rank : 39) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q07563, Q99LK8 | Gene names | Col17a1, Bp180, Bpag2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
DVL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.007387 (rank : 62) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.005830 (rank : 68) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
PP1RA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.023832 (rank : 36) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96QC0, O00405 | Gene names | PPP1R10, CAT53, FB19, PNUTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (Phosphatase 1 nuclear targeting subunit) (MHC class I region proline- rich protein CAT53) (FB19 protein) (PP1-binding protein of 114 kDa) (p99). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.006898 (rank : 66) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SYN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.031446 (rank : 14) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.007236 (rank : 64) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
PANK1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8TE04, Q7RTX6, Q7Z495, Q8TBQ8 | Gene names | PANK1, PANK | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 1 (EC 2.7.1.33) (Pantothenic acid kinase 1) (hPanK1) (hPanK). | |||||
|
PANK1_MOUSE
|
||||||
| NC score | 0.990989 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K4K6, Q9D3K1, Q9QXM8 | Gene names | Pank1, Pank | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 1 (EC 2.7.1.33) (Pantothenic acid kinase 1) (mPank1) (mPank). | |||||
|
PANK3_HUMAN
|
||||||
| NC score | 0.985887 (rank : 3) | θ value | 3.16172e-154 (rank : 4) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H999 | Gene names | PANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 3 (EC 2.7.1.33) (Pantothenic acid kinase 3) (hPanK3). | |||||
|
PANK3_MOUSE
|
||||||
| NC score | 0.985624 (rank : 4) | θ value | 2.26584e-152 (rank : 5) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R2W9 | Gene names | Pank3 | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 3 (EC 2.7.1.33) (Pantothenic acid kinase 3) (mPanK3). | |||||
|
PANK2_HUMAN
|
||||||
| NC score | 0.984801 (rank : 5) | θ value | 1.28067e-163 (rank : 3) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BZ23, Q5T7I2, Q8N7Q4, Q8TCR5, Q9BYW5, Q9HAF2 | Gene names | PANK2, C20orf48 | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 2, mitochondrial precursor (EC 2.7.1.33) (Pantothenic acid kinase 2) (hPANK2). | |||||
|
PANK4_MOUSE
|
||||||
| NC score | 0.844610 (rank : 6) | θ value | 5.55816e-50 (rank : 6) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80YV4, Q7M751, Q8BQE9 | Gene names | Pank4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pantothenate kinase 4 (EC 2.7.1.33) (Pantothenic acid kinase 4) (mPanK4). | |||||
|
PANK4_HUMAN
|
||||||
| NC score | 0.840245 (rank : 7) | θ value | 2.58187e-47 (rank : 7) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NVE7, Q53EU3, Q5TA84, Q7RTX3, Q9H3X5 | Gene names | PANK4 | |||
|
Domain Architecture |

|
|||||
| Description | Pantothenate kinase 4 (EC 2.7.1.33) (Pantothenic acid kinase 4) (hPanK4). | |||||
|
TRRAP_MOUSE
|
||||||
| NC score | 0.077006 (rank : 8) | θ value | 0.0961366 (rank : 9) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80YV3, Q8C0Z5, Q8K104 | Gene names | Trrap | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (Tra1 homolog). | |||||
|
TRRAP_HUMAN
|
||||||
| NC score | 0.075315 (rank : 9) | θ value | 0.0961366 (rank : 8) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
VASP_HUMAN
|
||||||
| NC score | 0.048646 (rank : 10) | θ value | 0.125558 (rank : 13) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P50552, Q6PIZ1, Q93035 | Gene names | VASP | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
VASP_MOUSE
|
||||||
| NC score | 0.048395 (rank : 11) | θ value | 0.0961366 (rank : 10) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P70460, Q3TAP0, Q3TCD2, Q3U0C2, Q3UDF1, Q91VD2, Q9R214 | Gene names | Vasp | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.045229 (rank : 12) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.042006 (rank : 13) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
SYN1_MOUSE
|
||||||
| NC score | 0.031446 (rank : 14) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88935, Q62279, Q8QZT8 | Gene names | Syn1, Syn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I). | |||||
|
CITE4_MOUSE
|
||||||
| NC score | 0.030730 (rank : 15) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WUL8, Q66JX0, Q8R176, Q9CZV4 | Gene names | Cited4, Mrg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cbp/p300-interacting transactivator 4 (MSG1-related protein 2) (MRG- 2). | |||||
|
CO4A3_HUMAN
|
||||||
| NC score | 0.030382 (rank : 16) | θ value | 0.125558 (rank : 11) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
PP1RA_MOUSE
|
||||||
| NC score | 0.030143 (rank : 17) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80W00, Q811B6, Q8C6T7, Q8K2U8 | Gene names | Ppp1r10, Cat53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (MHC class I region proline-rich protein CAT53). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.028483 (rank : 18) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.028318 (rank : 19) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
CO9A1_MOUSE
|
||||||
| NC score | 0.028091 (rank : 20) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
HNRPK_HUMAN
|
||||||
| NC score | 0.027899 (rank : 21) | θ value | 1.38821 (rank : 18) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61978, Q07244, Q15671, Q60577, Q922Y7, Q96J62 | Gene names | HNRPK, HNRNPK | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K (hnRNP K) (Transformation up-regulated nuclear protein) (TUNP). | |||||
|
HNRPK_MOUSE
|
||||||
| NC score | 0.027899 (rank : 22) | θ value | 1.38821 (rank : 19) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61979, Q07244, Q15671, Q60577, Q8BGQ8, Q922Y7, Q96J62 | Gene names | Hnrpk, Hnrnpk | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein K. | |||||
|
COBA1_MOUSE
|
||||||
| NC score | 0.027289 (rank : 23) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61245, Q64047 | Gene names | Col11a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
COBA1_HUMAN
|
||||||
| NC score | 0.027041 (rank : 24) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P12107, Q14034, Q9UIT4, Q9UIT5, Q9UIT6 | Gene names | COL11A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
CO1A1_HUMAN
|
||||||
| NC score | 0.026989 (rank : 25) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
CO9A1_HUMAN
|
||||||
| NC score | 0.026749 (rank : 26) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CO4A2_HUMAN
|
||||||
| NC score | 0.026476 (rank : 27) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
SYN1_HUMAN
|
||||||
| NC score | 0.026141 (rank : 28) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P17600, O75825, Q5H9A9 | Gene names | SYN1 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-1 (Synapsin I) (Brain protein 4.1). | |||||
|
CO1A1_MOUSE
|
||||||
| NC score | 0.025984 (rank : 29) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P11087, Q53WT0, Q60635, Q61367, Q61427, Q63919, Q6PCL3, Q810J9 | Gene names | Col1a1, Cola1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
CO3A1_MOUSE
|
||||||
| NC score | 0.025676 (rank : 30) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 466 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P08121, Q61429, Q9CRN7 | Gene names | Col3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
DBPA_MOUSE
|
||||||
| NC score | 0.025531 (rank : 31) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JKB3, Q80WG4, Q9EQF7, Q9EQF8 | Gene names | Csda, Msy4, Ybx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-binding protein A (Cold shock domain-containing protein A) (Y-box protein 3). | |||||
|
SYN2_HUMAN
|
||||||
| NC score | 0.025459 (rank : 32) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92777 | Gene names | SYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
CO3A1_HUMAN
|
||||||
| NC score | 0.025398 (rank : 33) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P02461, Q15112, Q16403, Q6LDB3, Q6LDJ2, Q6LDJ3, Q7KZ56 | Gene names | COL3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(III) chain precursor. | |||||
|
ZNHI4_HUMAN
|
||||||
| NC score | 0.025038 (rank : 34) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9C086 | Gene names | ZNHIT4, HMGA1L4, PAPA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger HIT domain-containing protein 4 (PAP-1-associated protein 1) (PAPA-1) (High mobility group AT-hook 1-like 4). | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.024907 (rank : 35) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
PP1RA_HUMAN
|
||||||
| NC score | 0.023832 (rank : 36) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96QC0, O00405 | Gene names | PPP1R10, CAT53, FB19, PNUTS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 1 regulatory subunit 10 (Phosphatase 1 nuclear targeting subunit) (MHC class I region proline- rich protein CAT53) (FB19 protein) (PP1-binding protein of 114 kDa) (p99). | |||||
|
SYN2_MOUSE
|
||||||
| NC score | 0.023092 (rank : 37) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q64332, Q6NZR0, Q9QWV7 | Gene names | Syn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
COFA1_MOUSE
|
||||||
| NC score | 0.022467 (rank : 38) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35206, Q9EQD9 | Gene names | Col15a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV)]. | |||||
|
COHA1_MOUSE
|
||||||
| NC score | 0.022358 (rank : 39) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q07563, Q99LK8 | Gene names | Col17a1, Bp180, Bpag2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVII) chain (Bullous pemphigoid antigen 2) (180 kDa bullous pemphigoid antigen 2). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.019490 (rank : 40) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
COCA1_MOUSE
|
||||||
| NC score | 0.018251 (rank : 41) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q60847, P70322 | Gene names | Col12a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
COCA1_HUMAN
|
||||||
| NC score | 0.017981 (rank : 42) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99715, Q99716 | Gene names | COL12A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XII) chain precursor. | |||||
|
CN004_MOUSE
|
||||||
| NC score | 0.017956 (rank : 43) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K3X4, Q3USE5, Q8BUS1, Q8C011 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4 homolog. | |||||
|
DAXX_MOUSE
|
||||||
| NC score | 0.016895 (rank : 44) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35613, Q9QWT8, Q9QWV3 | Gene names | Daxx | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx). | |||||
|
K0460_MOUSE
|
||||||
| NC score | 0.015822 (rank : 45) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.015730 (rank : 46) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.015616 (rank : 47) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
RAPSN_MOUSE
|
||||||
| NC score | 0.015006 (rank : 48) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P12672 | Gene names | Rapsn | |||
|
Domain Architecture |

|
|||||
| Description | 43 kDa receptor-associated protein of the synapse (RAPsyn) (Acetylcholine receptor-associated 43 kDa protein) (43 kDa postsynaptic protein). | |||||
|
IRS2_HUMAN
|
||||||
| NC score | 0.014721 (rank : 49) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
LRRF1_MOUSE
|
||||||
| NC score | 0.013585 (rank : 50) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
PPAC3_HUMAN
|
||||||
| NC score | 0.013563 (rank : 51) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NBV4, Q5T6P0, Q96SS7, Q9BRC3 | Gene names | PPAPDC3, C9orf67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable lipid phosphate phosphatase PPAPDC3 (EC 3.1.3.-) (Phosphatidic acid phosphatase type 2 domain-containing protein 3). | |||||
|
WDR33_HUMAN
|
||||||
| NC score | 0.013101 (rank : 52) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 638 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9C0J8 | Gene names | WDR33, WDC146 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 33 (WD repeat protein WDC146). | |||||
|
CYH1_HUMAN
|
||||||
| NC score | 0.011501 (rank : 53) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15438, Q9P123, Q9P124 | Gene names | PSCD1, D17S811E | |||
|
Domain Architecture |

|
|||||
| Description | Cytohesin-1 (SEC7 homolog B2-1). | |||||
|
DIAP2_HUMAN
|
||||||
| NC score | 0.009821 (rank : 54) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60879, O60878, Q8WX06, Q8WX48, Q9UJL2 | Gene names | DIAPH2, DIA | |||
|
Domain Architecture |

|
|||||
| Description | Protein diaphanous homolog 2 (Diaphanous-related formin-2) (DRF2). | |||||
|
EPS8_HUMAN
|
||||||
| NC score | 0.009795 (rank : 55) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12929, Q8N6J0 | Gene names | EPS8 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor kinase substrate 8. | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.009359 (rank : 56) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
IRX5_HUMAN
|
||||||
| NC score | 0.008741 (rank : 57) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P78411, P78416, Q7Z2E1 | Gene names | IRX5, IRX2A, IRXB2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-5 (Iroquois homeobox protein 5) (Homeodomain protein IRXB2) (IRX-2A). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.008354 (rank : 58) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.008316 (rank : 59) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
3BP1_HUMAN
|
||||||
| NC score | 0.008292 (rank : 60) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y3L3, Q6IBZ2, Q6ZVL9, Q96HQ5, Q9NSQ9 | Gene names | SH3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 1 (3BP-1). | |||||
|
PTPRV_MOUSE
|
||||||
| NC score | 0.007482 (rank : 61) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70289 | Gene names | Ptprv, Esp | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase V precursor (EC 3.1.3.48) (Embryonic stem cell protein-tyrosine phosphatase) (ES cell phosphatase). | |||||
|
DVL2_HUMAN
|
||||||
| NC score | 0.007387 (rank : 62) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14641 | Gene names | DVL2 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-2 (Dishevelled-2) (DSH homolog 2). | |||||
|
KCNH6_HUMAN
|
||||||
| NC score | 0.007349 (rank : 63) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.007236 (rank : 64) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
ADA19_MOUSE
|
||||||
| NC score | 0.006936 (rank : 65) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35674 | Gene names | Adam19, Mltnb | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 19 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 19) (Meltrin beta). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.006898 (rank : 66) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.006542 (rank : 67) | θ value | 0.125558 (rank : 12) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.005830 (rank : 68) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
EVX1_HUMAN
|
||||||
| NC score | 0.005094 (rank : 69) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49640 | Gene names | EVX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox even-skipped homolog protein 1 (EVX-1). | |||||
|
FGRL1_HUMAN
|
||||||
| NC score | 0.003965 (rank : 70) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N441, Q6PJN1, Q9BXN7, Q9H4D7 | Gene names | FGFRL1, FGFR5, FHFR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor-like 1 precursor (FGF receptor-like protein 1) (Fibroblast growth factor receptor 5) (FGFR-like protein) (FGF homologous factor receptor). | |||||
|
FOXD1_MOUSE
|
||||||
| NC score | 0.003458 (rank : 71) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61345 | Gene names | Foxd1, Fkhl8, Freac4, Hfhbf2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D1 (Forkhead-related protein FKHL8) (Forkhead- related transcription factor 4) (FREAC-4) (Brain factor 2) (HFH-BF-2). | |||||
|
KLF16_MOUSE
|
||||||
| NC score | 0.002146 (rank : 72) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P58334, Q3U3Y4, Q8C8S2 | Gene names | Klf16, Bteb4, Drrf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 16 (Transcription factor BTEB4) (Basic transcription element-binding protein 4) (BTE-binding protein 4) (Dopamine receptor-regulating factor). | |||||
|
KLF16_HUMAN
|
||||||
| NC score | 0.001764 (rank : 73) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BXK1 | Gene names | KLF16, BTEB4, NSLP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 16 (Transcription factor BTEB4) (Basic transcription element-binding protein 4) (BTE-binding protein 4) (Novel Sp1-like zinc finger transcription factor 2) (Transcription factor NSLP2). | |||||
|
DYR1B_HUMAN
|
||||||
| NC score | 0.000278 (rank : 74) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 831 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y463, O75258, O75788, O75789 | Gene names | DYRK1B, MIRK | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity tyrosine-phosphorylation-regulated kinase 1B (EC 2.7.12.1) (Mirk protein kinase) (Minibrain-related kinase). | |||||
|
ZBT16_HUMAN
|
||||||
| NC score | -0.000816 (rank : 75) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 75 | Target Neighborhood Hits | 1010 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q05516, Q8TAL4 | Gene names | ZBTB16, PLZF, ZNF145 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 16 (Zinc finger protein PLZF) (Promyelocytic leukemia zinc finger protein) (Zinc finger protein 145). | |||||